Abstract
Owing to the intimate linkage of sequence and structure in nucleic acids, DNA is an extremely attractive molecule for the development of molecular devices, in particular when a combination of information processing and chemomechanical tasks is desired. Many of the previously demonstrated devices are driven by hybridization between DNA ‘effector’ strands and specific recognition sequences on the device. For applications it is of great interest to link several of such molecular devices together within artificial reaction cascades. Often it will not be possible to choose DNA sequences freely, e.g. when functional nucleic acids such as aptamers are used. In such cases translation of an arbitrary ‘input’ sequence into a desired effector sequence may be required. Here we demonstrate a molecular ‘translator’ for information encoded in DNA and show how it can be used to control the release of a protein by an aptamer using an arbitrarily chosen DNA input strand. The function of the translator is based on branch migration and the action of the endonuclease FokI. The modular design of the translator facilitates the adaptation of the device to various input or output sequences.
INTRODUCTION
In the past few years, the information encoding and strand recognition capabilities of DNA have been utitilized for the realization of a variety of nanoscale DNA-based devices. DNA conformational changes have been shown to result in rotatory (1,2), stretching (3–12), and even translatory (13–18) movements. DNA devices have also been combined with functional nucleic acids such as ribozymes (9,17,19) and aptamers (20,21). Apart from chemomechanical action, DNA has also been demonstrated to be capable of a variety of information processing tasks (22–28). One of the more recent developments here is the realization of autonomous molecular automata performing, e.g. simple logical computations (29–36).
To implement more complex functions into molecular systems, it is of great interest to combine the computational and chemomechanical capabilities of these devices by linking them together into artificial reaction networks. As one example, we demonstrated recently how the operation of a DNA nanomechanical device can be controlled by mRNA signals transcribed from regulatory ‘genes’ (37,38). However, in this case both the ‘gene’ and the DNA device were artificially constructed and the sequences could be chosen freely. In a more realistic application one would like to trigger the action of a DNA-based device with an arbitrary DNA or RNA input signal. As an example, consider the release of a specific molecule bound to a DNA aptamer in response to the presence of a particular mRNA molecule. In general, the sequence of the mRNA (indicating the expression of a gene) will be completely unrelated to the DNA aptamer sequence. There are several possible solutions to this problem. One solution commonly found in nature is ‘allosteric regulation’. Here two spatially separated binding sites on an enzyme communicate with each other via a conformational change triggered by the binding of an effector to one of the binding sites. Such a principle has been utilized previously for the construction of allosteric ribozymes or aptazymes [e.g. (39–41)], and also recently for the DNA-controlled release of a small molecule by an aptamer (21). In some cases, however, allosteric regulation may not be an option—e.g. when the binding capacity of an aptamer or the activity of a ribozyme is affected too strongly by sequence modifications and the introduction of a regulatory part is not feasible. For ribozyme regulation, one approach is the ‘expansive regulation’ strategy developed by Wang et al. (42,43). Here the activity of a ribozyme is regulated by an effector molecule stabilizing the ribozyme–substrate complex. To regulate protein release by an aptamer, in the present work an alternative approach is taken in which a DNA or RNA input signal is translated into the desired output signal by a molecular translation device. One possible realization of such a translator is the molecular automaton recently introduced by Benenson et al. (31) which is based on the action of the restriction endonuclease FokI. As a type II-S endonuclease, FokI cuts 9 and 13 nt away from its 5 nt recognition sequence, and therefore the 4 nt sticky end created by this process is unrelated to the recognition sequence itself. In Ref. (31), the recognition sequence for FokI was hidden within an internal loop of a DNA double strand. In the presence of input strands (‘disease indicators’), the FokI recognition site was completed and a DNA ‘drug’—the product of cleavage by FokI—was administered. We here use the FokI system as a DNA signal translator to trigger the release of a protein by an aptamer device using an arbitrarily chosen DNA input signal. In contrast to Ref. (29), the DNA signal translator used here consists of two hairpin structures which contain the FokI recognition site and the effector sequence, respectively. The FokI site is inactivated by initially hybridizing the hairpin to a ‘protection’ strand. The translator is activated through the removal of the protection strand by a DNA input strand with a sequence completely unrelated to the effector or aptamer sequence.
MATERIALS AND METHODS
Materials
FokI (4 U/µl) was obtained from New England Biolabs, human α-thrombin in solution (200 µM) from Cell Systems, Germany. SYBR Gold nucleic acid stain was from Molecular Probes (Invitrogen), all other chemicals mentioned from Sigma–Aldrich. DNA strands were synthesized by biomers.net, Germany. The sequences of the oligonucleotides for the construction of the device (IN, CO, PR, OUT, APT) are given in Table 1. For fluorescence measurements a doubly labeled aptamer specific for the protein thrombin and extended by a 12 nt toehold was used (APT), similar to the aptamer device reported before (20,44). DNA strands were designed to minimize cross-hybridization and to have favorable ratios of melting temperatures in the different states of the device. The sequence generator software DNASequenceCompiler (45) was used for creating the sequences according to these specifications, the nucleic acids folding program RNAstructure (46) for checking the secondary structures.
