Abstract
Latency-associated transcripts of human herpesvirus 6 (H6LTs) (K. Kondo et al. J. Virol. 76:4145-4151, 2002) were maximally expressed at a fairly stable intermediate stage between latency and reactivation both in vivo and in vitro. H6LTs functioned as sources of immediate-early protein 1 at this stage, which up-regulated the viral reactivation.
Human herpesvirus 6 (HHV-6) belongs to the subfamily Betaherpesvirinae (30), which is represented by Human cytomegalovirus (HCMV). Both HHV-6 and HCMV establish latency in the monocyte/macrophage lineage (9, 17-19, 25, 33, 38), and during latent infection HHV-6 and HCMV express latency-associated transcripts that show similar features: (i) both transcripts contain open reading frames (ORFs) encoding immediate-early proteins IE1 and IE2 (20, 21) and (ii) both transcripts are expressed in a small proportion of latently infected cells (19, 20, 32). The function and expression profile of these transcripts have been unknown (24, 39).
In the present study, we first developed a sensitive method to detect the latency-associated transcripts of HHV-6 (H6LTs). Because productive-phase IE1 and IE2 transcripts share their entire sequences with H6LTs (Fig. 1), we previously used the 5′ rapid amplification of cDNA ends (RACE) method to distinguish H6LTs from IE1 and IE2 transcripts (20). To increase the sensitivity, we designed primer sets and probes to amplify the H6LTs or IE1 and IE2 transcripts (Fig. 1). We performed reverse transcription-PCR (RT-PCR) on the RNAs collected from the experimental latent-infection system (19, 20) (Fig. 2A and B).
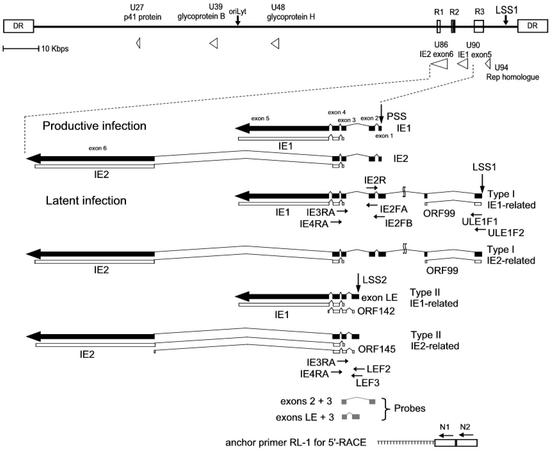
FIG. 1.
RT-PCR design for detecting latent and productive transcripts. The upper portion shows the positions and arrangements of the major repeat elements, R1, R2, and R3, the origin of replication (oriLyt), and the structure of the direct repeat (DR) termini. Positions of the H6LTs and other genes related to the present study are shown. Structures of the productive-phase IE1 and IE2 transcripts and H6LTs, primer positions, and the probes for Southern blot hybridization are shown. The drawings of the mRNAs are all in the same orientation relative to the HHV-6 genome. Thin lines, introns; thick arrows, exons. All exons and introns are drawn to scale. Latency-associated exons starting from latent start site 1 (LSS1) and LSS2 are depicted. The position of the productive start site (PSS) is also shown. Two additional exons of the type I H6LTs are located approximately 7.8 and 9.7 kb upstream from the productive transcription start site. The ORFs encoding IE1 and IE2 and small uORFs are also depicted. Translation for the IE1 and IE2 proteins encoded by H6LTs is suppressed (19, 20); suppression is thought to be caused by the uORF control mechanism (23).
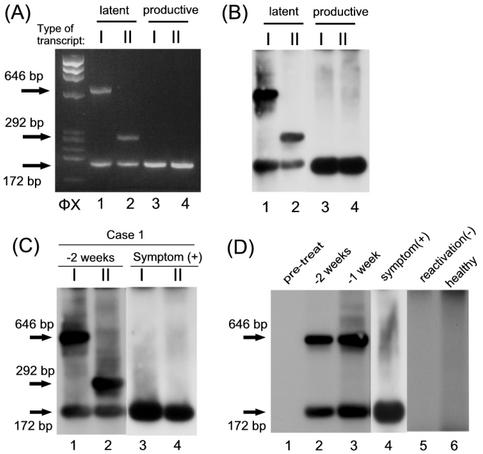
FIG. 2.
