Abstract
DNA methylation, various DNA repair mechanisms, and possibly early events in the opening of DNA as required for transcription and replication are initiated by flipping of a DNA base out of the DNA double helix. The energetics and structural mechanism of base flipping in the presence of the DNA-processing enzyme, cytosine 5-methyltransferase from HhaI (M.HhaI), were obtained through molecular dynamics based upon free-energy calculations. Free-energy profiles for base flipping show that, when in the closed conformation, M.HhaI lowers the free-energy barrier to flipping by 17 kcal/mol and stabilizes the fully flipped state. Flipping is shown to occur via the major groove of the DNA. Structural analysis indicates that flipping is facilitated by destabilization of the DNA double-helical structure and substitution of DNA base-pairing and base-stacking interactions with DNA–protein interactions. The fully flipped state is stabilized by DNA–protein interactions that are enhanced upon binding of coenzyme. This study represents an atomic detail description of the mechanism by which a protein facilitates specific structural distortion in DNA.
DNA methylation is involved in gene regulation in eukaryotes (1), protection of “self” DNA in prokaryotes (2), and it plays a role in the etiology of some cancers (3–5). For chemical modification of the target base being methylated to occur, as catalyzed by methyltransferases, it is necessary for the base to be flipped out of the DNA double helix (Fig. 1A) and into the enzyme's active site (6, 7). Base flipping is also important for other enzymes that chemically alter DNA, including DNA-mismatch repair enzymes, such as uracil glycosylase and endonucleases (8). Base flipping may even be linked to early events in the opening and unwinding of DNA for transcription and replication processes (9, 10). Understanding the mechanism by which an enzyme can facilitate base flipping is, therefore, an important first step in elucidating more complex enzyme-catalyzed DNA processing involved in transcription and replication.
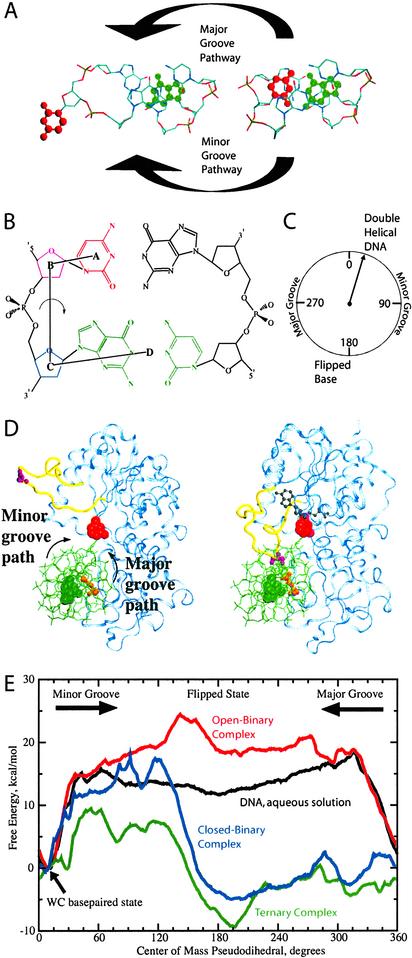
Figure 1.
(A) DNA double helix central three bases (tribase) showing flipping of the target C base (red) from the DNA double-helical conformation (Right) to a flipped conformation (Left) via the major or minor grooves. Orphan guanine is shown in green. (B) Schematic diagram of the pseudodihedral used to describe the flipping process. The pseudodihedral is defined by the center of masses of (i) the target C (red, A), (ii) its sugar moiety (purple, B), (iii) the adjacent 3′ sugar moiety (blue, C), and (iv) the 3′ GC base atoms (green, D). Hydrogens were omitted for clarity. (C) Diagram relating the periodic pseudodihedral to structural changes, where the DNA double-helical state corresponds to 10°, the flipped state to ≈195°, minor groove flipping as increasing from 10° to 195°, and major groove flipping as decreasing from 10° through 0–360° to 195°. (D) Open-binary DNA-M.HhaI complex (Left) and the closed-ternary DNA-M.HhaI-SAH complex (Right). DNA (green), target C base (red spheres), orphan G base (green spheres), catalytic and recognition domains of M.HhaI (blue), active-site loop (yellow), Ser-87 (purple), Gln-237 (gold), and SAH (gray) are shown. Arrows indicating the a priori minor and major groove-flipping pathways are shown on the open-binary complex structure. (E) Free-energy surfaces for base flipping for DNA in aqueous solution (black), the open DNA-M.HhaI binary complex (red), the closed DNA-M.HhaI binary complex (blue), and the DNA-M.HhaI-SAH ternary complex (green). Free-energy profiles have been offset to 0 kcal/mol at 10° (WC base-paired state). Energy barriers at 40° (minor groove) and 315° (major groove) for DNA in aqueous solution are in agreement with previous studies (14). Molecular images were generated with vmd (26).