Table 1.
Sequences of the DNA strands used
| Strand | Sequence (5′–3′) |
|---|---|
| IN | GATTGCGGAAAGAAGGTATGAGATAATGTCAC |
| CO | GCTTGCATCCGATTGCGGAAAGAAGGTATGAGATCGGATG |
| PR | GTGACATTATCTCATACCTTCTTTCCGCAATCGGA |
| OUT-1 | CAAGCAAGTCGCATTCATCCACCAACCGAGATGAATGCGACTT |
| OUT-1* | CGCATTCATCCACCAACCGAGATGAA |
| OUT-2 | CAAGCAGTGCGACTAACCACACCAACCGAGATGAACTTAGTCGCACT |
| OUT-2* | CGACTAACCACACCAACCGAGATGAACTTA |
| OUT-3 | CAAGCTTAGCGACCAACCGAGATTTGGTCGCTAA |
| OUT-3* | CGACCAACCGAGATTTG |
| APT | TAAGTTCATCTCGGTTGGTGTGGTTGG |
Device preparation and gel electrophoresis
For gel electrophoresis experiments, DNA strands were diluted to 1 µM in the hybridization buffer (50 mM potassium acetate, 20 mM Tris–acetate, 10 mM magnesium acetate, 1 mM DTT, pH 7.9 at 25°C). The concentration of the strands was determined from their OD260.
A typical reaction mixture contained 0.5 µl CO, 2.5 µl of PR, 4 µl of OUT and 4 U/10 µl FokI. The addition of 3 µl of IN started the reaction. All samples were adjusted to the same volume prior to the FokI reaction. Samples were incubated at 20°C for 2 h and heated to 65°C for 15 min to denature FokI and terminate the reaction. All assays were run on native 18% polyacrylamide gels in TBE (Tris–borate–EDTA, pH 8.3) buffer at 100 V/cm for 1.5–2 h and stained with SYBR Gold using the manufacturer's protocol and imaged under UV illumination.
Fluorescence measurements
Fluorescence resonance energy transfer (FRET) experiments were performed with the doubly labeled DNA aptamer strand APT. In its folded G quadruplex state (compare Figure 5) its two ends are in close proximity and energy transfer from fluorescein to TAMRA occurs efficiently. When APT is stretched due to hybridization with the output strand, FRET between the fluorophores is less efficient and fluorescein fluorescence is increased. The fluorescence signal therefore directly monitors the progress of the hybridization reaction of the output strand with the target strand APT. For the fluorescence experiments, all DNA strands were initially diluted to 10 µM. For the enzyme reaction, buffer composition, ratio of the strands, time and temperature was the same as for the gel assays. The FokI reaction was performed in a volume of 50 µl containing 20 U of FokI and terminated before the FRET measurements.
For the measurements, the aptamer strands were diluted in 1 ml aptamer buffer (20 mM Tris–HCl, 150 mM NaCl, 10 mM MgCl2 and 10 mM KCl, pH 8.0) to a final concentration of 75 nM.
FRET measurements were performed on a spectrofluorometer (Fluorolog-3, Jobin-Yvon, Munich, Germany). Fluorescence was excited at 490 nm and recorded at 515 nm, with excitation and emission bandwidths of 10 nm. The temperature was kept constant at 23°C.
Thrombin–aptamer complexes
For experiments with thrombin, the aptamer strand was mixed with thrombin in a ratio of 1:2. Aptamer and thrombin were incubated for 30 min at 23°C in a reaction volume of 50 µl before FRET measurements were conducted in a sample volume of 1 ml.
RESULTS AND DISCUSSION
Operation principle of the DNA transducer
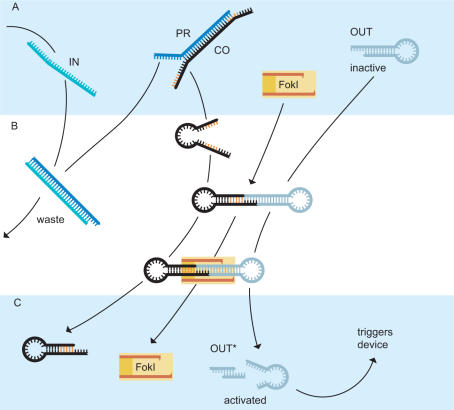
The operation principle of the transducer is schematically depicted in Figure 1. It is based on three processes which have been utilized previously for the operation of other DNA nanodevices and automata: (i) DNA branch migration (3); (ii) the specific DNA cleaving properties of the restriction endonuclease FokI (29); and (iii) the inhibition of hybridization between complementary strands by secondary structure formation (47). As shown in Figure 1A, the DNA output signal (OUT) initially is in an inactive state—it is forced into a small hairpin loop in which hybridization with its complement is inhibited (31,47). The stem of the hairpin has a sticky end which is complementary to the sticky end of a ‘connector strand’ (CO) which can also assume a hairpin conformation. However, initially the connector strand is forced into a duplex structure by a ‘protection strand’ (PR) in which it cannot hybridize to the inactive output strand. The protection strand in turn is equipped with a single-stranded ‘toehold’ (3) at which the input DNA strand (IN)—which is to be translated into an active output strand—can attach. The input strand can displace the protection strand from the connector strand by branch migration, producing a double-stranded waste product (IN-PR, Figure 1B). The released connector strand can now fold into its hairpin structure and hybridize to the inactive output signal. The sequence of the connector strand CO is chosen in such a way that in the folded state it contains the FokI recognition sequence (5′-GGATG-3′/3′-CCTAC-5′). FokI binds to the complex formed by connector and inactive output and cleaves the output strand 9 and 13 bases downstream from the FokI recognition site. As has been shown previously by Benenson et al. (30), FokI can cleave such a construct even without ligation of the two substrate duplexes. The result of the cleavage reaction is shown in Figure 1C. The connector strand leaves the reaction unaltered, whereas the stem of the output signal strand has been shortened (OUT becomes OUT*). The loop conformation of the output strand is therefore less stable and breaks open which makes the output DNA sequence available for hybridization with a downstream target sequence. In the present example, the activated output signal can bind to an aptamer-based DNA device and trigger the release of the protein thrombin. The release of the protein is achieved by the competition for binding between the output strand and the protein to the aptamer. The output strand sequence therefore necessarily has to be chosen (at least partly) complementary to the aptamer sequence. Using the DNA translator, an arbitrarily chosen DNA input strand can be translated into this specific output strand.