Transcription during latency and reactivation. mRNAs for type I (I) or type II (II) H6LTs were amplified by RT-PCR with the primers shown in Fig. 1. RNA from 105 latently infected macrophages (lanes 1 and 2) and 105 reactivation-induced macrophages (lanes 3 and 4) (A and B) from 1 ml of the blood samples from patients with SCT (C) was amplified by the RT-PCR. (C) Blood samples were collected at the onset of symptoms [symptom (+)] or 2 weeks (−2 weeks) before the onset of the symptoms. (D) Samples were collected before the chemotherapy for the SCT (pretreat), 2 weeks (−2 weeks) or 1 week (−1 week) before the onset of symptoms, or at the onset of symptoms [symptom(+)]. Samples from the patient who did not show viral reactivation [reactivation(−)] and a healthy control (healthy) are also shown. (A) Electrophoresis gel stained with ethidium bromide. (B to D) Southern blot hybridization. In panels C and D, exposure time for the samples from patients at the onset of symptoms (C, lanes 3 and 4; D, lane 4) is one-third of that for the other samples. Arrows, predicted sizes of the PCR products. HaeIII-digested φX174 DNA fragments were used as size markers (ΦX).
The latent-infection system of HHV-6 was established as described previously (19). Briefly, peripheral blood macrophages were cultured in RPMI 1640 supplemented with 25% horse serum on plastic plates coated with collagen (Sumitomo Bakelite Co., Ltd.). Macrophages were infected with HHV-6 strain HST on day 7 and were cultured for 4 weeks. At 4 weeks postinfection, no macrophages showed signs of viral replication, such as viral protein expression or infectious-virus production. Viral reactivation was induced by treatment with tetradecanoyl phorbol acetate (TPA; 20 ng/ml) for 7 days and was detected by cocultivation with phytohemagglutinin (PHA)-stimulated umbilical cord blood cells for 7 days.
For the type I H6LTs, the cDNA was amplified with primers IE4RA (5′-GACACATTCTTGGAAGCGATGTCG-3′), ULE1F2 (5′-GCATATCCTGGAGTGGCTGCGCTACC-3′), and IE2FB (5′-CATCCCATCAATTATTGGATTGCTGG-3′) and then with primers IE3RA (5′-GGATTCCATGTTGTTTCCAGAGG-3′), ULE1F1 (5′ CGTTACCGAAGATTACTTCGTGCTG-3′), and IE2FA (5′-GAAACCACCACCTGGAATCAATCTCC-3′). As expected from the structures of the transcripts (Fig. 1), two kinds of amplified products (646 and 172 bp) from type I H6LTs were obtained from latently infected macrophages (latent pattern in Fig. 2A and B). On the other hand, a single amplified product (172 bp) from productive-phase IE1 and IE2 transcripts was obtained from macrophages that were treated with TPA for 7 days to induce viral reactivation (productive pattern in Fig. 2A and B). Type II H6LTs were examined with primers IE4RA, LEF2 (5′-CGTCACAGAATCTAAAAACAAACCATCCGTG-3′), and IE2FB and then with primers IE3RA, LEF3 (5′-CCATCCGTGATTTTTTCCATTCTTAAGG-3′), and IE2FA. The amplified products from the type I (172 bp) and type II (292 bp) H6LTs were observed in latent-phase macrophages, and the product from IE1 and IE2 transcripts (172 bp) was detected in the reactivation phase macrophages. These data indicated that this system was useful for distinguishing between latent-phase and the productive-phase transcripts.
We then applied this method to analyze the RNA from 1 ml of peripheral blood from hematopoietic stem cell transplant (SCT) patients, who are known to show severe complications at the time of HHV-6 reactivation (5, 6, 8, 10, 12, 40). Informed consent was obtained from the blood donors for participation in the study. The RNA was purified as described previously (3) and treated with DNase. Sixteen SCT recipients (mean age, 7 years 2 months; range, 9 months to 16 years 6 months) were examined once a week for active HHV-6 infection. Nine of the patients showed symptoms associated with HHV-6 reactivation, such as fever and rash. Viral reactivation of HHV-6 was confirmed by sequential quantification of the viral DNA in the peripheral blood mononuclear cells (37). Blood samples collected at the onset and those collected 1 to 3 weeks prior to the reactivation were examined. One case that showed both type I and type II transcripts is shown in Fig. 2C, the time course of a representative case is shown in Fig. 2D, and the results are summarized in Table 1. All the blood samples from the above nine patients displayed the productive pattern (IE1 and IE2 expression) when the patients showed symptoms, and samples from two patients showed the productive pattern 1 week prior to the onset of the HHV-6 reactivation. Of the nine reactivation-positive patients, six (66%) showed expression of the type I H6LTs 1 to 3 weeks before the onset of the viral reactivation. Three negative cases were due to a shortage of lymphocytes in the samples (data not shown).