The cytosine 5-methyltransferase from the HhaI bacterium (M.HhaI) is the most studied enzyme among the DNA methyltransferases. The ternary complex of M.HhaI with DNA and the cofactor product S-adenosylhomocysteine (SAH) was the first system in which the structural phenomenon of base flipping was observed (11). Based on the this crystal structure, it was suggested that flipping of the target cytosine (target C) occurs via the minor groove of the DNA duplex. In uracil glycosylase, on the other hand, experimental evidence indicates that flipping occurs via the major groove (12, 13). Recent theoretical studies indicate that, in solution, flipping of the target C base out of either the minor or major groove of DNA have similar energetic barriers (14, 15). In addition, it is known that M.HhaI stabilizes the flipped conformation of the target C in the ternary complex (16). In the present work, the energetic consequences and structural events associated with base flipping in the presence of M.HhaI are studied to develop an atomistic structural model of enzymatic facilitation of the flipping process.
Structural information available from experimental studies on base flipping in M.HhaI is mostly restricted to the endpoint states because of the higher energies and resulting short lifetimes of the intermediate states (17–19). Computational methods based on molecular dynamics (MD) simulations can access these structural intermediates and identify accompanying energetic changes at an atomic level of detail (20). However, the current standard of MD simulations does not allow access to the millisecond time scales at which these events occur (21, 22). To overcome this limitation, the method of umbrella sampling (23) was applied in conjunction with MD simulations to drive flipping of the base out of the DNA double helix. In this approach, an external (i.e., umbrella) potential (24) is included in an MD simulation to maintain the base in high energy regions of the flipping free-energy surface not normally sampled. In the present work, the umbrella potential was based on a previously developed pseudodihedral angle, x, that allows the base flipping to be treated in a periodic fashion (Fig. 1 B and C; ref. 14), where x = 10° represents the DNA double-helical base-paired state, and x = 195° represents the fully flipped state. By performing MD simulations in the presence of the umbrella potential with different values of the pseudodihedral, the free-energy surface (or potential of mean force) for the entire 360° flipping process shown in Fig. 1 can be determined. Importantly, this approach allows for explicit investigation of both the minor and major groove flipping pathways as well as of the fully flipped conformation.
Umbrella sampling was applied to study the flipping of the target C base (underlined) from the GTCAGCGCATGG DNA sequence (25). Base flipping was studied for four distinct environments of the DNA: (i) in aqueous solution; (ii) complexed to the methyltransferase, M.HhaI, in the “open” conformation in which the active-site loop (residues 80–99) is in an extended conformation (open-binary complex); (iii) complexed to M.HhaI in the “closed” conformation in which the loop region is closed around the DNA (closed-binary complex); and (iv) complexed to M.HhaI along with the cofactor SAH in the closed conformation (ternary complex). The open-binary and closed-ternary complexes with the target C in the flipped orientation are shown in Fig. 1D, with the active-site loop in yellow. Comparison of the two structures shows the significant structural change the system undergoes where the active-site loop “pinches” the DNA upon formation of the closed conformation. Included on the image of the open-binary complex are arrows indicating the two a priori possible paths for flipping of the target base via the minor or major groove.
Methods
Calculations were performed with the program CHARMM (27, 28) using the all-hydrogen protein (29) and nucleic acid (30, 31) parameters, the TIP3P water model (32), and published sodium parameters (33). The M.HhaI-SAM binary complex crystal structure (34) was used to model the open-binary complex, and the ternary complex structure (M.HhaI-DNA-SAH; ref. 35) was used for both the closed-binary and ternary complexes. The initial DNA structure was the canonical B form overlaid with a 35-Å water sphere that contained ions, yielding an electrically neutral system, with the 72 flipped orientations of the base (every 5° from 0 to 360°) obtained by quickly flipping the base out of the DNA double helix, as described (14). Modeling of the flipped DNA intermediates onto the ternary complex was performed by least-squares fitting the DNA structures to that of the DNA in the crystallographic ternary complex, with the least-squares fitting excluding the flipped base region. The closed-binary complex was modeled in a manner identical to the ternary complex by omitting the SAH coenzyme. The open binary complexes were prepared by least-squares fitting the crystallographic binary protein structure, excluding the flexible loop, to the previously modeled ternary crystal structures, from which the orientation of the DNA relative to the binary protein structure was obtained. The modeled binary and ternary systems were then overlaid with a 35-Å water sphere, oriented with respect to protein residues 123 and 254, and sodium ions were added to obtain electrical neutrality.
