Abstract
Amyloid β peptide (Aβ) generated from amyloid precursor protein (APP) is central to Alzheimer's disease (AD). Signaling pathways affecting APP amyloidogenesis play critical roles in AD pathogenesis and can be exploited for therapeutic intervention. Here, we show that sumoylation, covalent modification of cellular proteins by small ubiquitin-like modifier (SUMO) proteins, regulates Aβ generation. Increased protein sumoylation resulting from overexpression of SUMO-3 dramatically reduces Aβ production. Conversely, reducing endogenous protein sumoylation with dominant-negative SUMO-3 mutants significantly increases Aβ production. We also show that mutant SUMO-3, K11R, which can only be monomerically conjugated to target proteins, has an opposite effect on Aβ generation to that by SUMO-3, which can form polymeric chains on target proteins. In addition, SUMO-3 immunoreactivity is predominantly detected in neurons in brains from AD, Down's syndrome, and nondemented humans. Therefore, polysumoylation reduces whereas monosumoylation or undersumoylation enhances Aβ generation. These findings provide a regulatory mechanism in APP amyloidogenesis and suggest that components in the sumoylation pathway may be critical in AD onset or progression.
Amyloid precursor protein (APP) is an integral transmembrane glycoprotein of multiple isoforms including the neuronal form of 695 amino acids. Proteolytic cleavage of APP by β-secretase or β-site APP cleavage enzyme (BACE) (1–6) initiates the amyloidogenic pathway with the liberation of N-terminal fragment (β-NTF) and the formation of β-C-terminal fragment, which is then further processed by γ-secretase (7, 8) to release amyloid β peptide (Aβ). A nonamyloidogenic pathway by α-secretase cleavage leads to the secretion of α-NTF and precludes the formation of full-length Aβ (9, 10). Various familial Alzheimer's disease (AD)-associated mutations in both APP and presenilins cause increased production or deposition of Aβ, which is believed to induce neuronal cell death because of its neurotoxic and/or inflammatory effects (for a review, see ref. 11). These and other observations support that molecular mechanisms involved in regulating APP amyloidogenesis are obligatory players in AD pathogenesis.
Sumoylation has begun to be recognized as an important posttranslational modification capable of influencing protein function (for reviews, see refs. 12–14). Sumoylation is biochemically similar to but functionally distinct from ubiquitination, the latter being involved in several neurodegenerative diseases but whose role in AD is still unclear (15–17). In mammalian cells, the small ubiquitin-like modifier (SUMO) protein family consists of three members, SUMO-1, -2, and -3, all displaying the conserved diglycine residues located in the C terminus. SUMO-2 and SUMO-3 are 95% identical at the amino acid level and together seem to be distinct from SUMO-1 in several respects. SUMO-2/3 seem to contribute more to total protein sumoylation than SUMO-1 does, and they may have distinct protein substrates (18). SUMO-2/3, but not SUMO-1, are capable of forming poly(SUMO) chains, which is similar to polyubiquitination (19). Biochemically, SUMOs are first processed at the diglycine motif via SUMO hydrolase/isopeptidase to generate a free glycine residue at the C terminus, which is covalently attached to their protein substrates through the formation of an isopeptide bond between the liberated glycine residue and a lysine residue on the target protein. The biological function of sumoylation is not fully understood, but it seems to regulate protein localization and stability, the latter through antagonism of ubiquitin-mediated proteolysis (20–22). Here, we report a previously unknown function for sumoylation in regulating APP processing.
Materials and Methods
Cell Culture.
Human embryonic kidney 293T cells and human neuroblastoma SK-N-MC cells were grown in high glucose-containing DMEM supplemented with 10% heat-inactivated FBS.
Normalized Library and Primary Screening.
A human fetal brain cDNA library purchased from Life Technologies (Rockville, MD) was normalized by the method of Soares et al. (23). The normalization was confirmed by randomly selecting ≈300 clones for sequencing, which showed minimal to no redundancy. The normalized library was plated for single colonies (≈100 colonies per plate) and scraped, and DNA preparations were made and assembled in 96-well microtiter plates. For initial screening, the cDNA library was cotransfected with an APP expression plasmid, followed by assays for Aβ, β-NTF, and α-NTF. After comparing the levels of these three APP products with the average of each plate, cDNA pools showing altered patterns of APP processing were deconvoluted and rescreened.