TABLE 1.
Summary of RT-PCR resultsa
| Sample | No. of samples showing detectable transcript/no. tested for:
|
|||
|---|---|---|---|---|
| Type I H6LTs
|
Type II H6LTs
|
|||
| Latentb | Productivec | Latent | Productive | |
| Pretreat | 0/9 | 0/9 | 0/9 | 0/9 |
| 2-3 wk | 5/9 | 0/9 | 1/9 | 0/9 |
| 1 wk | 6/9 | 2/9d | 1/9 | 2/9d |
| Onset | 0/9 | 9/9 | 0/9 | 9/9 |
| No reactivation | 0/7 | 0/7 | 0/7 | 0/7 |
| Healthy | 1/30 | 0/30 | 2/30 | 0/30 |
Samples obtained from the SCT patients were examined by RT-PCR with the primers shown in Fig. 1 before the chemotherapy for the SCT (pretreat), 2 to 3 weeks before the viral reactivation (2-3 wk), 1 week before the reactivation (1 wk), and at the onset of the reactivation (onset). Samples from patients who did not show HHV-6 reactivation (no reactivation) and from healthy age-matched controls (healthy) were also examined. RT-PCR for the type I and type II H6LTs was performed. Samples from the patients with no reactivation did not show detectable transcripts during the observation time period. The expression of the H6LTs was confirmed by H6LT-specific double-nested RT-PCR as described in our previous study (20).
Latent, sample that showed the latent pattern in the RT-PCR shown in Fig. 2.
Productive, sample that showed the productive expression pattern.
Two samples showed both type I H6LTs and productive transcripts, and one of the two samples showed type II H6LTs and productive transcripts.
The expression of H6LTs or IE1 and IE2 transcripts was barely detectable in the samples from the SCT patients before the chemotherapy for the SCT, in those from healthy controls, or in those from reactivation-negative patients (Table 1). The failure to detect the H6LTs is consistent with our previous finding that the H6LTs are not very abundant, and therefore are difficult to detect, in small blood samples from healthy donors (20). Reactivation-positive patients showed significantly higher levels of H6LT expression than the reactivation-negative patients and healthy controls before they showed viral reactivation. A significant increase in HHV-6 DNA was not observed in the reactivation-positive patients (data not shown); therefore, the observed enhancement of the H6LT expression was not thought to be caused by an increase in latently infected cells.
This in vivo study indicated that the expression level of H6LTs might be increased in the period shortly preceding viral reactivation. To investigate the regulation of H6LTs, latently infected macrophages were stimulated with TPA and the H6LT expression was examined. Latent HHV-6 can be reactivated in latently infected macrophages by a 7-day treatment with TPA followed by cocultivation with PHA-stimulated cord blood cells for another 7 days (19). The percentage of H6LT-positive cells was estimated by cell dilution RT-PCR analyses before and after TPA treatment as described in our previous study (20). Briefly, latently infected macrophages were detached from culture dishes, serially diluted, and cultured with a feeder layer of uninfected macrophages. The cells were treated with TPA for 3, 5, or 7 days. H6LTs were amplified by the RT-PCR technique discussed above to distinguish the latent transcripts from productive-phase transcripts. The percentage of transcription-positive cells and the copy number of the transcripts were estimated by a dilution method described previously (20). Briefly, latently infected macrophages were detached from the plates as described previously (19) and were serially diluted (104 to 10 cells) into sample tubes by using four tubes for each dilution. RNA isolated from each sample tube was evaluated by the RT-PCR technique discussed above. To estimate the copy number of the transcripts, mRNAs from the latent macrophages were reverse transcribed and serially diluted into sample tubes by using tRNA as a carrier (four tubes for each dilution) and RT-PCR was performed in each tube. The numbers of transcript-positive cells and copy numbers of the transcripts were calculated by the Reed-Muench method (29), and the copy numbers of the transcripts in each H6LT-positive cell were estimated using these data.
