Abstract
Human mtDNA shows striking regional variation, traditionally attributed to genetic drift. However, it is not easy to account for the fact that only two mtDNA lineages (M and N) left Africa to colonize Eurasia and that lineages A, C, D, and G show a 5-fold enrichment from central Asia to Siberia. As an alternative to drift, natural selection might have enriched for certain mtDNA lineages as people migrated north into colder climates. To test this hypothesis we analyzed 104 complete mtDNA sequences from all global regions and lineages. African mtDNA variation did not significantly deviate from the standard neutral model, but European, Asian, and Siberian plus Native American variations did. Analysis of amino acid substitution mutations (nonsynonymous, Ka) versus neutral mutations (synonymous, Ks) (ka/ks) for all 13 mtDNA protein-coding genes revealed that the ATP6 gene had the highest amino acid sequence variation of any human mtDNA gene, even though ATP6 is one of the more conserved mtDNA proteins. Comparison of the ka/ks ratios for each mtDNA gene from the tropical, temperate, and arctic zones revealed that ATP6 was highly variable in the mtDNAs from the arctic zone, cytochrome b was particularly variable in the temperate zone, and cytochrome oxidase I was notably more variable in the tropics. Moreover, multiple amino acid changes found in ATP6, cytochrome b, and cytochrome oxidase I appeared to be functionally significant. From these analyses we conclude that selection may have played a role in shaping human regional mtDNA variation and that one of the selective influences was climate.
Numerous previous surveys of aboriginal populations have demonstrated that the branches of the mtDNA tree (composed of groups of related haplotypes or haplogroups) are continent-specific, with virtually no mixing of mtDNA haplogroups from the different geographic regions (1). In Africa, the three most ancient mtDNA haplogroups (L0, L1, and L2), which make up macrohaplogroup L, are specific for sub-Saharan Africa. African macrohaplogroup L radiated to form the Africa-specific haplogroup L3 as well as the Eurasian macrohaplogroups M and N. M and N arose in northeastern Africa and individuals bearing M and N mtDNAs subsequently left Africa to colonize Europe and Asia (1, 2).
Among Europeans, haplogroups H, I, J, N1b, T, U, V, W, and X make up >98% of the mtDNAs. These haplogroups were derived primarily from macrohaplogroup N.
In Asia, macrohaplogroups N and M contributed equally to mtDNA radiation, with a plethora of derivative mtDNA lineages being generated within southeastern and central Asia (3). However, in Siberia, northward from the Altai Mountains and the Amur River, only six mtDNA haplogroups (A, C, D, G, Z, and Y) make up >75% of the mtDNAs. In contrast, south of Tibet and Korea, haplogroups A, C, D, and G represent only 14% of the mtDNAs, and haplogroups Y and Z are rare. Thus there is a 5-fold enrichment of A, C, D, and G mtDNAs between central Asia and Siberia (4, 5).
In Native American populations, only five Old World mtDNA haplogroups (A, B, C, D, and X) encompass 100% of the mtDNA variation (1). Haplogroups A, C, and D, which represent 58% of Siberian mtDNAs, came to the Americas from northern Siberia across the Bering land bridge. Haplogroup B may have arrived later, because it is virtually absent in Siberia and rare in northern North America, and its sequence diversity in Native Americans is less than that of A, C, or D. In Asia, B is found primarily along the Asian coast and out into the Pacific. Hence, it might have come to the Americas via a coastal route, thus bypassing the extreme north. Finally, Native American haplogroup X is concentrated in north central North America and is distantly related to European X. Hence it probably also arrived in the Americas via a northern route.
Thus, extensive global population studies have shown that there are striking differences in the nature of the mtDNAs found in different geographic regions. Previously, these marked differences in mtDNA haplogroup distribution were attributed to founder effects, specifically the colonizing of new geographic regions by only a few immigrants that contributed a limited number of mtDNAs. However, this model is difficult to reconcile with the fact that northeastern Africa harbors all of the African-specific mtDNA lineages as well as the progenitors of the Eurasia radiation, yet only two mtDNA lineages (macrohaplogroups M and N) left northeastern Africa to colonize all of Eurasia (1, 2) and also that there is a striking discontinuity in the frequency of haplogroups A, C, D, and G between central Asia and Siberia, regions that are contiguous over thousands of kilometers. Rather than Eurasia and Siberia being colonized by a limited number of founders, it seems more likely that environmental factors enriched for certain mtDNA lineages as humans moved to the more northern latitudes.