Plasmids.
APP695wt, APP695sw, and BACE expression plasmids are in pcDNA3 vectors. SUMO-3 expression plasmid is in pCMV.SPORT2 vector. HisG-tagged Nedd8 and ubiquitin-expression plasmids were constructed by PCR and confirmed by sequencing analysis. All mutant clones were generated by using the QuikChange (Stratagene) mutagenesis method and were confirmed by sequencing analysis.
Transfection Studies.
For determining the role of SUMO in APP processing, SUMO-3 plasmids were cotransfected with APP695 expression plasmid, followed by ELISAs for Aβ, β-NTF, and α-NTF. Plasmid transfections were performed with Lipofectamine 2000 (Life Technologies) in 24-well plates. For each transfection, 4 × 105 cells were incubated with a total of 800 ng of plasmid DNA combined with 2 μl of Lipofectamine 2000 according to the vendor's protocol. The total DNA contained 400 ng of APP695wt or 160 ng of APP695sw plasmid and various amounts of other plasmids. The empty vector pcDNA3.1 was supplemented to make up a total of 800 ng of DNA whenever necessary. Each transfection was done in triplicate. Forty-eight hours after transfection, the medium was collected and assayed for Aβ, β-NTF, and α-NTF.
ELISAs.
Determinations of Aβ, β-NTF, and α-NTF concentrations in growth medium used standard sandwich ELISA methods with the following antibodies. For total Aβ, the capture antibody was 266.1 raised against Aβ domain residues 13–29, and the detection antibody was biotinylated 3D6 raised against Aβ domain residues 1–5. For β-NTF, the capture antibody was 8E5 raised against residues 444–592 of APP protein, and the detection antibody (specific for free C terminus of β-NTF) was 192wt for wild-type APP and AF20 for Swedish APP, respectively, raised against residues 593–596. For α-NTF, the capture antibody was 8E5, and the detection antibody was biotinylated 2H3 raised against Aβ domain residues 1–12.
Standard ELISA procedure was followed. With the appropriate amount of sample, this assay procedure accurately measures concentrations of Aβ, β-NTF, and α-NTF, all compared with purified standards. Final results are expressed relative to controls or APP transfection alone.
Western Blot Analysis.
Cells were treated according to experimental designs and collected directly in lysis buffer according to the method of Desterro et al. (21). Equal amounts of protein from cell lysates were electrophoretically resolved on Tris-glycine SDS gels and processed by standard method. APP, SUMO-3, and SUMO-1 antibodies were obtained from Zymed, BACE antibody was from Chemicon, and anti-HisG antibody was from Invitrogen.
Pulse–Chase Experiments.
293T cells were transfected with APP695 and SUMO-3 plasmids in six-well plates and 48 h later were incubated with serum- and methionine/cysteine-free medium containing 200 μCi (1 Ci = 37 GBq) of Promix L (35S) in vitro cell-labeling mix (Amersham Pharmacia) for 20 min. Medium was replaced, and the cells were then chased in complete nonradioactive medium for up to 240 min. Cell lysates were prepared and immunoprecipitated with an APP C-terminal-specific antibody 369 (24). Immunoprecipitates were subjected to SDS/PAGE and analyzed by radiography.
Immunohistochemistry.
Brain tissues of AD and Down's syndrome patients or age-matched nondemented tissues were obtained from NIA/Stanford Brain Bank. Sections (7 μm thick) were preblocked and then probed with anti-SUMO-3 antibody at 1:500 dilution overnight at 4°C. Further processing was done following standard protocol. Tissue sections were finally visualized with DAB or VIP staining. In control staining, nonimmune serum was used in place of the anti-SUMO-3 antibody.
Results
SUMO-3 Sumoylation Regulates APP Processing.
To identify modulators of APP processing, we screened a normalized human brain cDNA expression library consisting of 1,001 pools, each with ≈100 plasmids. DNA from each pool was cotransfected with an APP expression plasmid into 293T cells, and 48 h later the growth medium was assayed by ELISA for the level of Aβ, β-NTF, and α-NTF (Fig. 1A). This allowed us to identify a number of plasmid pools that alter patterns of APP processing, including one that down-regulates the level of β-NTF and Aβ but up-regulates that of α-NTF (data not shown). After deconvolution, a single plasmid from this pool was identified capable of altering APP processing as the original pool. Sequence analysis of this clone revealed that it contains a full-length cDNA encoding SUMO-3, the 95-aa ubiquitin-like protein.