Equilibration of each modeled structure was done by energy minimization followed by a 60-ps MD simulation in the presence of the umbrella potential (14). In all simulations, the terminal base pairs of the DNA were harmonically constrained to their starting positions by using force constants of 2 kcal/mol/Å, and protein residues outside of the water sphere were fixed, whereas those residues with one or more atoms in the range of 31–35 Å from the center of the water sphere were subjected to harmonic constraints of 2 kcal/mol/Å. Water density in the simulation systems was maintained by using the miscellaneous mean field potential solvent boundary potential (33). All calculations used an atom-based truncation scheme updated heuristically with a list cutoff of 14 Å, a nonbond cutoff of 12 Å, and with the Lennard-Jones (LJ) smoothing function initiated at 10 Å. Electrostatic interactions were smoothed by using a force shift; LJ interactions were force switched (36). MD simulations used a 2-fs integration time step, SHAKE of covalent bonds involving hydrogens (37), and were in the canonical ensemble using the Nosé-Hoover temperature coupling scheme (38).
Determination of the free-energy profiles was performed with an umbrella potential, wi(x) = ki(x – xi)2, where ki is the force constant of 1,000 kcal/mol/rad, x is the value of the pseudodihedral, and xi is the constrained value of the pseudodihedral (Fig. 1 B and C). The pseudopotential was applied in 5° increments from 0 to 360° yielding a total of 72 windows in the free-energy profile. MD simulations in each window were 160 ps in duration. The simulations in the presence of the umbrella potential allows for all accessible conformations of the flipping base to be sampled, yielding a biased probability distribution, W(x)′. The probability distribution is then corrected by accounting for the umbrella potential, yielding the unbiased probability distribution, W(x), from which the free-energy surface can be extracted via ΔG = −kBT lnW(x). The unbiased free-energy profile was extracted by using a modified version of the weighted histogram procedure (39), in which the symmetry of the free-energy surface associated with the use of the periodic pseudodihedral constraint was enforced (40). Test of convergence of the free-energy profiles was performed by calculating the surfaces over incremental 20-ps windows and analyzing the change in the free-energy profiles.
Results and Discussion
Free-energy profiles for target C base flipping for the four environments of the DNA are presented in Fig. 1E. In aqueous solution, base flipping via either the minor or major groove involves a rapid rise in the free energy upon moving away from the Watson–Crick (WC) paired state at x = 10°. Clearly, spontaneous base flipping in DNA in aqueous solution involves large energetic penalties of 18 kcal/mol or more. Binding of the DNA to M.HhaI with the active-site loop in the open conformation, however, does not remedy the situation. In the open-binary complex, the energetic penalties associated with flipping through either groove are maintained, with the fully flipped state of the base (x ≈ 195°) less stable as compared with the corresponding state for DNA in aqueous solution. However, the conformational change in the active-site loop of M.HhaI leading to the closed form of the enzyme (Fig. 1D) has a dramatic impact on the free energy of base flipping. In the closed-binary complex, the WC state is strongly destabilized with the fully flipped state, x ≈ 195°, of the target C base becoming the free-energy minimum. This thermodynamic stabilization of the flipped base structure vs. the WC state by −5.1 kcal/mol is accompanied by large decreases in the barriers to base flipping, with the major groove barrier being significantly lowered. The result of this decrease is that base flipping through the major groove now has a very small barrier of 2.5 kcal/mol at x ≈ 285°. Upon addition of SAH, yielding the ternary complex, these changes are enhanced, with the major groove barrier to flipping now being 0.4 kcal/mol at x ≈ 285° relative to the WC state, x ≈ 10°. This change is accompanied by a −9.4 kcal/mol stabilization of the flipped state, in which the cytosine base (x ≈ 195°) is in the enzyme active site. The difference of ≈4 kcal/mol in the free energies for the flipped conformation upon going from the closed-binary to the ternary complex is in good agreement with the experimental evidence that the binding of SAH causes an increase in DNA binding by −4 kcal/mol (18).
Experimental investigations of base flipping in DNA bound to M.HhaI have concluded that flipping occurs via the minor groove (11), in stark contrast to the major groove pathway indicated by the present results. The previous conclusions appear to be based primarily on steric arguments motivated by the static crystal structures of the M.HhaI-SAM binary (34) and M.HhaI-DNA-SAH ternary (11) complexes. In the experimental ternary complex, in which the target C base is flipped into the active site of M.HhaI, Gln-237 accesses the DNA through the major groove, hydrogen bonding to the orphan G base (i.e., base pair partner of the target C). In the absence of any structural relaxation of the protein, this orientation of Gln-237 would block the major groove flipping pathway, requiring the target C base to flip out via the minor groove. This scenario is consistent with the conformational change in the active-site loop upon going from the open to closed conformation (Fig. 1D), where it moves toward the DNA, ultimately interacting with the minor groove in the closed conformation. This structural change further suggests that base flipping may occur in the open conformation, when the active-site loop does not block the minor groove pathway to flipping. Clearly, the present results yield a significantly different picture of the flipping process. How can the major groove pathway proposed be justified?