Natural selection has been hypothesized to explain anomalies in the branch lengths of certain European (6) and African (7) mtDNA lineages. The mtDNA encodes 13 polypeptides of oxidative phosphorylation (OXPHOS) including ND1, ND2, ND3, ND4, ND4L, ND5, and ND6 of complex I (NADH dehydrogenase); cytochrome b (cytb) of complex III (bc1 complex); COI, COII, and COIII of complex IV (cytochrome c oxidase); and ATP6 and ATP8 of complex V (ATP synthase). Hence, the genes of the mtDNA are central to energy production, both to generate ATP to perform work and to generate heat to maintain body temperature.
We now hypothesize that natural selection may have influenced the regional differences between mtDNA lineages. This hypothesis is supported by our demonstration of striking differences in the ratio of nonsynonymous (nsyn)/synonymous (syn) nucleotide changes in mtDNA genes between geographic regions in different latitudes. We speculate that these differences may reflect the ancient adaptation of our ancestors to increasingly colder climates as Homo sapiens migrated out of Africa and into Europe and northeastern Asia.
Materials and Methods
Sampling Strategy and Sequencing.
Fifty-six mtDNA sequences were available from the literature (8–12) encompassing individuals sampled from African, European, and Asian populations based on their language groups and geographic distribution. We analyzed 48 additional individuals from African, Asian, European, Siberian, and Native American populations to complete a global survey of mtDNA variation (13).
Sequence Quality Control and Haplogroup Assignment.
The quality of the sequences generated by automated DNA sequencing system (Applied Biosystems 377) was ensured, and nucleotide changes were analyzed by SEQUENCHER 4.0.5 software. Nucleotide changes were noted in comparison with the revised Cambridge Reference Sequence (9, 10). Assignment of sequences to specific haplogroups was performed according to criteria as discussed (1).
Phylogenetic Analysis.
A consensus neighbor-joining tree of the 104 complete human mtDNA sequences was constructed, and bootstrap values (percentage of 500 total bootstrap replicates) were calculated by using phylip (37). DNA sequence divergence was analyzed by using the diverge program (Wisconsin Package version 10.0, GCG).
Coalescence Dates for Haplogroups.
The times to the most recent common ancestor (MRCA) were calculated by using only the mtDNA coding region (nucleotide positions 577–16023), because of the high probability of reverse mutations in the control region. The average sequence evolution rate was estimated by using the HKY85 model (14). Standard errors were calculated from the inverse hessian at the maximum of the likelihood, do not include any uncertainty in the calibration point, and were calculated by using the delta method. The coalescence times of the various haplogroups may well be underestimated because of the small sample size.
Ka/Ks Analysis.
To detect the influence of selection, the ratio of the number of substitutions causing amino acid replacements (nsyn sites) per total possible nsyn sites (ka) was divided by the number of silent substitutions (syn sites) per possible syn sites (ks). ks and ka were calculated by using dnasp software (15). The ka/ks ratio for all 13 human mtDNA polypeptide genes from the 104 complete human mtDNA sequences (16) was calculated by adding a constant of the ks for one syn mutation to avoid dividing by 0 [ka/(ks + constant)]. As an alternative to adding a constant, we also calculated the ka/(ka + ks) ratio. This also yielded some zero values, which were dropped from subsequent calculations. The significance (P values) for comparisons of the distributions of ka/(ks + constant) or ka/(ka + ks) values for haplogroups of tropical (L0-L3) to temperate (H, V, U, J, T, I, X, N1b, W) or arctic (A, C, D, G, X, Y, Z) zones were calculated by using the Wilcoxon rank-sum test. Both ka/(ks + constant) and ka/(ka + ks) calculations gave similar results.
The data used in this paper are available at our MITOMAP web site, www.mitomap.org.
Results
Phylogenetic Analysis and Calculation of Coalescence Times.