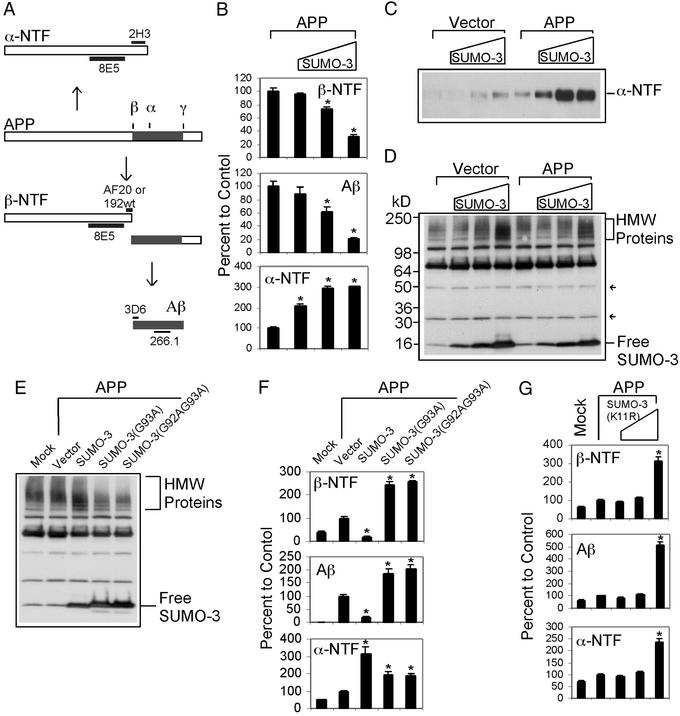
Figure 1.
SUMO-3 sumoylation regulates APP processing in 293T cells. (A) Diagram showing APP processing and antibody epitopes used in ELISA: 266.1 and 3D6 for Aβ, 8E5 and AF20 or 192wt for β-NTF, and 8E5 and 2H3 for α-NTF. Drawing not to scale. (B) Dose–effect of SUMO-3 expression on Aβ, β-NTF, and α-NTF generation from cells cotransfected with APP and without or with increasing amounts of SUMO-3 plasmids. All transfections contained 160-ng APP plasmids. The amount of SUMO-3 plasmids used were 0, 40, 160, and 640 ng in transfections corresponding to the columns from left to right. Vector DNAs were supplemented to bring the total amount of DNA to 800 ng for each transfection. A similar dilution scheme was used in other figures as well. The results were expressed as relative to the amount of APP-processing products indicated in each panel to the control. The control was APP transfection alone. (C) Western blot showing dose–effect of SUMO-3 on α-NTF production. Equal amounts of growth media from cells transfected as indicated were probed with 8E5 antibody. (D) Western blot showing SUMO-3 expression and protein sumoylation. Equal amounts of protein from lysates of cells transfected as indicated above were probed with affinity-purified SUMO-3 antibody. Arrows indicate SUMO dimer and trimer. HMW Proteins, high molecular weight proteins. (E) Western blot showing expression and sumoylation/desumoylation of SUMO-3 and mutants. Cells were mock-transfected or transfected with APP and SUMO plasmids, and lysates were probed with anti-SUMO-3 antibody. (F) Effect of SUMO-3 and mutants on Aβ, β-NTF, and α-NTF generation from transfected cells as determined by ELISA. (G) Dose–effect of SUMO-3(11R) on Aβ, β-NTF, and α-NTF generation as determined by ELISA. Each of the above experiments was carried out more than three times with equivalent results as those presented. *, Significantly different from APP transfection alone (P < 0.005, ANOVA post hoc tests).
We examined the dose-response of SUMO-3 expression on APP processing in 293T cells. The level of β-NTF was significantly lowered with increasing amounts of SUMO-3 plasmid transfected (Fig. 1B). The total level of secreted Aβ was also similarly reduced to ≈25% of the control at the highest SUMO-3 condition (Fig. 1B). Conversely, the level of α-NTF was increased in a dose-dependent fashion, which was also evident for endogenous APP (Fig. 1 B and C). When the extracts of the transfected cells were subjected to Western blot analysis by using a SUMO-3-specific antibody, both free SUMO-3 and sumoylated proteins were detected in a SUMO-3 plasmid dose-dependent manner (Fig. 1D). The increased sumoylated proteins were detected mainly as high molecular weight species, typical of what has been observed previously (18). Free monomers, dimers, and trimers of SUMO-3 were also evident. Unconjugated multimers of SUMO have been reported in yeast (25). These data indicate that increased SUMO-3 expression and/or sumoylation down-regulate amyloidogenic but up-regulate nonamyloidogenic processing of APP.