The energetics of base flipping from DNA in aqueous solution (Fig. 1E) shows that movement of the target C out of the helix and into an aqueous environment leads to a drastic increase in the free energy, thus disfavoring flipping. If flipping occurs via the minor groove in an open conformation, solvation of the base should occur in a manner similar to DNA in aqueous solution, thereby disfavoring flipping. Accordingly, it may be hypothesized that to facilitate flipping, the protein must supply an environment where the unfavorable energetics associated with flipping into aqueous solution are eliminated. Such an environment could be supplied by the protein via a major groove flipping pathway. In addition, it may be expected that the enzyme would also have to initially perturb the WC base pairing and stacking interactions that stabilize the double-helical structure of DNA (13, 16).
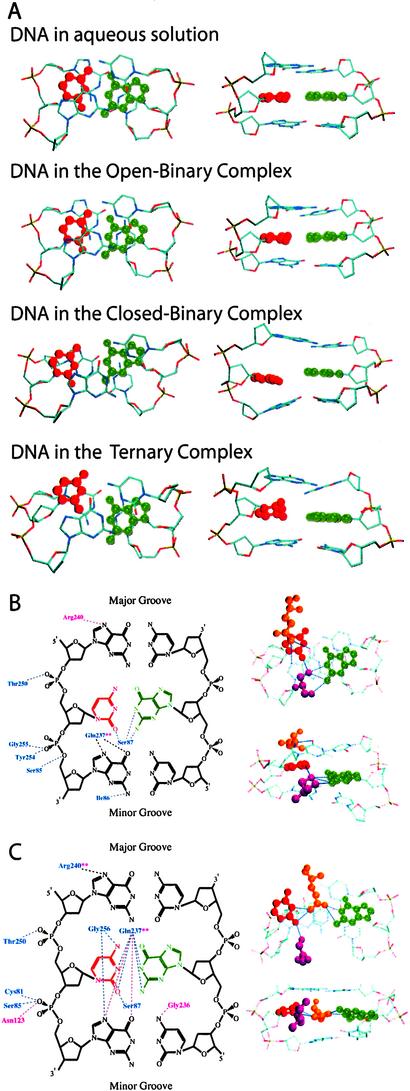
Inspection of the structures corresponding to the WC base-paired conformation (x = 10°) for the four systems studied gives initial insights into the mechanism by which the enzyme facilitates flipping (Fig. 2A). In DNA in aqueous solution and in the open-binary complex, the WC base-pairing and stacking interactions are maintained. However, in the closed-binary and ternary complexes, both the WC base-pairing and stacking interactions are severely disrupted. Indeed, inspection of the ternary complex shows the target C base to have already partially moved into the major groove. Supporting this is the average target C N3 to orphan G N1 distances reported in Table 1. Consistent with Fig. 2A, the N3-N1 distance is significantly longer in both the closed-binary and ternary complexes, as compared with DNA in aqueous solution or in the open-binary complex. Moreover, increased base-pairing distances also occur during the initial stages of major groove flipping; when DNA in aqueous solution and in the open binary complex have attained ≈50% of their maximum free energy at x = 340°, the N3-N1 distances are less then 4 Å, while at the same stage of flipping, the distances are significantly longer for the two closed complexes. It is evident that the closed conformation of the protein leads to significant destabilization of the DNA double-helical conformation in the vicinity of the target C base.
Figure 2.
(A) Central three DNA base pairs at the DNA WC base-paired state (x = 10°, Fig. 1C) for DNA in aqueous solution, in the open-binary complex, in the closed-binary complex, and in the ternary complex. Target C base (red) and orphan G base (green) are indicated. The two images for each complex are the same structures rotated ≈90°. (B and C, Left) Protein residues that interact with the central DNA tribase for the open-binary (purple) and closed-ternary complexes (blue) in the WC paired state (x = 10°) (B) and at 50% of the major groove barrier (x = 340°) (C). Residues interacting in both complexes are indicated by the purple **; the interactions are shown as purple and blue dashed lines for the open-binary and ternary complexes, respectively. Black dashed lines indicate interactions that occur in both complexes. Hydrogen bonds are defined as acceptor-to-donor distances ≤4 Å. (B and C, Right) DNA tribase from the ternary complex at B (x = 10°) and C (x = 340°). Ser-87 (purple), Gln-237 (gold), and protein–DNA hydrogen bonds (blue dashed lines) are shown. Structures were the minimized average Cartesian coordinates from the selected windows of the free-energy profile.