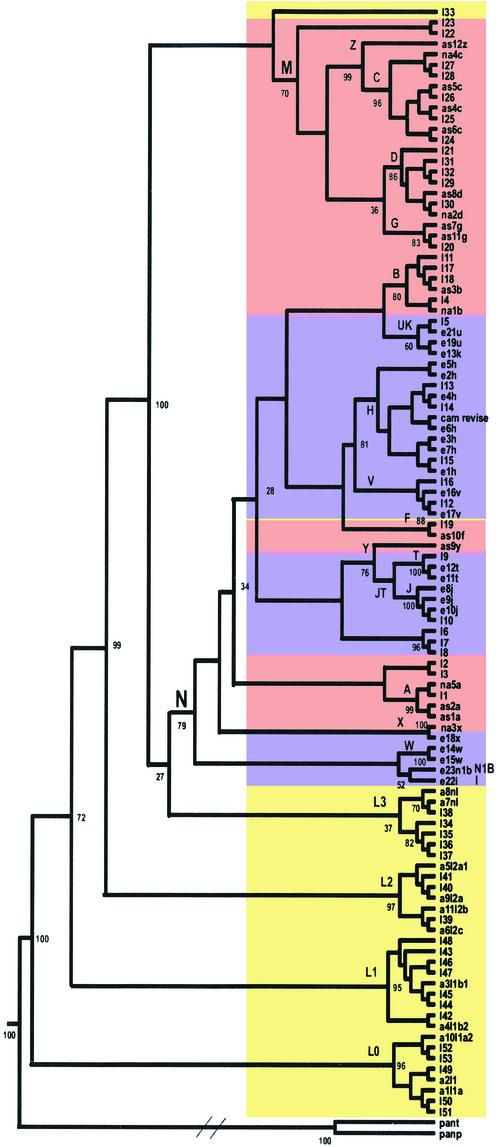
The complete sequences of 104 human mtDNAs were analyzed for the extent and nature of their variation. Analysis of the collective mtDNA sequences reaffirmed that all human mtDNAs belong to a single tree, rooted in Africa, that was derived from the sequential accumulation of mutations along radiating maternal lineages (1, 8, 13, 17, 18) (Fig. 1). Assuming that the mtDNA sequence evolution rate is constant, the mtDNA sequence diversity suggests that the MRCA of the human mtDNA phylogeny occurred ≈200,000 years before present (YBP), when calibrated by using the human-chimpanzee divergence time of 6.5 million YBP (19) (Table 1).
Figure 1.
Consensus neighbor-joining tree of 104 human mtDNA complete sequences. Numbers correspond to bootstrap values (percentage of 500 total bootstrap replicates). Because this is a consensus tree, based on bootstrapping, the branch length is not proportional to the mutation numbers. Diagonal lines are drawn in the chimp lineage to denote the much greater genetic distance between human and chimp than among the various human mtDNAs. Maximum likelihood and unweighted pair group method with arithmetic mean methods yielded the same branching orders with respect to the geographically delimited mtDNA haplogroups. Sequences are: I1–53, GenBank accession nos. AF346963–AF347015, numbered according to figure 2 in Ingman et al. (8), starting from the top of that figure; e21u, GenBank accession no. X93334; a1l1a, GenBank accession no. D38112; cam revise, GenBank accession no. NC_001807 corrected according to ref. 10; the rest are 48 sequences generated by us by using Applied Biosystems 377. Colors correspond to the continental origin of the individuals chosen for this analysis: yellow, Africans; purple, European; pink, Asians and Native Americans. Specific mutations in patient samples that have been implicated in disease were excluded from this analysis, as were gaps and deletions, with the exception of the 9-bp deletion (nucleotide positions 8272–8280). Haplogroup names are designated with capital letters. Pan paniscus and Pan troglodytes mtDNA sequences were used as outgroups. Haplogroups L0 and L1 replace the previously defined haplogroups L1a and L1b, respectively (35).
Table 1.
Coalescence dates for haplogroups
| Haplogroup | Sample sizes | Time to MRCA ± SE, ×10−4 mutations per nucleotide position* | Time to MRCA ± SE, ×103 years |
|---|---|---|---|
| Chimp+human | 1 + 104 | 818.05 ± 0.75 | 6,500 |
| Humans | 104 | 24.88 ± 0.90 | 198 ± 19 |
| L0 | 8 | 17.92 ± 1.87 | 142 ± 17 |
| L1 | 9 | 17.81 ± 1.77 | 142 ± 17 |
| L2 | 7 | 11.57 ± 1.30 | 91.9 ± 11.8 |
| N | 50 | 8.09 ± 0.53 | 64.3 ± 5.8 |
| A | 4 | 4.06 ± 0.92 | 32.3 ± 7.6 |
| R | 37 | 7.66 ± 0.51 | 60.9 ± 5.5 |
| HV | 15 | 3.61 ± 0.73 | 28.7 ± 6.1 |
| H | 11 | 2.40 ± 0.40 | 19.1 ± 3.4 |
| V | 3 | 1.71 ± 0.60 | 13.6 ± 4.8 |
| JT | 7 | 6.29 ± 0.74 | 50.0 ± 6.7 |
| J | 4 | 4.33 ± 0.87 | 34.4 ± 7.2 |
| T | 3 | 1.40 ± 0.55 | 11.1 ± 4.4 |
| U | 4 | 6.51 ± 0.66 | 51.7 ± 6.2 |
| M | 22 | 8.15 ± 0.74 | 64.8 ± 7.1 |
| CZ | 10 | 5.91 ± 0.87 | 47.0 ± 7.6 |
| C | 9 | 3.56 ± 0.65 | 28.3 ± 5.5 |
| D | 6 | 4.19 ± 0.67 | 33.3 ± 5.7 |
Based on these data we estimated the average sequence evolution rate as 1.26 ± 0.08 × 10−8 base substitutions per nucleotide per year, using the HKY85 model (14).