Figure 1.
Operation principle of the DNA translator. (A) Initially, the output signal (OUT) is present in an inactive stem–hairpin loop conformation. Hybridization with the sequence hidden in the loop is kinetically inhibited. Also present is a ‘connector strand’ (CO) partially hybridized to a protection strand (PR) which in turn is complementary to the input signal strand. When added to the reaction, the input strand (IN) binds to the unhybridized ‘toehold’ section of the protection strand and displaces the connector through a branch migration process. (B) The connector is now able to fold into its hairpin configuration, the two halves of the FokI recognition site are united and are recognized by the enzyme. Together with the protected output signal strand, the connector forms a dumbbell shaped dimer and the FokI enzyme cuts the output strand 9 and 13 bases downstream of the recognition site, across the unligated nicks. The waste product formed by the input and protection strand is very stable and does not participate in the further reaction. (C) The stability of the remaining short stem of the output strand is too low to keep the loop closed and so the activated output signal OUT* can bind to other DNA-based devices to start or inhibit a reaction.
Design of the input section
The various sections of the transduction device are indicated in Figure 2. In order to emphasize the modularity and generality of our approach, we here describe the design of the input and output sections in terms of an ‘algorithm’. (i) Given is the input signal strand IN which is to be transduced into a different sequence. This input signal can be of any sequence with length of >20 nt, it does not have to be extended or altered. (ii) The protection strand PR is the complement of strand IN and has to be augmented at the 3′ end with the three bases ‘GGA’ contained in the FokI recognition sequence. This prevents the binding of FokI to dimers which may be formed by two protected connector strands in the inactive state of the device. (iii) The connector hairpin loop CO can be derived from IN in the following way: cut off 8–10 bases from the 3′ end of IN, as they are used as toehold sequence for the displacement of PR from CO. (iv) Add the reverse complement of the first three bases of the 5′ end to the 3′ end, this acts as a spacer between the FokI binding site and the loop (compare section B of Figure 2). We experimentally verified that binding of FokI is obstructed if <3 bp are located upstream between the recognition sequence and the first bases of the loop. (v) Next add the FokI recognition sequences to the ends: CATCC to the 5′ end and GGATG to the 3′ end (C in Figure 2). (vi) As a last step, a sticky end has to be added to the connector hairpin (D in Figure 2). Since FokI cleaves asymmetrically with an overhang of 4 bases at the 5′ end, it is best to attach the sticky end at the 5′ end as well. The lower limit of the length of the sticky is set by the temperature at which the intermediate dumbbell structure (Figures 1B and 2) has to be stable. The upper limit is given by the distance between the recognition and the restriction site of FokI on the 5′ end, i.e. 13 bases. Reasonable values are between 4 and 8 bases (compare next paragraph).
Figure 2.
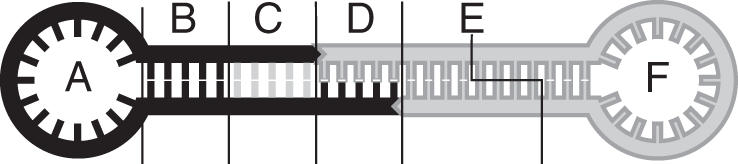
Core of the device composed of strands CO and OUT from Figure 1. Section A contains the part that is complementary to the protection strand PR. C is the FokI recognition sequence, spacer section B is needed to ensure that the binding of the enzyme is not disturbed by the loop A. Section D connects the two loops, it should be short and rich in GC pairs to improve FokI performance across the nicks. Line E indicates the FokI cleavage site, with the output F as result of the translation process.