Before TPA treatment, only a small percentage of the latently infected macrophages expressed detectable latent transcripts; however, 3 and 5 days after the treatment with TPA the distribution of cells that expressed the type I H6LTs significantly increased, and then decreased at day 7 (Table 2). These findings suggested that the expression of the H6LTs might be up-regulated at an early stage of viral reactivation and down-regulated after the viral reactivation started. The amount of each H6LT in one cell also increased at days 3 and 5 and then decreased at day 7 (Table 2). Transcription of productive-phase IE1 and IE2 mRNA was not detected at days 0, 3, and 5 by the 5′ RACE method used in our previous report (20) (Fig. 3).
TABLE 2.
Percentage of H6LT-expressing cells during viral latency and reactivationa
| Transcript | % of positive cells (copy no. of transcript in 1 H6LT positive cell) at indicated no. of days poststimulation
|
|||
|---|---|---|---|---|
| 0 | 3 | 5 | 7 | |
| Type I H6LTs | 4.3 ± 1.7 (79 ± 48) | 27.3 ± 13.2 (228 ± 53) | 20.7 ± 12.4 (191 ± 142) | 1.9 ± 1.4 (43 ± 7.5) |
| Type II H6LTs | 5.2 ± 1.0 (65 ± 11) | 11.0 ± 4.6 (131 ± 94) | 8.8 ± 3.7 (104 ± 75) | 1.6 ± 0.48 (34 ± 17) |
| Productive IE1 and IE2 | ND | ND | 0.07 ± 0.06 | 6.5 ± 6.1 (159 ± 118) |
Percentages of H6LT-positive cells during viral reactivation were estimated by the Reed-Muench method (29). The copy numbers of each type of transcript in one H6LT-positive cell are shown in the parentheses. The percentage of cells that showed the productive pattern is also shown. The average of four independent cultures ± the standard deviation is shown for each latent transcript. Expression of the H6LTs was confirmed by the double-nested RT-PCR method described previously (20) (data not shown). Day 0, cells not stimulated with TPA. ND, not detected.
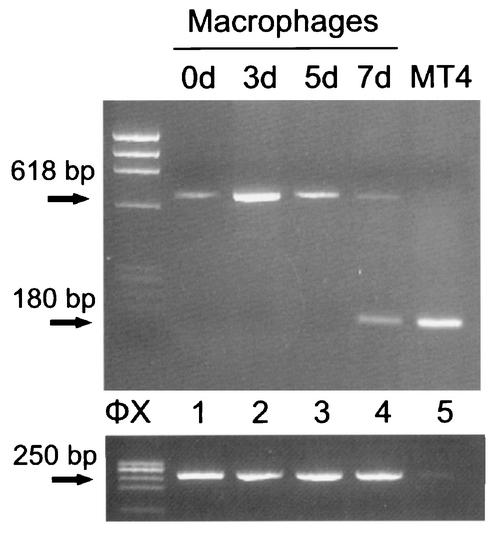
FIG. 3.
5′ RACE amplification of the latent and reactivation-induced cells. RNA from 105 latently infected macrophages (lane 1), 105 reactivation-induced macrophages (lanes 2 to 4), and 102 productively infected MT4 cells (lane 5) was analyzed by the 5′ RACE method. The number of days poststimulation is indicated above each lane. The RACE method used was the same as in our previous study (20). The 5′ end of the transcript was tailed with deoxyadenosine and annealed with the anchor primer RL-1 (Fig. 1) and was amplified first with primers N2 and IE4RA and then with primers N1 and IE2R. The 5′ ends of the productive IE1 and IE2 transcripts (180 bp) and type I H6LTs (618 bp) were detected. Expression of elongation factor 1a mRNA was examined as a standard by RT-PCR, as described previously (amplified product, 250 bp) (42). HaeIII-digested φX174 DNA fragments were used as size markers (ΦX).
We next examined the expression of the IE1 protein (11, 13, 26, 27, 31) in the latent cells that were treated with TPA for 3 and 5 days. Although H6LTs contain the ORF encoding IE1, its translation is significantly suppressed, probably by the translational control mechanism mediated by a small upstream ORF (uORF) (23) (Fig. 1). We used an anti-IE1 mouse monospecific antibody (36) combined with biotin-avidin immunofluorescence systems (Vector Laboratories). The IE1 protein was detectable in approximately 5% of the cells; however, glycoproteins B (3, 12) and H (17, 23, 29) and the p41 protein (2, 7, 16, 41), which are known to be expressed during productive infection, were not detectable (Fig. 4). Infectious virus was not detected at day 3 or 5 (data not shown). Because productive-phase IE1 mRNAs were not detectable in these cells, the H6LTs, which contain the IE1 ORF (Fig. 1), were considered to be translated into the IE1 protein at this novel, intermediate stage. For some mRNAs that have small uORFs that restrict the translation of the downstream ORFs, it has been reported that modulation of both translational regulation and the mRNA level is important for release from the uORF control (14, 28). An alteration in the regulation of translation as well as the increase of the H6LTs might have contributed to the increased IE1 protein expression (Table 2).