Although all of the mtDNA macrolineages are represented in northeastern Africa, only two macrolineages (M and N), sharing the same approximate date of origin (65,000 years before present), subsequently left Africa to colonize Europe and Asia. All European, Asian, and Native American mtDNA lineages are derived from these two founder M and N lineages (1, 2) (Fig. 1, Table 1). Our phylogenetic analysis and MRCA calculations are in agreement with previous studies based on D-loop sequences, PCR-restriction fragment length polymorphism variation, and mtDNA sequences analysis of samples collected by using different sampling strategies (1, 8, 13, 17, 18).
Nonrandom Continental mtDNA Variation.
To determine whether the observed regional transitions of mtDNA haplogroups represented a deviation from the standard neutral model, we analyzed the distribution of mtDNA sequence variants between geographic regions by using the Tajima's D and the Fu and Li D* tests (20, 21). We also calculated the continental frequency distribution of pairwise mtDNA sequence differences to test for rapid population expansion (22) (Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org).
For all of the African mtDNA sequences belonging to the L haplogroups (L0–L3) (n = 32), the Tajima's D and the Fu and Li D* test results did not significantly deviate from the standard neutral model. Similarly, these parameters did not significantly deviate from neutrality for each of the haplogroup L lineages alone: L0 (n = 8), L1 (n = 9), L2 (n = 7), or L3 (n = 7). Moreover, the frequency distribution of pairwise sequence difference counts for all haplogroup L mtDNAs revealed a broad and ragged distribution (Fig. 4). All of these results are consistent with the African mtDNA population having been relatively stable for a long time.
By contrast, the non-African macrohaplogroup M and N mtDNAs (n = 72) showed a highly significant deviation from neutrality (Tajima's D = −2.43, P < 0.01; Fu and Li D* = −5.09, P < 0.02). This was also true for macrohaplogroup M mtDNAs (Tajima's D = −2.01, P < 0.05) and Fu and Li D* = −3.35 (P < 0.02) and macrohaplogroup N mtDNAs (Tajima's D = −2.54 (P < 0.001) and Fu and Li D* = −4.38 (P < 0.02) when analyzed separately as well. Furthermore, analysis of the pairwise sequence differences of the macrohaplogroup M and N mtDNAs gave a bell-shaped frequency distribution. These results are consistent with population expansions out of Africa having distorted the frequency distribution of mtDNA variation (6, 23).
To further define the regional distribution of these demographic influences, we divided Eurasian samples into European and Asian plus Native American mtDNAs (4, 24). Analysis of all European mtDNAs (n = 31) also revealed significant deviations from the standard neutral model (Tajima's D = −2.19, P < 0.01; Fu and Li D* = −3.31, P < 0.02) and a bimodal distribution of pairwise comparisons (Fig. 4).
Similarly, analysis of the aggregated Asian and Native American mtDNAs (n = 41) also revealed significant deviations from the standard neutral model (Tajima's D = −2.28, P < 0.01; Fu and Li D* = −4.31, P < 0.02) as well as a broad, bell-shaped distribution of pairwise differences consistent with rapid population expansion (Fig. 4). When the Siberian and Native American haplogroup A, B, C, D, G, Z, Y, and X mtDNAs (n = 32) were analyzed separately, they also showed significant deviation from neutrality (Tajima's D = −1.94, P < 0.05; Fu and Li D* = −3.41, P < 0.02). When these same Siberian and Native American haplogroup A, C, D, G, Z, Y and X mtDNAs were analyzed without B (n = 26), they also showed significant deviation from neutrality for the Fu and Li D* test (D* = −2.96, P < 0.05), although not for the Tajima's D test (D = −1.77, not significant). The distribution of the pairwise sequence differences for these mtDNAs was also unimodal. Thus, population expansions have distorted mtDNA variation as people moved into Siberia and Beringia and on into North Americas.
Variable Replacement Mutation Rates in Human mtDNA Genes.
If natural selection were an important factor in shaping regional human mtDNA sequence variation, then selection would principally act through amino acid variants in the mtDNA OXPHOS polypeptides. The effect of selection on a particular mtDNA protein gene can be assessed by determining the frequency of amino acid changes (nsyn, ka) versus silent base changes (syn, ks). An increase in the ratio of ka/ks would then reflect an increased amino acid substitution rate in that protein.