Design of the output section
The complement of the sequence of the target strand (e.g. an aptamer-based device) has to be incorporated into the stem and/or the loop of the output section (E and F in Figure 2). In the inactive state (before cleavage by FokI), hybridization between the output loop and the target strand should be kinetically inhibited, whereas in the active state this reaction should be fast. The stem of the output loop (E in Figure 2) therefore has to be sufficiently long to be stable in its uncleaved form and short enough to break open after restriction by FokI. Furthermore, the opening of the loop (F in Figure 2) has to be kept small enough to prevent the complementary strand from threading through (47). Several design variations based on these considerations are described below. The design of the sticky end connecting the input and output section of the device (D in Figure 2) has to ensure efficient cleavage by FokI. As described in Ref. (30), cleavage without prior ligation is possible for short sticky ends with a high CG-content. In experiments at various temperatures we found that the restriction reaction was successful with a 5 nt sticky end for temperatures up to 20°C. It has been found previously (30) that with increasing length of the sticky ends, FokI tends to cut only one strand of the helix. This reaction results in an additional band in the gel assay (band OUT# in Figure 3). For efficient cleavage at relatively high temperatures, 4 or 5 bases are the best choices for the length of the sticky ends.
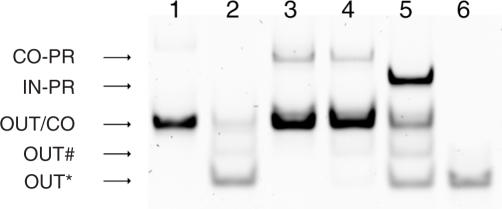
Figure 3.
Operation of the device characterized by gel electrophoresis. Lane 1 contains output (OUT) and connector (CO) strand in the absence of FokI. Lane 2 contains OUT and CO with FokI added, leading to the appearance of the cleavage products OUT*, OUT#. The latter is due to incomplete cleavage of the output strand (see text). Lane 3 contains protected connector strand (CO-PR) and OUT in the absence of FokI; lane 4 contains the same strands plus FokI added. Almost no cleavage reaction occurs in this situation. In lane 5, the input strand IN was added to the mixture contained in lane 4, leading to cleavage of the output strand. A new band appears for the ‘waste’ product IN-PR. Lane 6 contains the pre-cleaved output strand OUT* for comparison.
Experimental proof of the mechanism
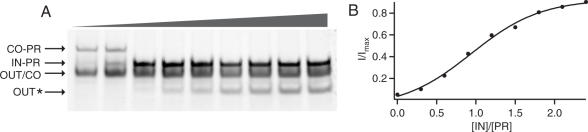
The basic operation of the DNA translator was characterized in gel electrophoresis experiments. The gel image displayed in Figure 3 clearly demonstrates that FokI can cleave the output loop OUT in the presence of the connector loop CO even without ligation. It also shows that cleavage is inhibited when the connector strand CO is bound to the protection strand PR. When a removal strand IN is added to the reaction mixture, the protection strand is displaced from the input loop by branch migration and the restriction reaction starts. This can be monitored by the appearance of a band for the ‘waste’ duplex (IN-PR) and for the restricted output strand OUT*. The intensity of these bands can also be used to obtain the transfer function of the device which relates the concentration of the output strand to that of the input strand (Figure 4). For the strand concentrations used in this particular experiment (compare Materials and Methods), the I/O relation is slightly sigmoidal (Figure 4B). The linear relationship between band intensity and DNA concentration in the relevant concentration range used was verified in a separate experiment.
Figure 4.
The response of the device depending on the input strand concentration. (A) Gel analysis of the device with different input signal amounts, ranging from 0 to 6 µl of 1 µM concentration. This highest value corresponds to 2.4 times the concentration of the protection strand, which was kept constant in all lanes. (B) A plot of the intensity of the product band as a function of input strand concentration.
Influence of design parameters
As described above, several design parameters are critical for the operation of the DNA transducer: On the input side, the length of the spacer sequence (section B in Figure 2) is important. When shorter than 2 bp, the distance of the FokI recognition site and the loop is too small to accommodate the FokI enzyme. When larger than 4 bp, the formation of dimers between protected input strands leads to false positive reactions. The ideal length therefore turns out to be 3 bp. To reduce side reactions due to dimer formation, it is necessary that the protection strand not only overlaps with one side of the spacer sequence (B in Figure 2), but also with part of the recognition site (C in Figure 2). The length of the linker sequence (D in Figure 2) influences the temperature at which the device performs correctly. To be able to work at room temperature, an overlap of 5 bases was used in all cases.