We next examined the effect of lowering endogenous SUMO-3 sumoylation on APP processing. Because SUMO-3 sumoylation requires a C-terminal diglycine motif generated through enzymatic removal of the terminal two proximal amino acids, we assessed mutants of the diglycine motif for the ability to act in a dominant negative manner to down-regulate intracellular SUMO-3 sumoylation. Two SUMO-3 variants, SUMO-3(G93A) and SUMO-3(G92AG93A), where one or both of the conserved glycine residues was mutated to alanine, were generated. These mutants were transfected into 293T cells, and the SUMO-3 sumoylation was examined by Western blot analysis. Compared with endogenous levels of sumoylation by SUMO-3 (mock- and vector-transfected controls), both mutants significantly decreased the total level of protein sumoylation as judged by a reduction in the intensity of the high molecular weight sumoylated species (Fig. 1E). Concomitantly with lower sumoylation levels, increased unconjugated free SUMO-3 was detected with each mutant compared with wild-type SUMO-3. When the culture medium was assayed for APP derivatives, we found that cells cotransfected with SUMO-3 mutants and APP secreted significantly more β-NTF and Aβ than transfection with only APP, which was opposite to that caused by transfection with wild-type SUMO-3 (Fig. 1F). Therefore, the level of β-NTF and Aβ is inversely associated with the level of intracellular SUMO-3 sumoylation. In addition, the level of α-NTF was also higher in the presence of SUMO-3 mutants (Fig. 1F), suggesting that the effect of sumoylation and desumoylation on β-secretase and α-secretase pathways is uncoupled.
We also determined the effect of SUMO-3-mediated mono- vs. polysumoylation on APP amyloidogenesis. To do so, we examined the effect of SUMO-3(K11R), a SUMO-3 variant with arginine replacing lysine at position 11. Arginine 11 is required for the formation of poly(SUMO) chains; SUMO-3(K11R) is capable of conjugating to its target, although its ability to form poly(SUMO) chains is impaired (19). The influence of the SUMO-3(K11R) mutant was assessed on APP processing. When 293T cells were cotransfected with APP and SUMO-3(K11R), the generation of Aβ and β-NTF, as well as that of α-NTF, were all up-regulated compared with APP transfection alone (Fig. 1G). Therefore, monosumoylation has a positive effect on Aβ production as opposed to polysumoylation, which negatively regulates Aβ production.
Effect of Sumoylation on APP and BACE Expression.
As a step toward elucidating the mechanistic details for these observations, we examined two obvious targets of sumoylation, APP and BACE. Because sumoylation can stabilize proteins in contrast to ubiquitin-mediated proteolysis, we asked whether APP is directly sumoylated and whether the half-life of APP is affected directly or indirectly by sumoylation. We cotransfected APP and SUMO-3 into 293T cells, cell extracts were immunoprecipitated with an APP antibody, and the immunoprecipitates were subjected to Western blot analysis with a SUMO-3 antibody. No sumoylated APP was detected based on the lack of labeled bands 100–130 kDa or larger (data not shown). This is not unexpected as the APP sequence does not contain the consensus sumoylation motif, ψKxE/D, where ψ is an aliphatic residue (26, 27). It should be noted that because of the paucity of known SUMO-3 substrates, the SUMO-3 sumoylation motif is not yet definitively established. However, SUMO-3 can be conjugated at the same site (ψKxE/D) as that used by SUMO-1 (28). We next performed a pulse–chase analysis to determine the half-life of APP in the presence or absence of exogenous SUMO-3 or SUMO-3(G93A). Although SUMO-3 seemed to alter the maturation of APP by retarding or reducing glycosylation, the half-life of APP was not affected by SUMO-3 or SUMO-3(G93A) (Fig. 2A). In particular, the kinetics of APP metabolism was indistinguishable between 293T cells transfected with APP and SUMO-3(G93A) and that with APP alone. These data suggest that the effect of sumoylation on Aβ generation is not simply caused by an altered rate of APP turnover.