To determine the overall amino acid diversities of the 13 mtDNA proteins among humans, we calculated the ka/ks ratios for all pairwise combinations of the 104 human mtDNA sequences by using the method of Nei and Gojobori (16). The ka/ks ratios were calculated in two ways: ka/(ks + constant) and ka/(ka + ks) (Fig. 2). Both of these analyses revealed a surprising result. ATP6, which is generally found to be one of the more highly conserved genes in comparisons between distant species (25–27), had the highest amino acid sequence variation of any human mtDNA gene (Fig. 2).
Figure 2.
Distribution of the relative selective constraints [ka/(ks + constant)] of the 13 human mtDNA polypeptide genes calculated from the 104 complete human mtDNA sequences (16). For each gene, the bottom and top of the line indicates the minimum and maximum values, respectively. The bottom, intermediate, and top horizontal lines in the boxes represent the 25th, 50th (median), and 75th percentile values, respectively. The dot indicates the mean. We have also calculated ka/(ks + ka), dropping those values that were 0/0. This calculation gave essentially the same results (Fig. 5, which is published as supporting information on the PNAS web site).
Moreover, ATP8, the companion of ATP6 in the ATP synthase, is one of the least conserved mtDNA protein coding genes in comparisons between distant species (25, 26). Yet it proved to be one of the most highly conserved genes in the human mtDNAs (Fig. 2).
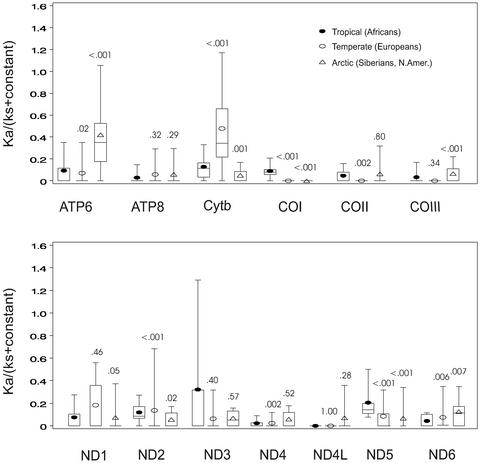
To determine whether the increased ATP6 variation in the human mtDNA comparisons (Fig. 2) also correlates with regional transitions, we compared the ka/ks ratios of the 13 mtDNA genes for mtDNA haplogroup lineages from three different geographic regions: the African-specific mtDNA haplogroups L0–L3 representing the tropical and subtropical zones; the European-specific mtDNA haplogroups H, V, U, J, T, I, X, N1b, and W representing the temperate zone; and the Siberian and Native American haplogroups A, C, D, G, Z, Y, and X representing the subarctic and arctic zones. Comparison of the distribution of the ka/ks ratios (16) for each climatic zone revealed dramatic differences in the amino acid sequences of particular mtDNA genes (Fig. 3).
Figure 3.
Distribution of the relative selective constrains [ka/(ks + constant)] calculated for the human mtDNA lineages associated with different climatic zones: tropical and subtropical (African), temperate (European), and arctic and subarctic (Siberian and Native American). Calculation of ka/(ks + constant) and distribution of values are as presented in Fig. 2. Numbers above plots represent P values (Wilcoxon rank-sum test) for the comparison of the distribution of ka/(ks + constant) values for tropical (L0–L3) to temperate (H, V, U, J, T, I, X, N1b, and W) or arctic (A, C, D, G, X, Y, and Z) zones. Very similar distributions and P values were obtained for the arctic whether or not haplogroup B mtDNAs were included in the calculation. Similar results have been obtained by calculating ka/(ks + ka) where significant differences (P ≤ 0.01) were found between tropical (Africans) and arctic (Siberians and Native Americans) for the ND1, ND3, ND5, ND6, COI, COIII, ATP6, and ATP8 genes and between tropical and temperate (Europeans) for the ND1, ND2, ND5, ND6, cytb, COI, COII, and ATP8 genes (Table 2, which is published as supporting information on the PNAS web site). To control for the possibility that the observed differences in the distribution of ka/(ks + constant) ratios were simply an artifact of pairwise calculations, we also compared the raw number of nsyn and syn mutations for each lineage. Using ATP6 as an example, the nsyn/syn ratio for the tropics was 3/15 (0.20), temperate 5/6 (0.83), and arctic 7/5 (1.4). By two-tailed Fisher's exact test, the tropical to arctic ratios were significantly different (P ≤ 0.05). To determine the importance of the distribution of nsyn and syn mutations along individual mtDNA lineages on ka/(ks + constant), we used paml (36) to chart the locations of nsyn and syn variants for ATP6 in the arctic A–D and X and the tropical L0–L3 haplogroups. This process revealed that nsyn and syn mutations were relatively uniformly distributed across the A–D and X lineages, whereas the few African ATP6 variants were located near the ends of the L0 and L3 branches.