Table 1.
WC base-pairing distances and solvent accessibilities of the four systems at selected points along the base-flipping pathway
| Extent of flipping | Aqueous | Binary, open | Binary, closed | Ternary |
|---|---|---|---|---|
| N1-N3 hydrogen bonding distances for the target C-orphan G base pair | ||||
| Double-helical WC | 2.97 ± 0.09 | 2.95 ± 0.09 | 4.33 ± 0.92 | 4.71 ± 0.28 |
| 50% barrier | 3.77 ± 0.21 | 3.74 ± 0.20 | 4.74 ± 0.48 | 7.34 ± 0.65 |
| 100% barrier | 9.30 ± 2.11 | 7.98 ± 0.56 | 8.24 ± 0.50 | 9.60 ± 0.59 |
| Flipped state | 15.53 ± 0.75 | 18.04 ± 0.33 | 18.53 ± 0.55 | 18.56 ± 0.36 |
| Minor barrier | 5.27 ± 0.29 | 6.36 ± 1.02 | 5.28 ± 0.21 | 2.98 ± 0.11 |
| Solvent accessibilities of the DNA central tribase | ||||
| Double-helical WC | 866 ± 12 | 656 ± 25 | 427 ± 20 | 472 ± 16 |
| 50% barrier | 869 ± 17 | 607 ± 17 | 492 ± 12 | 487 ± 13 |
| 100% barrier | 976 ± 47 | 618 ± 21 | 485 ± 14 | 423 ± 21 |
| Flipped state | 1152 ± 31 | 604 ± 17 | 385 ± 16 | 367 ± 12 |
| Minor barrier | 934 ± 17 | 656 ± 40 | 459 ± 26 | 439 ± 13 |
Distances are given in Å, solvent accessibilities are given in Å2. Errors represent rms fluctuations. Double-helical WC corresponds to a pseudodihedral value of x = 10° in Fig. 1E; 50% barrier corresponds to 340°; 100% barrier corresponds to the maxima at 315° for DNA in aqueous solution and the open-binary complex for major groove flipping; the flipped state corresponds to 195°, and the minor groove barrier is the approximate location of the barrier for DNA in aqueous solution and the open-binary flipping profiles at 40°.
But how is M.HhaI in the closed conformation destabilizing the double-helical conformation? Two possibilities exist: desolvation of the DNA (41) or protein–DNA interactions. Desolvation effects can be addressed by measuring the solvent accessibility of the DNA in the vicinity of the target C for the four systems at different points along the flipping surface (Table 1). In aqueous solution, there is a gradual increase in the solvent accessibility as flipping occurs, as expected. The solvent accessibilities are smaller in the DNA–protein complexes, with the decrease being larger in the closed complexes. A significant decrease in solvent accessibility is evident for DNA in the open-binary complex as compared to that in aqueous solution, yet no facilitation of base flipping occurs. A dominant role of desolvation in destabilization of the WC state is, therefore, not supported. Further support for this conclusion is obtained from analysis of the solvent accessibilities at the barrier to minor groove flipping in Table 1. For all three complexes, the decrease in the solvent accessibility is similar to that at the barrier to major groove flipping, supporting the hypothesis that desolvation is not dominating destabilization of the DNA double-helical conformation. Thus, a low-dielectric environment around the DNA associated with desolvation is, of itself, not responsible for destabilization of the double-helical DNA, although some contribution from this term should not be excluded.
Alternatively, are protein–DNA interactions making significant contributions to destabilization of the WC paired DNA? To identify such interactions, all possible hydrogen bonds between the DNA and the protein for the open-binary and ternary complexes in the WC state (x = 10°) structures were identified and are shown in Fig. 2B. As expected, in the ternary complex several new interactions not present in the open-binary complex occur between residues in the now closed active-site loop (residues 85–87) and the DNA. Beyond the active-site loop, additional interactions occur with residues Thr-250, Tyr-254, and Gly-255, whereas in the binary complex, the only unique interaction involves Arg-240. The observed interactions between M.HhaI and DNA in the WC state of the ternary complex are suggested, therefore, to distort the local DNA structure, leading to destabilization of this conformation and, concomitantly, facilitation of base flipping.