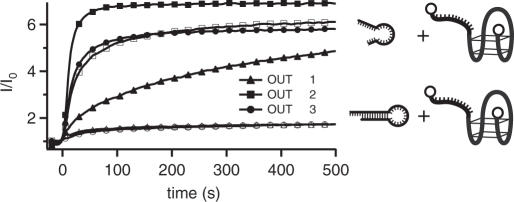
The details of the design of the output end strongly influence the hybridization behavior of the device in the presence of the target DNA sequence. Ideally, in the inactive state, no hybridization between output loops and target DNA should occur. On the other hand, after activation of the translator by an input strand, hybridization of the target with the FokI cleavage product should occur efficiently. Experimentally, three different designs for the output section with different stem lengths and loop sizes were studied [OUT-1, 14 bp stem before, 6 bp stem after cleavage/10 nt loop; OUT-2, 11 bp (3 bp) stem/20 nt loop; OUT-3, 11 bp (3 bp) stem, 7 nt loop, (for sequences see Table 1)]. To study the hybridization kinetics, we first performed experiments with synthesized output strands with appropriately shortened sequences to mimic the behavior of the FokI reaction products. The hybridization kinetics of the three different output loops in their cleaved and uncleaved form was determined in FRET measurements. In all cases the target sequence for hybridization was the doubly labeled aptamer strand APT. Hybridization between the output strand and APT results in a strong increase in fluorescence, as the FRET donor (fluorescein) and acceptor (TAMRA) are spatially more separated from each other in the duplex form than in the folded single-stranded G quadruplex form of APT. As can be judged from Figure 5, the three different designs for the output section result in completely different hybridization kinetics. This is due mainly to two effects. First, hybridization of a DNA strand with a stable DNA hairpin loop is extremely slow. To form a double helix, the DNA strand has to form a few initial base pairs with bases in the loop and then wind through the loop hole. Depending on the size of the loop, this can be very slow or even impossible on an experimental time scale. It also has to be considered that hybridization of a DNA strand to a hairpin loop structure is reduced even further when no end of the strand is able to bind within the loop and break open the stem through branch migration (47).
Figure 5.
Binding kinetics of processed (closed symbols) and unprocessed (open symbols) output strands to the aptamer beacon APT, measured by FRET. The three strand designs differ in stem length and loop sizes (triangles, OUT-1, 14 bp stem/10 nt loop, OUT-1*, 6 bp stem/10 nt loop; squares, OUT-2, 11 bp stem/20 nt loop, OUT-2*, 3 bp stem/20 nt loop; circles, OUT-3, 11 bp stem/7 nt loop, OUT-3*, 3 bp stem/7 nt loop). Larger loop sizes lead to stronger unwanted interactions with the aptamer. The stem size further regulates the hybridization rate after output strand cleavage.
The second important parameter—the stem length—has influence on the stability of the loop conformation. A short stem will break open occasionally and make the DNA loop sequence more accessible for hybridization with its complement. Similar effects have been studied extensively in the context of molecular beacons and also as a concept for fuelling free-running DNA nanomachines.
OUT-2 and OUT-3 have the same stem lengths and different loop sizes, whereas OUT-1 and OUT-3 have different stem lengths and comparable loop sizes. As can be seen in Figure 5, owing to its 20 nt long loop region, OUT-2 already strongly hybridizes with the target sequence in the ‘inactive’ state before cleavage by FokI. In contrast OUT-1 and OUT-3—with the small loops—only show a low level of unwanted hybridization with the target. After cleavage, all output strands hybridize well with the target sequence. Here, OUT-1* displays slower hybridization kinetics than OUT-2* and OUT-3* as it still has a considerable stem length (6 bp) after cleavage.
Performance of the device
In Figure 6, the hybridization kinetics of the full translation device containing FokI, PR, CO, OUT-3 with the target strand APT is shown. Before the addition of the input signal IN, the spurious hybridization reactions are at roughly the same level as in a ‘clean’ system containing only OUT-3. After addition of the input signal strand IN, the degree of hybridization to the target is enhanced several fold, showing the successful performance of the DNA translation device. The efficiency of hybridization is comparable with that with the pre-cleaved strands OUT-3*. Also shown is a test experiment, in which only the arbitrarily chosen strand IN is added to the aptamer device APT. This input strand in fact does not display any interaction with the target at all. This demonstrates that the device in fact works as a signal translator. By the action of the translator an otherwise non-reactive input strand can influence the hybridization with a target sequence.
Figure 6.
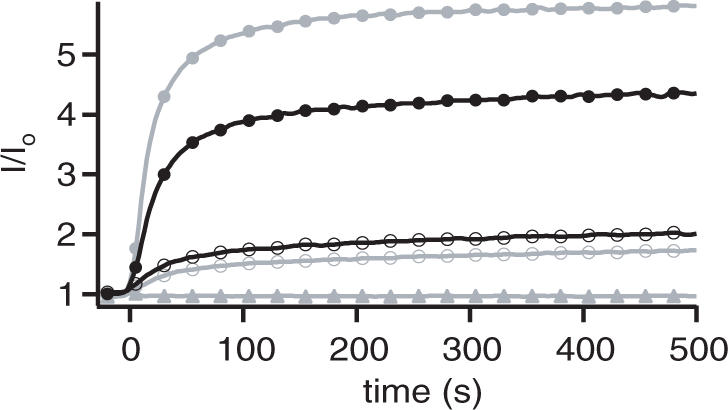
Comparison of the action of an output signal released by the translator device and a synthesized analogous output strand (OUT-3*). The closed circles represent the interaction between the aptamer beacon and the products of the device (solid, active; empty, idle). To examine the deviation from optimal performance, the same experiment was done with the inactive output loops only (gray open circles) and with pre-synthesized output loops (gray solid circles). Also shown is the direct interaction of the input signal with the aptamer beacon (gray triangles). As desired, the addition of the input strand leads to a strong enhancement of the hybridization signal (closed compared with open circles). The performance of the full translator device is almost as good as when only the pre-synthesized OUT-3* is hybridized to APT (gray solid circles).