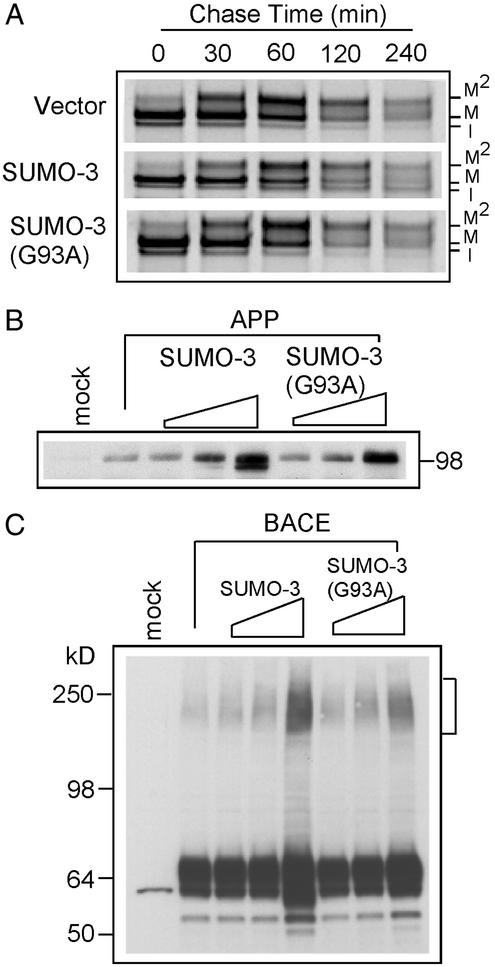
Figure 2.
Effect of SUMO-3 and SUMO-3(G93A) on APP and BACE expression. (A) Pulse–chase study of the kinetics of APP metabolism in 293T cells cotransfected with equal amounts of APP and the plasmid indicated. Immature (I) and mature (M, M2) APP species are indicated. (B) Dose–effect of SUMO-3 and mutant on APP expression. Equal amounts of proteins from lysates of cells transfected with the indicated plasmids for 48 h were probed with an anti-C-terminal APP antibody. (C) Western blot showing dose–effect of SUMO-3 and mutant on BACE expression. Equal amounts of proteins from cells transfected with indicated plasmids for 48 h were probed with an anti-BACE antibody.
We also examined the effect of wild-type and mutant SUMO-3 on the level of intracellular APP by Western blot analysis. Increased expression of either SUMO-3 or SUMO-3(G93A) increased the steady-state level of APP (Fig. 2B). No additional high molecular weight APP species were observed (data not shown), which is consistent with the above conclusion that APP is not directly sumoylated. The increased total APP by either SUMO-3 or SUMO-3(G93A) may be correlated with overall alterations in cellular metabolism when wild-type or mutant SUMO-3 is expressed at very high levels.
Because SUMO-3 affects APP processing, we next assessed its influence on BACE. BACE levels were unchanged at the lower doses of exogenous SUMO-3. Similar to APP, BACE levels were slightly elevated when a BACE plasmid was expressed with high levels of exogenous SUMO-3 or SUMO-3(G93A) as detected by Western blot analysis (Fig. 2C). This effect was somewhat more dramatic in the presence of wild-type SUMO-3 than the mutant. Overexpression of wild-type SUMO-3 also resulted in a faster migrating BACE species, which might reflect immature BACE as observed with APP. At high doses of SUMO-3, high molecular weight immunoreactive BACE proteins at ≈250 kDa were also detected on the blot (Fig. 2C). Because SUMO-3(G93A) also increased the level of high molecular weight BACE species (Fig. 2C), it is unlikely that these represent sumoylated BACE. BACE does contain a sequence, LKMD (positions 274–277), which conforms to a sumoylation motif. However, mutation of the acceptor lysine to arginine on BACE did not eliminate or reduce the amount of high molecular weight BACE species when coexpressed with SUMO-3 (data not shown). The level of the high molecular weight BACE seems to be correlated with high BACE expression levels and may simply represent aggregation. Together, these observations indicate that overexpression of both wild-type SUMO-3 and SUMO-3(G93A) increases intracellular APP and BACE levels and that high levels of wild-type SUMO-3 seem to retard or reduce posttranslational modification of APP and BACE. Also, the data show that the effects of sumoylation on APP processing are indirect.