The amino acid sequence variation of mtDNA proteins from the arctic and subarctic zone Siberians and Native Americans revealed that ATP6 was extremely highly variable. Amino acid sequence variation was also increased in COIII and ND6 among arctic mtDNA lineages.
Analysis of amino acid sequence variation of the temperate-zone Europeans revealed strikingly high amino acid sequence variation in the cytb protein (Fig. 3). Lastly, analysis of the protein sequence variation of the tropical- and subtropical-zone Africans revealed heightened amino acid sequence variation for COI and ND5. Increased amino acid variation was also observed in the tropics for ATP6, cytb, COII, ND1, and ND2 (Fig. 3). Thus, there is a dramatic correlation between increased amino acid substitutions in particular genes of the mtDNA and climatic zone.
Climatically Delimited Amino Acid Substitutions in ATP6, Cytb, and COI.
To investigate the possible functional significance of the ATP6 amino acid variation in the arctic and subarctic zones, in cytb for the temperate zone, and in COI for the tropical and subtropical zones we examined the evolutionary conservation of the variable amino acids. Amino acid conservation was determined by comparing the mtDNA proteins of 39 animal species (12 primates, 22 other mammals, four nonmammalian vertebrates, and Drosophila). Many of the substitutions did alter evolutionarily conserved amino acids.
For the ATP6 gene, striking amino acid substitutions were found throughout Eurasia and the Americas. Among the haplogroups of macrohaplogroup M, the related Siberian-Native American haplogroups C and Z share an alanine to threonine substitution at codon 20 (A20T). A nonpolar amino acid is found in this position in all animal species except for Macaca, Papio (baboon), Balaenoptera (whale), and Drosophila.
The ATP6 genes of macrohaplogroup N are separated from those of the rest of the world by a T59A substitution. The polar T at position 59 is conserved in all great apes and some Old World monkeys. Similarly, the macrohaplogroup N, non-R, N1b lineage harbors two distinctive amino acid substitutions: M104V and T146A. The M at position 104 is conserved in all mammals, whereas the T at position 146 is conserved throughout all animal mtDNAs. Moreover, the T146A substitution is within the same transmembrane α-helix as the pathogenic mutation L156R that alters the coupling efficiency of the ATP synthase and causes the neurogenic weakness, ataxia, and retinitis pigmentosa and Leigh syndromes (28).
Also in macrohaplogroup N, the ATP6 gene of the Siberian-Native American haplogroup A mtDNAs harbors a H90Y amino acid substitution. The H in this position is conserved in all placental mammals except Pongo (orangutan), Cebus (capuchin monkey), and Loxodonta (elephant) and occurs within a highly conserved region.
Analysis of the cytb gene variants in Europeans revealed that haplogroup HV is associated with an I7T variant, with a nonpolar amino acid being found in this position in chimpanzee, bonobo, and most humans, whereas a polar amino acid is found in this human mtDNA lineage and all other animals. Similarly, a L236I variant is found in the cytb genes of haplogroup JT, yet an L is found in this position in all other simians.
Examination of COI in Africans revealed Y496H substitution present in all haplogroup L1 mtDNAs, whereas an uncharged amino acid is found in this position in all primates. Likewise, an A415T substitution is present in virtually all African L0 and L1 mtDNAs, whereas a nonpolar amino acid is found in this position in chimpanzee, gorilla, orangutan, bonobo, gibbon, baboon, and tarsius, although not in Macaca and Cebus.
Because each of the mtDNA sequences used for the nonhuman mtDNA comparisons is derived from only one or two individuals it is possible that the deviant cases in these species may also be caused by adaptive mtDNA mutations. Hence, a number of the ATP6, cytb, and COI amino acid polymorphisms have the characteristics expected for evolutionarily adaptive mutations.
Discussion
Our data suggest that regional variation in mtDNA sequences is likely to have been shaped by natural selection. mtDNA variation deviates from neutrality in European, Central Asian, and Siberian plus Native American mtDNA lineages but not African lineages. The ATP6 gene is the most variable gene among human mtDNAs. ATP6, cytb, and COI are specifically variable in the arctic, temperate, and tropical zones, respectively; and a number of the amino acid substitutions of these genes alter evolutionarily conserved amino acids.
mtDNA Mutations May Permit Adaptation to Changes in Diet and Climate.
mtDNA variation would be the ideal method to foster adaptation to different environments. Mitochondrial oxidative phosphorylation (OXPHOS) uses dietary calories to generate ATP to do work and heat to maintain body temperature. The balance between these two functions is determined by the efficiency of coupling the mitochondrial inner membrane electrochemical gradient to synthesize ATP through the ATP synthase. Variants that reduce the coupling efficiency would reduce ATP production, but increase heat production. Such variants would be advantageous in the subarctic and arctic where survival of cold stress would be a major factor in survival. Partial uncoupling of the mitochondria would increase the basal metabolic rate of the individual and hence would require a higher caloric intake, such as that provided by a high-fat diet. Thus, mtDNA ATP6 variants that reduce coupling might partially account for the increased basal metabolic rate that has been observed in indigenous, circumpolar, human populations (29).