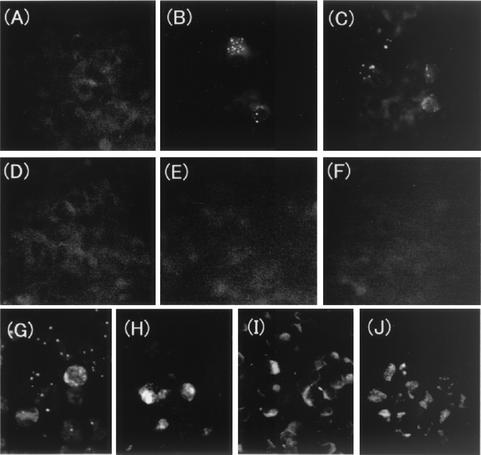
FIG. 4.
Protein expression in the H6LT-expressing macrophages. (A) Latently infected macrophages; (B to F) infected macrophages treated with TPA for 3 days (B) or 5 days (C to F); (G to J) productively infected MT4 cells. Cells were stained with the following antibodies: a monospecific antibody against IE1 (A to C, and G) (34), a monoclonal antibody (MAb) against glycoprotein B (D and H) (28), a MAb against glycoprotein H (E and I) (28), and a MAb against p41 (F and J) (Advanced Biotechnologies Inc.).
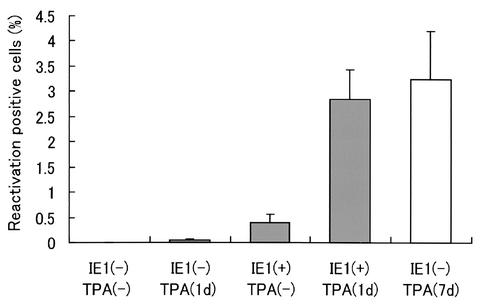
To study the function of the expressed IE1 protein, we generated the plasmid expression vector pcDNA3.1-IE1 by cloning the full-length cDNA for IE1 (20; K. Kondo, unpublished data). The latent macrophages were transfected with the pcDNA3.1-IE1 by using a Nucleofector electroporator (Amaxa Biosystems) according to the manufacturer's recommended protocol. Briefly, latently infected cells were detached from the culture dish with 1% EDTA as described previously (19). The cells were mixed with 5 μg of the plasmid and 100 μl of Nucleofector solution V, and electroporation was performed with Nucleofector by using the program T-20. The transfection efficiency was assessed by examining IE1 protein expression, which ranged from 2.1 to 4.0%. The transfected cells were serially diluted and cocultivated with an uninfected macrophage feeder layer. When TPA treatment was performed, viral reactivation was observed 4 days after the appearance of the IE1 protein (Fig. 4); therefore, PHA-stimulated cord blood cells were added to the macrophages 4 days after the transfection, and the appearance of the infectious virus was monitored. The efficiency of the viral reactivation was calculated by the Reed-Muench method (29), and the data were corrected for transfection efficiency (Fig. 5). Because the reactivation of another betaherpesvirus, HCMV, is known to depend on the differentiation and/or activation of monocytes/macrophages (33, 34), we induced the differentiation and/or activation of the latently infected macrophages by using a short TPA treatment (10 ng/ml for 24 h) (1, 35). We confirmed the differentiation and/or activation of the cells morphologically and by phagocytosis assay (4) (data not shown). The virus was efficiently reactivated by IE1 expression combined with the short TPA treatment, and the reactivation rate was similar to that induced by long TPA treatment (7 days), which was previously thought to be the most efficient way to induce reactivation in this system (Fig. 5) (19). The virus was not reactivated by the short TPA treatment alone (Fig. 5). These data suggested that IE1 protein expression was an important factor for HHV-6 reactivation and that there were other control points during the course of the viral reactivation, as has also been suggested for HCMV and murine cytomegalovirus (22, 34).
FIG. 5.