Effect of Ubiquitin and Nedd-8 on APP Processing.
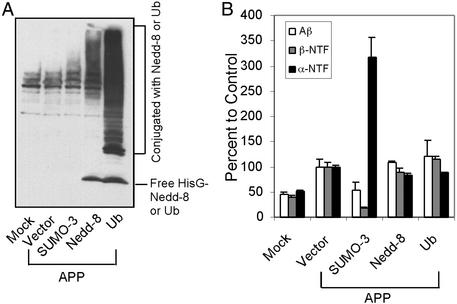
To determine whether ubiquitination potentially plays an antagonistic role to sumoylation, we examined whether ubiquitin and Nedd-8 also effect APP processing. Ubiquitin has been implicated in AD, but whether it effects APP processing is unknown. Nedd-8 is a ubiquitin homolog, and conjugation of Nedd-8 to its substrate promotes ubiquitin polymerization (29). Interestingly, the APP binding protein, APP-BP1 (30), is one of two components of the E1 activating enzyme required for Nedd-8 conjugation (31, 32). It is also unclear whether Nedd-8 affects APP processing. We transfected plasmids expressing HisG-tagged ubiquitin or Nedd-8 into 293T cells and examined their expression and substrate conjugation by Western blot analysis. Fig. 3A shows that both free ubiquitin and Nedd-8 and their conjugates were detected. Neither ubiquitination nor Nedd-8 altered APP processing (Fig. 3B), and both were without effect on levels of intracellular APP (data not shown). Therefore, ubiquitination plays no significant role in APP processing.
Figure 3.
Effect of ubiquitination and neddylation on APP processing in 293T cells. (A) Western blot showing expression and conjugation of HisG-tagged ubiquitin and HisG-tagged Nedd-8. Lysates from transfected cells were probed with an anti-HisG antibody. (B) Effect of ubiquitin and Nedd-8 on APP processing as determined by ELISA for Aβ, β-NTF, and α-NTF.
SUMO-3 in Brain and Neuronal Cells.
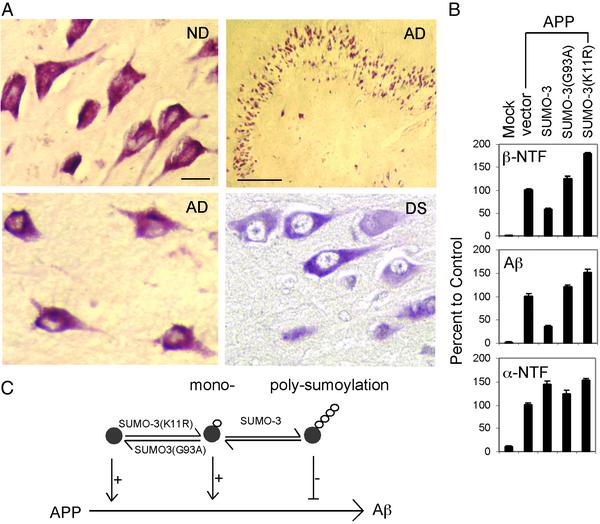
To determine the physiological relevance of the above observations, we examined whether SUMO-3 protein is present in brain and whether changes in sumoylation also effect amyloidogenesis in neuronal cells. We stained sections of brain from AD, Down's syndrome, and nondemented individuals with affinity-purified anti-SUMO-3 antibody. Fig. 4A shows that neuronal populations have positive immunoreactivity for SUMO-3. SUMO-3 staining is remarkable in hippocampal neurons, a population significantly targeted in AD. Four to five cases were examined for each group. Qualitatively, it seems that a larger percentage of neurons in AD and Down's syndrome brain have a SUMO-3 distribution limited to the neuronal soma in contrast to nondemented brains, where SUMO-3 is both somal and nuclear (Fig. 4A and data not shown). A rigorous analysis with additional cases is needed to confirm this preliminary observation. Nonetheless, we show that SUMO-3 is detectable in brain neurons.
Figure 4.