The high mutation rate of mtDNA and the central role of mitochondrial proteins in cellular energetics make the mtDNA an ideal system for permitting rapid human and animal adaptation to new climate and dietary conditions. The uniparental (maternal) inheritance of the mtDNA favors the rapid segregation, expression, and adaptive selection of new advantageous mtDNA alleles. The lack of recombination would mean that selection of beneficial mutants would increase the frequency of the entire mtDNA haplotype through hitchhiking. Hence, climatic selection would lead to the regional-specific haplogroups that are observed.
Evidence has already accumulated that different human mtDNA lineages are functionally different. Haplogroup T is associated with reduced sperm motility in European males (30), and the tRNAGln nucleotide position 4336 variant in haplogroup H is associated with late-onset Alzheimer's disease (31). Moreover, Europeans harboring the mild ND6 nucleotide position 14484 and ND4L nucleotide position 10663 Leber's hereditary optic neuropathy missense mutations are more prone to blindness if they also harbor the mtDNA haplogroup J (32, 33), and haplogroup J is associated with increased European longevity (34). Because haplogroup J mtDNAs harbor two missense mutations in complex I genes (Y304H in ND1 and A458T in ND5), in addition to the above-mentioned L236T variant in the cytb gene, these polymorphisms all could affect the efficiency of OXPHOS ATP production and thus exacerbate the energy defects of mildly deleterious new mutations.
Given that mtDNA lineages are functionally different, it follows that the same variants that are advantageous in one climatic and dietary environment might be maladaptive when these individuals are placed in a different environment. Hence, ancient regionally beneficial mtDNA variants could be contributing to modern bioenergetic disorders such as obesity, diabetes, hypertension, cardiovascular disease, and neurodegenerative diseases as people move to new regions and adopt new lifestyles.
If selection has played an important role in the radiation of human mtDNA lineages, then the rate of mtDNA molecular clock may not have been constant throughout human history. If this is the case, then conjectures about the timing of human migrations may need to be reassessed.
Supplementary Material
Acknowledgments
We thank Drs. Jeffrey T. Lell and Debra O. Prosser for their multiple intellectual and technical contributions to this investigation. This work was supported by National Institutes of Health Grants AG13154, HL64017, NS21328, and NS37167; an Ellison Foundation Senior Scientist Grant (to D.C.W.); Fogerty National Institutes of Health Grants TW01366 (to A.O. and D.C.W.) and TW01175 (to R.I.S. and M.D.B.); a Wellcome Trust Research Career Development fellowship (to V.M.); and a Bikura Fellowship from the Israel Science Foundation (to D.M.).
Abbreviations
- cytb
cytochrome b
- nsyn
nonsynonymous
- syn
synonymous
- MRCA
most recent common ancestor
Footnotes
References
- 1.Wallace D C, Brown M D, Lott M T. Gene. 1999;238:211–230. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 2.Quintana-Murci L, Semino O, Bandelt H J, Passarino G, McElreavey K, Santachiara-Benerecetti A S. Nat Genet. 1999;23:437–441. doi: 10.1038/70550. [DOI] [PubMed] [Google Scholar]
- 3.Schurr T G, Wallace D C. Hum Biol. 2002;74:431–452. doi: 10.1353/hub.2002.0034. [DOI] [PubMed] [Google Scholar]
- 4.Torroni A, Miller J A, Moore L G, Zamudio S, Zhuang J, Droma R, Wallace D C. Am J Phys Anthropol. 1994;93:189–199. doi: 10.1002/ajpa.1330930204. [DOI] [PubMed] [Google Scholar]
- 5.Schurr T G, Sukernik R I, Starikovskaya Y B, Wallace D C. Am J Phys Anthropol. 1999;108:1–39. doi: 10.1002/(SICI)1096-8644(199901)108:1<1::AID-AJPA1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 6.Excoffier L. J Mol Evol. 1990;30:125–139. doi: 10.1007/BF02099939. [DOI] [PubMed] [Google Scholar]