The roles of Ser-87 and Gln-237 in base flipping are of particular interest. Images of the interactions between those residues and the ternary complex DNA are shown on the right of Fig. 2B for the structure corresponding to the WC (x = 10°) conformation. Ser-87 forms three hydrogen bonds with the orphan G and one with the target C, whereas Gln-237 is stacked over the target C, forming two hydrogen bonds with the G base 3′ to the target C. This orientation contrasts with that observed in the ternary experimental crystal structure where the base is fully flipped, and Gln-237 hydrogen bonds with the orphan G. Although previously hypothesized to facilitate base flipping by “pushing” the target C out of the double helix and into the minor groove (42), the present work indicates that Gln-237 initially contributes to base flipping by (i) stacking with the target C base, and (ii) hydrogen bonding to the base adjacent to the target C, helping to distort the helical structure. Distortion of the helix is further facilitated by hydrogen bonding of Ser-87 to both the target C and orphan G bases, effectively competing for the normal C–G WC base-pair interactions. Thus, rather than the target C base being “pushed” out of the DNA helix, the helix is simply destabilized, facilitating flipping.
Although destabilization of the double-helical state via protein–DNA interactions initially facilitates base flipping, the protein must also provide an environment that promotes flipping along the major groove pathway. This is achieved, in part, by simply excluding the flipping base from aqueous solution (in Table 1, compare the open-binary and two closed-complex solvent accessibilities). Exclusion from solvent, however, is apparently not adequate, which is consistent with the limited role of desolvation discussed above. Shown in Fig. 2C are the protein–DNA interactions for the major groove 50% barrier, x = 340°, for the open-binary and ternary complexes along with the ternary complex tribase structure, including residues Ser-87 and Gln-237 for the same conformation. During major groove flipping, the protein forms numerous interactions with the target C base, with the number of interactions being larger in the ternary complex. These interactions act to replace the WC base-pairing and stacking interactions that are lost as the target C base moves out of the DNA double helix. Gln-237 is now interacting with the target C base as well as with the orphan G, lying approximately in the plane of both bases. This orientation is a significant structural change away from that observed in the x = 10° structure (Fig. 2B) and toward that observed in the fully flipped ternary complex. It should be noted that the present analysis does not address entropic contributions to the free-energy surface. It is possible, for example, that as the base and surrounding phosphates are exposed to water molecules, there is a significant loss of conformational freedom of those waters. Such a loss of conformational freedom may lead to a significant entropy loss, contributing to the larger free-energy barriers to flipping in DNA in aqueous solution and in the open-binary complex (Fig. 1E). Movement of the base through the protein would avoid such a loss.
Once the barriers to flipping are lowered when the enzyme assumes the closed conformation, the target C base may readily undergo the structural change to the fully flipped state. The structure of the fully flipped state in the ternary complex has been previously addressed in detail based on the experimental crystallographic structures; the majority of DNA–protein interactions in the x = 195° ternary complex structure are consistent with those observed experimentally (not shown). A significant observation in the present study is the stabilization of the flipped conformation in the ternary complex, as compared with the closed-binary complex (Fig. 1E). Investigation of the protein–DNA interactions showed an increase in the total number of interactions upon going from the closed-binary to the ternary complex. In the x = 195° closed-binary complex structure, there are 38 DNA tribase heteroatom–protein heteroatom interactions ≤4 Å vs. 49 in the ternary complex. Stabilization of the fully flipped state of the DNA upon binding of the coenzyme, therefore, is suggested to be caused by conformational changes induced in the protein surrounding the flipped target C leading to increased protein–DNA interactions.
Insights into the kinetic mechanism of M.HhaI (17–19), NMR, and electrophoresis experiments (16) may be obtained from the present observations. Kinetically, the enzyme primarily follows an ordered bi-bi mechanism with DNA binding followed by coenzyme binding, leading to the catalytically competent complex, although a partial random mechanism has been suggested (19). The free-energy surfaces (Fig. 1E) for the open and closed-binary complexes show that, upon formation of the binary complex, the active-site loop must close on the DNA for the protein to facilitate base flipping. At this stage, the target C base would move out of the helix, sampling different conformations along the major groove pathway. This motion is consistent with the flipped-out “Complex II,” as proposed in the NMR study for the binary complex. Upon formation of the ternary complex, the fully flipped conformation is stabilized, yielding the catalytically active complex, consistent with “Complex III” proposed in the NMR work. Once methylation occurs, the coenzyme first dissociates from the ternary structure, which is consistent with the ordered bi-bi mechanism. Coenzyme dissociation leads to the closed-binary complex, in which the fully flipped state is destabilized relative to the ternary complex, facilitating movement of the target C base back into the DNA double helix. Although not addressed in the present work, dissociation of the coenzyme may facilitate opening of the active-site loop, which would further favor movement of the target C back into the DNA as the free-energy surface shifts from that of the closed-binary to that of the open-binary complex (Fig. 1E). Indeed, if M.HhaI does function via a processive mechanism (18), where the protein does not dissociate from the DNA between methylation events, it is anticipated that once the coenzyme dissociates, the active-site loop moving away from the DNA may allow the protein to diffuse along the DNA.