SUMO-3 in human brain and its effect on APP processing in a human neuronal cell line. (A) SUMO-3 immunoreactivity in brain. ND, nondemented. Scale bars represent 20 and 100 μm for high and low magnifications, respectively. (B) Effect of SUMO-3 on Aβ, β-NTF, and α-NTF generation in SK-N-MC cells as determined by ELISA. (C) Schematic diagram summarizing effects of sumoylation on APP amyloidogenesis. Polysumoylation negatively (−) regulates but monosumoylation or undersumoylation positively (+) regulates Aβ production. SUMO proteins are indicated by small white circles.
We next examined whether regulation of APP processing by sumoylation is also operative in neuronal cells. An APP expression plasmid was transfected without or with various SUMO constructs into the human neuroblastoma cell line SK-N-MC. APP processing was assessed by ELISA. Fig. 4B shows that the effect of sumoylation on APP processing observed in 293T cells was replicated in SK-N-MC neuronal cells. Therefore, sumoylation plays a role in APP processing in neuronal cells and is likely to influence brain Aβ levels.
Discussion
These results bring a new perspective to the field of AD research, specifically as it pertains to APP processing and biology, which has been extensively characterized over the past decade. The effect of sumoylation on APP biogenesis and processing is multifaceted. We have shown that increased and decreased SUMO-3 sumoylation are correlated with the down- and up-regulation of Aβ peptide production, respectively. Hence, abnormal regulation of APP amyloidogenesis by sumoylation may occur in AD. In support of this notion, SUMO-3 immunoreactivity is present in neuronal populations in brain. Putative SUMO-3 sumoylation disturbances could be caused by a number of factors, including dysfunction of SUMO proteins or the enzymatic and regulatory components involved in protein sumoylation. Sumoylation is a reversible process, employing SUMO-specific proteases to remove SUMO moieties from proteins. Genes encoding enzymes or proteins involved in sumoylation are thus potential risk factors for AD. Environmental factors such as oxidative stress, which has been shown to increase SUMO-3 sumoylation in certain cells (18), may also affect APP amyloidogenesis by effecting sumoylation. From a therapeutic point of view, the sumoylation pathway may be exploited because, for example, a robust reduction in Aβ production could be achieved through increasing SUMO-3 sumoylation by inhibiting SUMO-3-specific proteases. A family of at least seven members of such proteases has been identified to date (13).
The above results also show the functional divergence for mono- and polysumoylation and the utility of dominant negative SUMO variants to the emerging field of protein sumoylation. The dependence of biological functionality, or APP amyloidogenesis in this case, on the SUMO chains is a vivid resemblance to ubiquitination. The length of ubiquitin chains can subserve different biological activities where polyubiquitination usually designates a protein to proteasome destruction, whereas monoubiquitination regulates other cellular processes such as endocytosis (33). The mechanism governing inhibition of APP amyloidogenesis by SUMO-3 polysumoylation warrants further investigation, particularly in light of the fact that both APP and BACE levels are increased by this regulatory process. The latter might be explained by the fact that sumoylation generally affects protein localization and transport, which is consistent with APP and BACE using the same trafficking route and perhaps the same transport compartment (34).
Although the sumoylation substrate(s) relevant to APP processing remains unknown, our observations show that sumoylation plays a critical role in APP amyloidogenesis (schematically summarized in Fig. 4C). Components of the sumoylation pathway should now be further investigated as not only potential AD risk factors but also therapeutic targets.
Acknowledgments
We thank Drs. J. McCarthy and F. Schimmoller for discussions, A. Lam and J. Miller for technical assistance, Dr. S. Gandy for the 369 antibody, Dr. G. Murphy for brain tissues, and Eli Lilly for funding this research.