Translator-controlled release of thrombin by the aptamer device
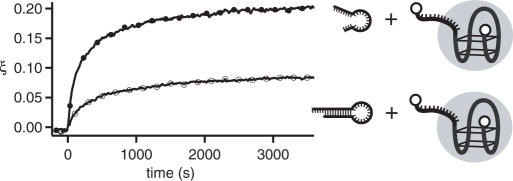
As an application of the signal translator concept, we demonstrate how the release of a protein by an aptamer-based DNA device can be controlled by an arbitrarily chosen DNA input signal. In Figure 7, the hybridization of the DNA translator reaction mixture with the DNA aptamer device APT bound to the protein thrombin is shown with and without the addition of the input strand IN. The initial fluorescence level is decreased to about 80% in comparison with Figure 6 (Ibound = 0.78 I0), as the folded G quadruplex structure of APT is now bound to the protein, leading to a stronger FRET effect—i.e. APT acts as a sensor for thrombin (48,49). Upon hybridization of the output strands with the aptamer device the fluorescence intensity increases. In a simplified two-state model, the total fluorescence signal is given as I(t) = ξ(t) Ireleased + [1 − ξ(t)] Ibound, where ξ(t) is the fraction of aptamers which have released their protein. Ireleased is the maximum fluorescence value obtained for aptamer beacons completely hybridized to the output strands and can be estimated from the limiting value Ireleased = 5.8 I0 (from Figure 6), where I0 is the initial fluorescence value for APT in the absence of protein. In Figure 7, the degree of protein release ξ(t) = [I(t) − Ibound]/(Ireleased − Ibound] is plotted. In the presence of the protein, this process is much slower than the hybridization of output strands with the aptamer without thrombin. The side reaction with the ‘inactive’ output strand OUT-3 already leads to a slight protein release. However, as desired the action of the DNA translator in response to the input strand IN leads to a much more efficient protein release. In principle, the side reaction could be completely suppressed if two distinct input signal strands were used and a correspondingly longer stem region for the inactive output strand (31).
Figure 7.
Fraction ξ of released thrombin controlled by the translation device. Closed circles, triggered device; open circles, idle device. The fraction ξ is determined indirectly from the fluorescence of the aptamer device as described in the text. The fact that the protein is initially bound to the aptamer results in a much slower kinetics of the change of the signal than in the absence of the protein.
CONCLUSIONS
We have demonstrated how a molecular signal translation device based on DNA hairpin loops and the action of the restriction endonuclease FokI can be used to release a specific DNA output strand in response to an arbitrarily chosen input strand. This translator can be used to uncouple nucleic acids-based signal and effector molecules and therefore link together molecular processes which produce or require specific DNA or RNA strands. Such situations may arise in DNA-based self-assembly, when the temporal or logical order of several self-assembly steps is to be controlled or when reaction networks of DNA-based nanodevices are to be constructed. Similar situations are of considerable interest in the context of intelligent biosensing or even drug delivery, when a specific compound is to be released in response to the presence of a certain nucleic acids molecule, e.g. mRNA. As a specific example, we demonstrated how the release of a protein by an aptamer-based nanodevice can be controlled by an arbitrarily chosen DNA strand which is translated by the molecular translator into a protein-releasing effector sequence.
Acknowledgments
Continuous support by Jörg P. Kotthaus is acknowledged. The authors acknowledge financial support by the Deutsche Forschungsgemeinschaft (Emmy Noether grant DFG SI 761/2-2). Funding to pay the Open Access publication charges for this article was provided by Nanion Technologies, GmbH, Munich, Germany.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mao C.D., Sun W.Q., Shen Z.Y., Seeman N.C. A nanomechanical device based on the B–Z transition of DNA. Nature. 1999;397:144–146. doi: 10.1038/16437. [DOI] [PubMed] [Google Scholar]
- 2.Yan H., Zhang X.P., Shen Z.Y., Seeman N.C. A robust DNA mechanical device controlled by hybridization topology. Nature. 2002;415:62–65. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 3.Yurke B., Turberfield A.J., Mills A.P., Simmel F.C., Neumann J.L. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 4.Simmel F.C., Yurke B. A DNA-based molecular device switchable between three distinct mechanical states. Appl. Phys. Lett. 2002;80:883–885. [Google Scholar]