Viral reactivation induced by IE1. Percentages of reactivation-positive cells in the following samples are shown: untreated cells [IE1(−) TPA(minus)], cells treated with a low concentration of TPA for 1 day [IE1(minus) TPA(1d)], cells transfected with an IE1-expressing plasmid without TPA [IE1(+) TPA(−)], and cells transfected with IE1 and treated with a low concentration of TPA for 1 day [IE1(−) TPA(1d)]. As a positive control cells were treated with a high concentration of TPA for 7 days and cocultivated with umbilical cord blood cells for 7 days [IE1(−) TPA(7d)], as described previously (19). The averages and standard deviations of three independent studies are shown.
Only a small proportion of latently infected cells (approximately 5 to 7%) contain detectable latent transcripts during HHV-6 latency (20) (Table 2); however, our findings have raised the possibility that the expression of the latent transcripts may correspond to a separate stage of latency. In the present study, we carefully analyzed the expression of the H6LTs over time in vivo and in vitro and found that the H6LTs might be expressed most abundantly just before the onset of viral reactivation (Tables 1 and 2). The abundantly expressed H6LTs could be the sources of the IE1 protein (Fig. 4), and IE1 enhanced the viral reactivation (Fig. 5). Importantly, this intermediate phase of viral infection is different from the reactivation phase, which is characterized by the expression of productive-phase IE1 and IE2 transcripts (15, 22, 33). The cellular differentiation and/or activation of the latently infected cells may an important factor in the induction of complete viral reactivation (Fig. 5) (34), and the differentiation and/or activation of the latently infected cells that are in the intermediate phase might cause viral reactivation. The intermediate stage seems to be relatively stable, because viral regulation did not occur during this stage (Fig. 4) and because it took another 4 days for the in vitro reactivation to occur (Table 2) and another 1 to 3 weeks for the in vivo reactivation to occur (Table 1).
Thus, we have recognized a novel intermediate stage in HHV-6 latency that may be a preparatory stage for reactivation and that is characterized by the abundant expression of H6LTs. Latent transcripts are detected in a small percentage cells that are latently infected with HCMV, as is observed in HHV-6 latency (18, 21, 32), and many other features of HCMV latency are similar to those of HHV-6 latency, as described above. Therefore, we hypothesize that the intermediate stage of latency might be common to HHV-6 and HCMV and that the expression of HCMV latent transcripts might be enhanced during this stage.
Acknowledgments
We thank Takako Yamada, Junichi Hara, and their colleagues for their kind help in obtaining blood samples.
This study was supported by Special Coordination Funds for Promoting Science and Technology and a grant-in-aid for general scientific research from the Ministry of Education, Culture, Sports, Science and Technology, the Japanese Government.
REFERENCES
- 1.Abrahm, J. L., and R. Smiley. 1981. Modification of normal human myelopoiesis by 12-0 tetradecanoylphorbol-13-acetate (TPA). Blood 58:1119-1126. [PubMed] [Google Scholar]
- 2.Agulnick, A. D., J. R. Thompson, S. Iyengar, G. Pearson, D. Ablashi, and R. P. Ricciardi. 1993. Identification of a DNA-binding protein of human herpesvirus 6, a putative DNA polymerase stimulatory factor. J. Gen. Virol. 74:1003-1009. [DOI] [PubMed] [Google Scholar]
- 3.Aono, T., K. Kondo, H. Miyoshi, K. Tanaka-Taya, M. Kondo, Y. Osugi, J. Hara, S. Okada, and K. Yamanishi. 1998. Monitoring of human cytomegalovirus infections in pediatric bone marrow transplant recipients by nucleic acid sequence-based amplification. J. Infect. Dis. 178:1244-1249. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, P. A., B. P. Canono, and D. A. Drevets. 1995. Measurement of bacterial ingestion and killing by macrophages, p. 14.6.1-14.6.3. In R. Coico (ed.), Current protocols in immunology. John Wiley and Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 5.Carrigan, D. R., and K. K. Knox. 1994. Human herpesvirus 6 (HHV-6) isolation from bone marrow: HHV-6-associated bone marrow suppression in bone marrow transplant patients. Blood 84:3307-3310. [PubMed] [Google Scholar]