In the NMR study, it was indicated that an increase in the base flipping rate did not occur in the M.HhaI-DNA binary complex (16), although the possibility that accelerated flipping may occur from a minimally populated species was suggested. It was also stated that the dominant species in the binary complex was Complex I, in which the base is stacked and the active-site loop is open. Those observations are consistent with the energetics of the open-binary complex (Fig. 1E), such that, if Complex I is the dominant species in the NMR experiments, no increase in the flipping rate would be expected.
One of the interesting phenomena with M.HhaI is the increased binding of mismatched bases, abasic moieties, and gaps at the target site in the binary complex (43, 44). Analysis of the free-energy profile for the closed-binary complex (Fig. 1E) shows that, although the WC state is destabilized, there is a local minimum at x = 10° as well as at 310° and at the flipped state. Accordingly, it may be hypothesized that, whereas the normal target C would sample all these conformations, mismatches or the absence of a base would hinder or eliminate sampling of the local minimum at 10°, thereby favoring the equilibrium toward the fully flipped state, leading to increased binding. However, upon formation of the ternary complex, the structure of the protein adjusts to maximize protein–DNA interactions, as evidenced by the decrease in free energy of the flipped state (Fig. 1E). These structural changes are suggested to lead to the experimentally observed preference for cytosine over mismatched bases at the target site in the ternary complex, and the suggestion that coenzyme binding is a contributing factor to specificity (43). The adjustment of the binding pocket to accommodate the target C base is consistent with the formation of the fully closed form of the enzyme with the normal C substrate, as evidenced by gel-shift experiments (43).
State of the art free-energy calculations combined with MD simulations have been used to study base flipping in DNA in the presence of M.HhaI. It is observed that the protein does facilitate the base-flipping process, leading to negligible free-energy barriers to flipping. This facilitation is caused by destabilization of the double-helical DNA conformation through protein–DNA interactions followed by interactions of the flipping base with the protein matrix, stabilizing the partially flipped states. Upon attaining the fully flipped state, the presence of the coenzyme is shown to stabilize this conformation by increasing the number of protein–DNA interactions and decreasing the solvent exposure of the DNA. Most notable is the observation that flipping occurs via the major groove of the DNA, in contrast to the current assumption that a minor groove pathway is involved. This observation, as well as the elucidation of structural details of the high-energy flipping states not readily accessible through experimental approaches, emphasizes the power of computational methods in elucidating structure–function relationship in DNA-protein systems. The approach used here, along with results showing that protein–DNA interactions and desolvation of the DNA facilitate a conformational change in DNA, are expected to be applicable to other protein–DNA complexes.
Acknowledgments
We acknowledge the National Institutes of Health for financial support, and the National Science Foundation Partnerships for Advanced Computational Infrastructure Program, Pittsburgh Supercomputing Center Terascale Computing, and Department of Defense Aeronautical Systems Center Major Shared Resource Center for computational support.
Abbreviations
- SAH
S-adenosylhomocysteine
- target C
target cytosine
- MD
molecular dynamics
- WC
Watson–Crick
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nakao M. Gene. 2001;278:25–31. doi: 10.1016/s0378-1119(01)00721-1. [DOI] [PubMed] [Google Scholar]
- 2.Roberts R J, Halford S S. In: Nucleases. Linn S M, Lloyd R S, Roberts R J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. [Google Scholar]
- 3.Plass C, Soloway P D. Eur J Hum Genet. 2002;10:6–16. doi: 10.1038/sj.ejhg.5200768. [DOI] [PubMed] [Google Scholar]
- 4.Goodman J I, Watson R E. Annu. Rev. Pharmacol. Toxicol. 2002. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M, Herman J G. J Pathol. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 6.Roberts R J. Cell. 1995;82:9–12. doi: 10.1016/0092-8674(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 7.Roberts R J, Cheng X. Annu Rev Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 8.Cheng X, Roberts R J. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel P H, Suzuki M, Adman E, Shinkai A, Loeb L A. J Mol Biol. 2001;308:823–837. doi: 10.1006/jmbi.2001.4619. [DOI] [PubMed] [Google Scholar]