- 5.Li J.W.J., Tan W.H. A single DNA molecule nanomotor. Nano Lett. 2002;2:315–318. doi: 10.1021/nl9011694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alberti P., Mergny J.L. DNA duplex-quadruplex exchange as the basis for a nanomolecular machine. Proc. Natl Acad. Sci. USA. 2003;100:1569–1573. doi: 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D.S., Balasubramanian S. A proton-fuelled DNA nanomachine. Angew. Chem. Int. Ed. Engl. 2003;42:5734–5736. doi: 10.1002/anie.200352402. [DOI] [PubMed] [Google Scholar]
- 8.Feng L.P., Park S.H., Reif J.H., Yan H. A two-state DNA lattice switched by DNA nanoactuator. Angew. Chem. Int. Ed. Engl. 2003;42:4342–4346. doi: 10.1002/anie.200351818. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y., Wang M.S., Mao C.D. An autonomous DNA nanomotor powered by a DNA enzyme. Angew. Chem. Int. Ed. Engl. 2004;43:3554–3557. doi: 10.1002/anie.200453779. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y., Lee S.H., Mao C.D. A DNA nanomachine based on a duplex–triplex transition. Angew. Chem. Int. Ed. Engl. 2004;43:5335–5338. doi: 10.1002/anie.200460789. [DOI] [PubMed] [Google Scholar]
- 11.Shen W.Q., Bruist M.F., Goodman S.D., Seeman N.C. A protein-driven DNA device that measures the excess binding energy of proteins that distort DNA. Angew. Chem. Int. Ed. Engl. 2004;43:4750–4752. doi: 10.1002/anie.200460302. [DOI] [PubMed] [Google Scholar]
- 12.Brucale M., Zuccheri G., Samori B. The dynamic properties of an intramolecular transition from DNA duplex to cytosine-thymine motif triplex. Org. Biomol. Chem. 2005;3:575–577. doi: 10.1039/b418353n. [DOI] [PubMed] [Google Scholar]
- 13.Sherman W.B., Seeman N.C. A precisely controlled DNA biped walking device. Nano Lett. 2004;4:1203–1207. [Google Scholar]
- 14.Yin P., Yan H., Daniell X.G., Turberfield A.J., Reif J.H. A unidirectional DNA walker that moves autonomously along a track. Angew. Chem. Int. Ed. Engl. 2004;43:4906–4911. doi: 10.1002/anie.200460522. [DOI] [PubMed] [Google Scholar]
- 15.Shin J.S., Pierce N.A. A synthetic DNA walker for molecular transport. J. Am. Chem. Soc. 2004;126:10834–10835. doi: 10.1021/ja047543j. [DOI] [PubMed] [Google Scholar]
- 16.Tian Y., Mao C.D. Molecular gears: a pair of DNA circles continuously rolls against each other. J. Am. Chem. Soc. 2004;126:11410–11411. doi: 10.1021/ja046507h. [DOI] [PubMed] [Google Scholar]
- 17.Tian Y., He Y., Chen Y., Yin P., Mao C.D. Molecular devices—a DNAzyme that walks processively and autonomously along a one-dimensional track. Angew. Chem. Int. Ed. Engl. 2005;44:4355–4358. doi: 10.1002/anie.200500703. [DOI] [PubMed] [Google Scholar]
- 18.Bath J., Green S.J., Turberfield A.J. A free-running DNA motor powered by a nicking enzyme. Angew. Chem. Int. Ed. Engl. 2005;44:4358–4361. doi: 10.1002/anie.200501262. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y., Mao C.D. Putting a brake on an autonomous DNA nanomotor. J. Am. Chem. Soc. 2004;126:8626–8627. doi: 10.1021/ja047991r. [DOI] [PubMed] [Google Scholar]
- 20.Dittmer W.U., Reuter A., Simmel F.C. A DNA-based machine that can cyclically bind and release thrombin. Angew. Chem. Int. Ed. Engl. 2004;43:3550–3553. doi: 10.1002/anie.200353537. [DOI] [PubMed] [Google Scholar]
- 21.Kolpashchikov D.M., Stojanovic M.N. Boolean control of aptamer binding states. J. Am. Chem. Soc. 2005;127:11348–11351. doi: 10.1021/ja051362f. [DOI] [PubMed] [Google Scholar]
- 22.Adleman L.M. Molecular computation of solutions to combinatorial problems. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 23.Ouyang Q., Kaplan P.D., Liu S.M., Libchaber A. DNA solution of the maximal clique problem. Science. 1997;278:446–449. doi: 10.1126/science.278.5337.446. [DOI] [PubMed] [Google Scholar]
- 24.Sakamoto K., Gouzu H., Komiya K., Kiga D., Yokoyama S., Yokomori T., Hagiya M. Molecular computation by DNA hairpin formation. Science. 2000;288:1223–1226. doi: 10.1126/science.288.5469.1223. [DOI] [PubMed] [Google Scholar]
- 25.Faulhammer D., Cukras A.R., Lipton R.J., Landweber L.F. Molecular computation: RNA solutions to chess problems. Proc. Natl Acad. Sci. USA. 2000;97:1385–1389. doi: 10.1073/pnas.97.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braich R.S., Chelyapov N., Johnson C., Rothemund P.W.K., Adleman L. Solution of a 20-variable 3-SAT problem on a DNA computer. Science. 2002;296:499–502. doi: 10.1126/science.1069528. [DOI] [PubMed] [Google Scholar]