- 7.Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, Coppa A, Calderon F L, Simionati B, Valle G, Richards M, et al. Am J Hum Genet. 2001;69:1348–1356. doi: 10.1086/324511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingman M, Kaessmann H, Paabo S, Gyllensten U. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 9.Anderson S, Bankier A T, Barrell B G, de Bruijn M H, Coulson A R, Drouin J, Eperon I C, Nierlich D P, Roe B A, Sanger F, et al. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 10.Andrews R M, Kubacka I, Chinnery P F, Lightowlers R N, Turnbull D M, Howell N. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 11.Horai S, Hayasaka K, Kondo R, Tsugane K, Takahata N. Proc Natl Acad Sci USA. 1995;92:532–536. doi: 10.1073/pnas.92.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnason U, Xu X, Gullberg A. J Mol Evol. 1996;42:145–152. doi: 10.1007/BF02198840. [DOI] [PubMed] [Google Scholar]
- 13.Wallace D C, Brown M D, Schurr T G, Chen E, Chen Y-S, Starikovskaya Y B, Sukernik R I. In: The Origin of Humankind. Aloisi M, Battaglia B, Carafoli E, Danieli G A, editors. Venice: IOS Press; 2000. pp. 9–11. [Google Scholar]
- 14.Hasegawa M, Kishino H, Yano T. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 15.Rozas J, Rozas R. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 16.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 17.Cann R L, Stoneking M, Wilson A C. Nature. 1987;325:31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- 18.Johnson M J, Wallace D C, Ferris S D, Rattazzi M C, Cavalli-Sforza L L. J Mol Evol. 1983;19:255–271. doi: 10.1007/BF02099973. [DOI] [PubMed] [Google Scholar]
- 19.Goodman M, Porter C A, Czelusniak J, Page S L, Schneider H, Shoshani J, Gunnell G, Groves C P. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- 20.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Y X, Li W H. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers A R, Harpending H. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 23.Merriwether D A, Clark A G, Ballinger S W, Schurr T G, Soodyall H, Jenkins T, Sherry S T, Wallace D C. J Mol Evol. 1991;33:543–555. doi: 10.1007/BF02102807. [DOI] [PubMed] [Google Scholar]
- 24.Torroni A, Bandelt H J, D'Urbano L, Lahermo P, Moral P, Sellitto D, Rengo C, Forster P, Savontaus M L, Bonne-Tamir B, Scozzari R. Am J Hum Genet. 1998;62:1137–1152. doi: 10.1086/301822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallace D C, Ye J H, Neckelmann S N, Singh G, Webster K A, Greenberg B D. Curr Genet. 1987;12:81–90. doi: 10.1007/BF00434661. [DOI] [PubMed] [Google Scholar]
- 26.Neckelmann N, Li K, Wade R P, Shuster R, Wallace D C. Proc Natl Acad Sci USA. 1987;84:7580–7584. doi: 10.1073/pnas.84.21.7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saccone C, Gissi C, Lanave C, Larizza A, Pesole G, Reyes A. Gene. 2000;261:153–159. doi: 10.1016/s0378-1119(00)00484-4. [DOI] [PubMed] [Google Scholar]
- 28.Trounce I, Neill S, Wallace D C. Proc Natl Acad Sci USA. 1994;91:8334–8338. doi: 10.1073/pnas.91.18.8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leonard W R, Sorensen M V, Galloway V A, Spencer G J, Mosher M J, Osipova L, Spitsyn V A. Am J Hum Biol. 2002;14:609–620. doi: 10.1002/ajhb.10072. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz-Pesini E, Lapena A C, Diez-Sanchez C, Perez-Martos A, Montoya J, Alvarez E, Diaz M, Urries A, Montoro L, Lopez-Perez M J, Enriquez J A. Am J Hum Genet. 2000;67:682–696. doi: 10.1086/303040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shoffner J M, Brown M D, Torroni A, Lott M T, Cabell M R, Mirra S S, Beal M F, Yang C, Gearing M, Salvo R, et al. Genomics. 1993;17:171–184. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- 32.Brown M D, Sun F, Wallace D C. Am J Hum Genet. 1997;60:381–387. [PMC free article] [PubMed] [Google Scholar]
- 33.Brown M D, Zhadanov S, Allen J C, Hosseini S, Newman N J, Atamonov V V, Mikhailovskaya I E, Sukernik R I, Wallace D C. Hum Genet. 2001;109:33–39. doi: 10.1007/s004390100538. [DOI] [PubMed] [Google Scholar]
- 34.Rose G, Passarino G, Carrieri G, Altomare K, Greco V, Bertolini S, Bonafe M, Franceschi C, De Benedictis G. Eur J Hum Genet. 2001;9:701–707. doi: 10.1038/sj.ejhg.5200703. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y S, Olckers A, Schurr T G, Kogelnik A M, Huoponen K, Wallace D C. Am J Hum Genet. 2000;66:1362–1383. doi: 10.1086/302848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 37.Felsenstein J. Phylogeny Inference Package 3.53c. Seattle: University of Washington; 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.