- 6.Caserta, M. T., D. J. Mock, and S. Dewhurst. 2001. Human herpesvirus 6. Clin. Infect. Dis. 33:829-833. [DOI] [PubMed] [Google Scholar]
- 7.Chang, C. K., and N. Balachandran. 1991. Identification, characterization, and sequence analysis of a cDNA encoding a phosphoprotein of human herpesvirus 6. J. Virol. 65:2884-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cone, R. W., M. L. Huang, L. Corey, J. Zeh, R. Ashley, and R. Bowden. 1999. Human herpesvirus 6 infections after bone marrow transplantation: clinical and virologic manifestations. J. Infect. Dis. 179:311-318. [DOI] [PubMed] [Google Scholar]
- 9.Dankner, W. M., J. A. McCutchan, D. D. Richman, K. Hirata, and S. A. Spector. 1990. Localization of human cytomegalovirus in peripheral blood leukocytes by in situ hybridization. J. Infect. Dis. 161:31-36. [DOI] [PubMed] [Google Scholar]
- 10.Dockrell, D. H., and C. V. Paya. 2001. Human herpesvirus-6 and -7 in transplantation. Rev. Med. Virol. 11:23-36. [DOI] [PubMed] [Google Scholar]
- 11.Dominguez, G., T. R. Dambaugh, F. R. Stamey, S. Dewhurst, N. Inoue, and P. E. Pellett. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73:8040-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautheret-Dejean, A., C. Manichanh, F. Thien-Ah-Koon, A. M. Fillet, N. Mangeney, M. Vidaud, N. Dhedin, J. P. Vernant, and H. Agut. 2002. Development of a real-time polymerase chain reaction assay for the diagnosis of human herpesvirus-6 infection and application to bone marrow transplant patients. J. Virol. Methods 100:27-35. [DOI] [PubMed] [Google Scholar]
- 13.Gompels, U. A., J. Nicholas, G. Lawrence, M. Jones, B. J. Thomson, M. E. Martin, S. Efstathiou, M. Craxton, and H. A. Macaulay. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29-51. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann, B., O. Valerius, M. Andermann, and G. H. Braus. 2001. Transcriptional autoregulation and inhibition of mRNA translation of amino acid regulator gene cpcA of filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 12:2846-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hummel, M., Z. Zhang, S. Yan, I. DePlaen, P. Golia, T. Varghese, G. Thomas, and M. I. Abecassis. 2001. Allogeneic transplantation induces expression of cytomegalovirus immediate-early genes in vivo: a model for reactivation from latency. J. Virol. 75:4814-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyengar, S., P. H. Levine, D. Ablashi, J. Neequaye, and G. R. Pearson. 1991. Sero-epidemiological investigations on human herpesvirus 6 (HHV-6) infections using a newly developed early antigen assay. Int. J. Cancer 49:551-557. [DOI] [PubMed] [Google Scholar]
- 17.Kempf, W., V. Adams, N. Wey, R. Moos, M. Schmid, E. Avitabile, and G. Campadelli-Fiume. 1997. CD68+ cells of monocyte/macrophage lineage in the environment of AIDS-associated and classic-sporadic Kaposi sarcoma are singly or doubly infected with human herpesviruses 7 and 6B. Proc. Natl. Acad. Sci. USA 94:7600-7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondo, K., H. Kaneshima, and E. S. Mocarski. 1994. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc. Natl. Acad. Sci. USA 91:11879-11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo, K., T. Kondo, T. Okuno, M. Takahashi, and K. Yamanishi. 1991. Latent human herpesvirus 6 infection of human monocytes/macrophages. J. Gen. Virol. 72:1401-1408. [DOI] [PubMed] [Google Scholar]
- 20.Kondo, K., K. Shimada, J. Sashihara, K. Tanaka-Taya, and K. Yamanishi. 2002. Identification of human herpesvirus 6 latency-associated transcripts. J. Virol. 76:4145-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo, K., J. Xu, and E. S. Mocarski. 1996. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc. Natl. Acad. Sci. USA 93:11137-11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurz, S. K., and M. J. Reddehase. 1999. Patchwork pattern of transcriptional reactivation in the lungs indicates sequential checkpoints in the transition from murine cytomegalovirus latency to recurrence. J. Virol. 73:8612-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovett, P. S., and E. J. Rogers. 1996. Ribosome regulation by the nascent peptide. Microbiol. Rev. 60:366-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lunetta, J. M., and J. A. Wiedeman. 2000. Latency-associated sense transcripts are expressed during in vitro human cytomegalovirus productive infection. Virology 278:467-476. [DOI] [PubMed] [Google Scholar]