- 27.Su X.P., Smith L.M. Demonstration of a universal surface DNA computer. Nucleic Acids Res. 2004;32:3115–3123. doi: 10.1093/nar/gkh635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt K.A., Henkel C.V., Rozenberg G., Spaink H.P. DNA computing using single-molecule hybridization detection. Nucleic Acids Res. 2004;32:4962–4968. doi: 10.1093/nar/gkh817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benenson Y., Paz-Elizur T., Adar R., Keinan E., Livneh Z., Shapiro E. Programmable and autonomous computing machine made of biomolecules. Nature. 2001;414:430–434. doi: 10.1038/35106533. [DOI] [PubMed] [Google Scholar]
- 30.Benenson Y., Adar R., Paz-Elizur T., Livneh Z., Shapiro E. DNA molecule provides a computing machine with both data and fuel. Proc. Natl Acad. Sci. USA. 2003;100:2191–2196. doi: 10.1073/pnas.0535624100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benenson Y., Gil B., Ben-Dor U., Adar R., Shapiro E. An autonomous molecular computer for logical control of gene expression. Nature. 2004;429:423–429. doi: 10.1038/nature02551. [DOI] [PubMed] [Google Scholar]
- 32.Adar R., Benenson Y., Linshiz G., Rosner A., Tishby N., Shapiro E. Stochastic computing with biomolecular automata. Proc. Natl Acad. Sci. USA. 2004;101:9960–9965. doi: 10.1073/pnas.0400731101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stojanovic M.N., Mitchell T.E., Stefanovic D. Deoxyribozyme-based logic gates. J. Am. Chem. Soc. 2002;124:3555–3561. doi: 10.1021/ja016756v. [DOI] [PubMed] [Google Scholar]
- 34.Stojanovic M.N., Stefanovic D. Deoxyribozyme-based half-adder. J. Am. Chem. Soc. 2003;125:6673–6676. doi: 10.1021/ja0296632. [DOI] [PubMed] [Google Scholar]
- 35.Stojanovic M.N., Stefanovic D. A deoxyribozyme-based molecular automaton. Nat. Biotechnol. 2003;21:1069–1074. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]
- 36.Soreni M., Yogev S., Kossoy E., Shoham Y., Keinan E. Parallel biomolecular computation on surfaces with advanced finite automata. J. Am. Chem. Soc. 2005;127:3935–3943. doi: 10.1021/ja047168v. [DOI] [PubMed] [Google Scholar]
- 37.Dittmer W.U., Simmel F.C. Transcriptional control of DNA-based nanomachines. Nano Lett. 2004;4:689–691. [Google Scholar]
- 38.Dittmer W.U., Kempter S., Radler J.O., Simmel F.C. Using gene regulation to program DNA-based molecular devices. Small. 2005;1:709–712. doi: 10.1002/smll.200500074. [DOI] [PubMed] [Google Scholar]
- 39.Robertson M.P., Ellington A.D. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechnol. 1999;17:62–66. doi: 10.1038/5236. [DOI] [PubMed] [Google Scholar]
- 40.Burgstaller P., Jenne A., Blind M. Aptamers and aptazymes: Accelerating small molecule drug discovery. Curr. Opin. Drug Discov. Dev. 2002;5:690–700. [PubMed] [Google Scholar]
- 41.Liu J.W., Lu Y. Adenosine-dependent assembly of aptazyme-functionalized gold nanoparticles and its application as a colorimetric biosensor. Anal. Chem. 2004;76:1627–1632. doi: 10.1021/ac0351769. [DOI] [PubMed] [Google Scholar]
- 42.Wang D.Y., Lai B.H.Y., Sen D. A general strategy for effector-mediated control of RNA-cleaving ribozymes and DNA enzymes. J. Mol. Biol. 2002;318:33–43. doi: 10.1016/S0022-2836(02)00046-3. [DOI] [PubMed] [Google Scholar]
- 43.Wang D.Y., Lai B.H.Y., Feldman A.R., Sen D. A general approach for the use of oligonucleotide effectors to regulate the catalysis of RNA-cleaving ribozymes and DNAzymes. Nucleic Acids Res. 2002;30:1735–1742. doi: 10.1093/nar/30.8.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beyer S., Dittmer W.U., Simmel F.C. Design variations for an aptamer-based DNA nanodevice. J. Biomed. Nanotechnol. 2005;1:96–101. [Google Scholar]
- 45.Feldkamp U., Saghafi S., Banzhaf W., Rauhe H. DNA computing. In: Jonoska N., Seeman N.C., editors. Proceedings of Seventh International Workshop on DNA-Based Computers, DNA7. Vol. 2340. Tampa, FL: Springer; 2001. pp. 23–32. [Google Scholar]
- 46.Mathews D.H., Disney M.D., Childs J.L., Schroeder S.J., Zuker M., Turner D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. PNAS. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turberfield A.J., Mitchell J.C., Yurke B., Mills A.P., Blakey M.I., Simmel F.C. DNA fuel for free-running nanomachines. Phys. Rev. Lett. 2003;90:118102. doi: 10.1103/PhysRevLett.90.118102. [DOI] [PubMed] [Google Scholar]
- 48.Nutiu R., Li Y.F. Structure-switching signaling aptamers. J. Am. Chem. Soc. 2003;125:4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 49.Hamaguchi N., Ellington A., Stanton M. Aptamer beacons for the direct detection of proteins. Anal. Biochem. 2001;294:126–131. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]