- 10.Schneider T D. Nucleic Acids Res. 2001;29:4881–4891. doi: 10.1093/nar/29.23.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klimasauskas S, Kumar S, Roberts R J, Cheng X. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 12.Slupphaug G, Mol C D, Kavli B, Arvai A S, Krokan H E, Tainer J A. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 13.Stivers J T, Pankiewicz K W, Watanabe K A. Biochemistry. 1999;38:952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 14.Banavali N K, MacKerell A D., Jr J Mol Biol. 2002;319:141–160. doi: 10.1016/S0022-2836(02)00194-8. [DOI] [PubMed] [Google Scholar]
- 15.Varnai P, Lavery R. J Am Chem Soc. 2002;124:7272–7273. doi: 10.1021/ja025980x. [DOI] [PubMed] [Google Scholar]
- 16.Klimasauskas S, Szyperski T, Serva S, Wuthrich K. EMBO J. 1998;17:317–324. doi: 10.1093/emboj/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J C, Santi D V. J Biol Chem. 1987;262:4778–4786. [PubMed] [Google Scholar]
- 18.Lindstrom W M, Jr, Flynn J, Reich N O. J Biol Chem. 2000;275:4912–4919. doi: 10.1074/jbc.275.7.4912. [DOI] [PubMed] [Google Scholar]
- 19.Vilkaitis G, Merkiene E, Serva S, Weinhold E, Klimasauskas S. J Biol Chem. 2001;276:20924–20934. doi: 10.1074/jbc.M101429200. [DOI] [PubMed] [Google Scholar]
- 20.Karplus M, Petsko G A. Nature. 1990;347:631–639. doi: 10.1038/347631a0. [DOI] [PubMed] [Google Scholar]
- 21.Boczko E M, Brooks C L., III Science. 1995;269:393–396. doi: 10.1126/science.7618103. [DOI] [PubMed] [Google Scholar]
- 22.Berneche S, Roux B. Nature. 2001;414:73–77. doi: 10.1038/35102067. [DOI] [PubMed] [Google Scholar]
- 23.McQuarrie D A. Statistical Mechanics. New York: Harper & Row; 1976. [Google Scholar]
- 24.McCammon J A, Karplus M. Proc Natl Acad Sci USA. 1979;76:3585–3589. doi: 10.1073/pnas.76.8.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheikhnejad G, Brank A, Christman J K, Goddard A, Alvarez E, Ford H, Jr, Marquez V E, Marasco C J, Sufrin J R, O'Gara M, Cheng X. J Mol Biol. 1999;285:2021–2034. doi: 10.1006/jmbi.1998.2426. [DOI] [PubMed] [Google Scholar]
- 26.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 27.Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 28.MacKerell A D, Jr, Brooks B, Brooks C L, III, Nilsson L, Roux B, Won Y, Karplus M. In: Encyclopedia of Computational Chemistry. Schleyer P v R, Allinger N L, Clark T, Gasteiger J, Kollman P A, Schaefer H F, III, Schreiner P R., editors. Vol. 1. New York: Wiley; 1998. pp. 271–277. [Google Scholar]
- 29.MacKerell A D, Jr, Bashford D, Bellott M, Dunbrack R L, Jr, Evanseck J, Field M J, Fischer S, Gao J, Guo H, Ha S, et al. J Phys Chem. 1998;102:B3586–B3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 30.Foloppe N, MacKerell A D., Jr J Comput Chem. 2000;21:86–104. [Google Scholar]
- 31.MacKerell A D, Jr, Banavali N. J Comput Chem. 2000;21:105–120. [Google Scholar]
- 32.Jorgensen W L. J Phys Chem. 1983;87:5304–5312. [Google Scholar]
- 33.Beglov D, Roux B. J Phys Chem. 1997;101:7821–7826. [Google Scholar]
- 34.Cheng X, Kumar S, Posfai J, Pflugrath J W, Roberts R J. Cell. 1993;74:299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- 35.O'Gara M, Roberts R J, Cheng X. J Mol Biol. 1996;263:597–606. doi: 10.1006/jmbi.1996.0601. [DOI] [PubMed] [Google Scholar]
- 36.Steinbach P J, Brooks B R. J Comput Chem. 1994;15:667–683. [Google Scholar]
- 37.Ryckaert J P, Ciccotti G, Berendsen H J C. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 38.Nosé S. J Chem Phys. 1984;81:511–519. [Google Scholar]
- 39.Kumar S, Bouzida D, Swendsen R H, Kollman P A, Rosenberg J M. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 40.Crouzy S, Baudry J, Smith J C, Roux B. J Comput Chem. 1999;20:1644–1658. [Google Scholar]
- 41.Elcock A H, McCammon J A. J Am Chem Soc. 1996;118:3787–3788. [Google Scholar]
- 42.Cheng X, Blumenthal R M. Structure (London) 1996;4:639–645. doi: 10.1016/s0969-2126(96)00068-8. [DOI] [PubMed] [Google Scholar]
- 43.Klimasauskas S, Roberts R J. Nucleic Acids Res. 1995;23:1388–1395. doi: 10.1093/nar/23.8.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang P, Nicklaus M C, Marquez V E, Brank A S, Christman J, Banavali N K, MacKerell A D., Jr J Am Chem Soc. 2000;122:12422–12434. [Google Scholar]