Abbreviations
- Aβ
amyloid β peptide
- AD
Alzheimer's disease
- APP
amyloid precursor protein
- BACE
β-site APP cleavage enzyme
- NTF
N-terminal fragment
- SUMO
small ubiquitin-like modifier
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Farzan M, Schnitzler C E, Vasilieva N, Leung D, Choe H. Proc Natl Acad Sci USA. 2000;97:9712–9717. doi: 10.1073/pnas.160115697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hussain I, Powell D, Howlett D R, Tew D G, Meek T D, Chapman C, Gloger I S, Murphy K E, Southan C D, Ryan D M, et al. Mol Cell Neurosci. 1999;14:419–427. doi: 10.1006/mcne.1999.0811. [DOI] [PubMed] [Google Scholar]
- 3.Sinha S, Anderson J P, Barbour R, Basi G S, Caccavello R, Davis D, Doan M, Dovey H F, Frigon N, Hong J, et al. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 4.Vassar R, Bennett B D, Babu-Kahn S, Kahn S, Mendiaz E A, Denis P, Teplow D B, Ross S, Amarante P, Loeloff R, et al. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 5.Yan R, Bienkowski M J, Shuck M E, Miao H, Tory M C, Pauley A M, Brashier J R, Stratman N C, Mathews W R, Buhl A E, et al. Nature. 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- 6.Lin X, Koelsch G, Wu S, Downs D, Dashti A, Tang J. Proc Natl Acad Sci USA. 2000;97:1456–1460. doi: 10.1073/pnas.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strooper B D, Annaert W. Nat Cell Biol. 2001;3:E221–E225. doi: 10.1038/ncb1001-e221. [DOI] [PubMed] [Google Scholar]
- 8.Wolfe M S, Haass C. J Biol Chem. 2001;276:5413–5416. doi: 10.1074/jbc.R000026200. [DOI] [PubMed] [Google Scholar]
- 9.Buxbaum J D, Liu K N, Luo Y, Slack J L, Stocking K L, Peschon J J, Johnson R S, Castner B J, Cerretti D P, Black R A. J Biol Chem. 1998;273:27765–27767. doi: 10.1074/jbc.273.43.27765. [DOI] [PubMed] [Google Scholar]
- 10.Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, Haass C, Fahrenholz F. Proc Natl Acad Sci USA. 1999;96:3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selkoe D J. Nature. 1999;399:23–31. doi: 10.1038/399a023. [DOI] [PubMed] [Google Scholar]
- 12.Muller S, Hoege C, Pyrowolakis G, Jentsch S. Nat Rev Mol Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- 13.Yeh E T, Gong L, Kamitani T. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- 14.Hochstrasser M. Cell. 2001;107:5–8. doi: 10.1016/s0092-8674(01)00519-0. [DOI] [PubMed] [Google Scholar]
- 15.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 16.Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein M J, Jonnalagada S, Chernova T, et al. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 17.Saigoh K, Wang Y L, Suh J G, Yamanishi T, Sakai Y, Kiyosawa H, Harada T, Ichihara N, Wakana S, Kikuchi T, et al. Nat Genet. 1999;23:47–51. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]
- 18.Saitoh H, Hinchey J. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 19.Tatham M H, Jaffray E, Vaughan O A, Desterro J M, Botting C H, Naismith J H, Hay R T. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 20.Matunis M J, Coutavas E, Blobel G. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desterro J M, Rodriguez M S, Hay R T. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 22.Kim Y H, Choi C Y, Kim Y. Proc Natl Acad Sci USA. 1999;96:12350–12355. doi: 10.1073/pnas.96.22.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soares M B, Bonaldo M F, Jelene P, Su L, Lawton L, Efstratiadis A. Proc Natl Acad Sci USA. 1994;91:9228–9232. doi: 10.1073/pnas.91.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buxbaum J D, Gandy S E, Cicchetti P, Erlich M E, Czernik A J, Fracasso R P, Ramabhadran T V, Unterbeck A J, Greengard P. Proc Natl Acad Sci USA. 1990;87:6003–6006. doi: 10.1073/pnas.87.15.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson E S, Gupta A A. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 26.Johnson E S, Blobel G. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sternsdorf T, Jensen K, Reich B, Will H. J Biol Chem. 1999;274:12555–12566. doi: 10.1074/jbc.274.18.12555. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann H, Floss S, Stamminger T. J Virol. 2000;74:2510–2524. doi: 10.1128/jvi.74.6.2510-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu K, Chen A, Pan Z Q. J Biol Chem. 2000;275:32317–32324. doi: 10.1074/jbc.M004847200. [DOI] [PubMed] [Google Scholar]
- 30.Chow N, Korenberg J R, Chen X N, Neve R L. J Biol Chem. 1996;271:11339–11346. doi: 10.1074/jbc.271.19.11339. [DOI] [PubMed] [Google Scholar]
- 31.Gong L, Yeh E T. J Biol Chem. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 32.Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickart C M. Mol Cell. 2001;8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- 34.Kamal A, Almenar-Queralt A, LeBlanc J F, Roberts E A, Goldstein L S. Nature. 2001;414:643–648. doi: 10.1038/414643a. [DOI] [PubMed] [Google Scholar]