- 25.Minton, E. J., C. Tysoe, J. H. Sinclair, and J. G. Sissons. 1994. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J. Virol. 68:4017-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicholas, J. 1994. Nucleotide sequence analysis of a 21-kbp region of the genome of human herpesvirus-6 containing homologues of human cytomegalovirus major immediate-early and replication genes. Virology 204:738-750. [DOI] [PubMed] [Google Scholar]
- 27.Nicholas, J., and M. E. Martin. 1994. Nucleotide sequence analysis of a 38.5-kilobase-pair region of the genome of human herpesvirus 6 encoding human cytomegalovirus immediate-early gene homologs and transactivating functions. J. Virol. 68:597-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nomura, A., Y. Iwasaki, M. Saito, Y. Aoki, E. Yamamori, N. Ozaki, K. Tachikawa, N. Mutsuga, M. Morishita, M. Yoshida, M. Asai, Y. Oiso, and H. Saito. 2001. Involvement of upstream open reading frames in regulation of rat V(1b) vasopressin receptor expression. Am. J. Physiol. 280:E780-E787. [DOI] [PubMed] [Google Scholar]
- 29.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493. [Google Scholar]
- 30.Roizman, B., R. C. Desrosiers, B. Fleckenstein, C. Lopez, A. C. Minson, and M. J. Studdert. 1992. The family Herpesviridae: an update. Arch. Virol. 123:425-449. [DOI] [PubMed] [Google Scholar]
- 31.Schiewe, U., F. Neipel, D. Schreiner, and B. Fleckenstein. 1994. Structure and transcription of an immediate-early region in the human herpesvirus 6 genome. J Virol 68:2978-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slobedman, B., and E. S. Mocarski. 1999. Quantitative analysis of latent human cytomegalovirus. J. Virol. 73:4806-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soderberg-Naucler, C., K. N. Fish, and J. A. Nelson. 1997. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 91:119-126. [DOI] [PubMed] [Google Scholar]
- 34.Soderberg-Naucler, C., D. N. Streblow, K. N. Fish, J. Allan-Yorke, P. P. Smith, and J. A. Nelson. 2001. Reactivation of latent human cytomegalovirus in CD14+ monocytes is differentiation dependent. J. Virol. 75:7543-7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svet-Moldavskaya, I. A., S. N. Zinzar, G. J. Svet-Moldavsky, P. E. Mann, J. G. Bekesi, J. F. Holland, B. D. Clarkson, Z. Arlin, and B. Koziner. 1979. Macrophage-like cell transformation and CFU(c) fluctuations in normal and leukemic human marrow cultures treated by phorbol diester. Biomedicine 31:252-257. [PubMed] [Google Scholar]
- 36.Takeda, K., N. Nakagawa, T. Yamamoto, R. Inagi, K. Kawanishi, Y. Isegawa, and K. Yamanishi. 1996. Prokaryotic expression of an immediate-early gene of human herpesvirus 6 and analysis of its viral antigen expression in human cells. Virus Res. 41:193-200. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka-Taya, K., T. Kondo, N. Nakagawa, R. Inagi, H. Miyoshi, T. Sunagawa, S. Okada, and K. Yamanishi. 2000. Reactivation of human herpesvirus 6 by infection of human herpesvirus 7. J. Med. Virol. 60:284-289. [DOI] [PubMed] [Google Scholar]
- 38.Taylor-Wiedeman, J., J. G. Sissons, L. K. Borysiewicz, and J. H. Sinclair. 1991. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J. Gen. Virol. 72:2059-2064. [DOI] [PubMed] [Google Scholar]
- 39.White, K. L., B. Slobedman, and E. S. Mocarski. 2000. Human cytomegalovirus latency-associated protein pORF94 is dispensable for productive and latent infection. J. Virol. 74:9333-9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zerr, D. M., D. Gupta, M. L. Huang, R. Carter, and L. Corey. 2002. Effect of antivirals on human herpesvirus 6 replication in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 34:309-317. [DOI] [PubMed] [Google Scholar]
- 41.Zhou, Y., C. K. Chang, G. Qian, B. Chandran, and C. Wood. 1994. trans-Activation of the HIV promoter by a cDNA and its genomic clones of human herpesvirus-6. Virology 199:311-322. [DOI] [PubMed] [Google Scholar]
- 42.Zou, P., Y. Isegawa, K. Nakano, M. Haque, Y. Horiguchi, and K. Yamanishi. 1999. Human herpesvirus 6 open reading frame U83 encodes a functional chemokine. J. Virol. 73:5926-5933. [DOI] [PMC free article] [PubMed] [Google Scholar]