Abstract
Class I aminoacyl-tRNA synthetases catalyze editing reactions that prevent ambiguity from entering the genetic code. Misactivated amino acids are translocated in cis from the active site for aminoacylation to the center for editing, located ≈30 Å away. Mutational analysis has functionally separated the two sites by creating mutations that disrupt the catalytic center for editing but not for aminoacylation and vice versa. What is not known is whether translocation per se can be disrupted without an effect on either catalytic center. Here we describe mutations in a presumptive “hinge region” of isoleucyl-tRNA synthetase that is situated between the two sites. Interstice mutations had little or no effect on either catalytic center. In contrast, the same specific mutations disrupted translocation. Thus, with these mutations all three functions, translocation, catalysis of aminoacylation, and editing, have been mutationally separated. The results are consistent with translocation involving a hinge-region conformational shift that does not perturb the two catalytic centers.
Considerable evidence supports the concept that the universal tree of life could not be generated without the editing activities of aminoacyl-tRNA synthetases (1–3). These ancient proteins catalyze aminoacylation reactions that are the basis of the genetic code, that is, the matching of amino acids with specific nucleotide triplets imbedded in transfer RNAs (4–8). Inherent limitations to the capacity of active sites to discriminate between closely similar amino acids (9) was compensated by the introduction of a second active center where misactivated amino acids are cleared (10–15). Without this active site for editing, ambiguity is introduced into the code as demonstrated by the extensive misincorporation of amino acids into cellular proteins when editing is disrupted (1).
Class I aminoacyl-tRNA synthetases such as isoleucyl-, valyl-, and leucyl-tRNA synthetase are characterized by a catalytic center based on a Rossmann nucleotide-binding fold of alternating β-strands and α-helices (6, 16–21). The fold is split by an insertion known as connective polypeptide 1 (CP1) that joins one half of the catalytic domain to the other (12, 21–23). This insertion contains the active site for editing, located ≈30 Å from the catalytic center for aminoacylation (11–13, 15, 21, 24). The editing reactions require specific tRNAs as cofactors (25, 26). The role of the tRNA is to trigger translocation of the misactivated residue from the catalytic center to the active site for editing (27–29).
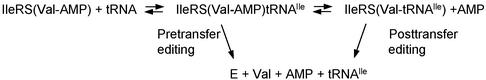
Amino acids are condensed with ATP to form aminoacyl adenylates bound to their cognate tRNA synthetases in the first step of protein synthesis. Occasional errors lead to activation of the wrong amino acid such as activation of valine by isoleucyl-tRNA synthetase (IleRS) or activation of threonine by valyl-tRNA synthetase (10, 30). Editing proceeds through two distinct pathways (Fig. 1). One is designated as the pretransfer step (10, 31). Here the misactivated amino acid (in the form of an aminoacyl adenylate) is translocated from the active site to the editing center. The reaction is strictly tRNA-dependent and, in the case of IleRS, requires specific nucleotides in the dihydrouridine loop of tRNAIle to effect translocation (25). The misactivated adenylate is then hydrolyzed, thus establishing a cycle of misactivation and hydrolysis that leads to the consumption of ATP. The second pathway is designated posttransfer editing. In this instance, the misactivated amino acid is transferred to the 3′ end of tRNA to create a mischarged tRNA [such as Val-tRNAIle (32), Ser-tRNAThr (33), Ile-tRNAPhe (34), Ile-tRNALeu (35), Ala-tRNAPro (36), and Gly-tRNAAla (3)]. The mischarged aminoacyl group is then cleared at the editing center. For IleRS the editing center is operationally divided into subsites, with one to clear the aminoacyl moiety of the misactivated adenylate and the other to clear the mischarged aminoacyl group from the tRNA (37).
Figure 1.
Schematic of the two pathways for editing. After valine is misactivated, it can be cleared either by the pretransfer pathway that acts directly on Val-AMP or by posttransfer clearance of the valyl moiety of Val-tRNAIle.
Previous work supported a model in which editing was initiated with a “priming” step, where the misactivated amino acid was linked to tRNA and then shifted to the editing center (29). In a way not understood, this priming step facilitates rounds of pretransfer editing, where the misactivated adenylate is translocated from the active site to the center for editing and then cleared away. The fraction of editing that is generated by pretransfer as opposed to posttransfer editing is determined by the number of cycles of pretransfer editing that occur before dissociation of the tRNA “primer.” Previous studies suggested that 70–90% of the editing by Escherichia coli IleRS proceeds through the pretransfer pathway (10).
Much of the understanding of editing by class I enzymes derives from extensive functional analysis and structural studies of IleRS. Through a series of mutational analyses, the centers for editing and the aminoacylation were clearly resolved even before a three-dimensional structure was available (11–13). Strikingly, the two sites are functionally independent in that mutations in one do not affect the other (12, 21, 29, 37, 38). However, the translocation step has been most difficult to resolve. Indeed, the only mutation that clearly blocks translocation also disrupts the catalytic site for editing (29). This mutation, D342A, is thought to prevent binding of the amino group (of the valyl-moiety of Val-tRNAIle) to the center for editing. As a consequence, the aforementioned priming step is blocked, and translocation cannot occur. At the same time, however, the mutation prevents hydrolysis of Val-tRNAIle. Thus, not clear is whether in fact translocation can be mutationally separated from the other two activities or whether it is intrinsically connected to the chemical steps of editing. It was with this perspective that we set out to explore the possibility of obtaining translocation-specific mutations of IleRS. Such mutations would provide clear evidence that translocation per se did not depend on the chemical steps.
Materials and Methods
Preparation of Val-AMP and Val-tRNAIle.
Val-AMP was chemically synthesized as described (28) and stored at −80°C. Val-tRNAIle was prepared as detailed (29). For this purpose, tRNAIle was overexpressed and purified as described by Glasfeld and Schimmel (39), and the editing-defective T242P IleRS (38) was used to generate mischarged Val-tRNAIle.
Plasmid Construction.
W421A and W421A/K183A IleRS were generated by the QuikChange (Stratagene) system on plasmid pVDC-433, which encodes WT IleRS (37). Sequences of the mutagenic primers are available upon request. Construction of K183A IleRS was accomplished by isolating a NcoI–KpnI restriction fragment from the plasmid encoding W421A/K183A IleRS. This fragment spans the region coding for the K183A substitution. It then was used to replace the NcoI–KpnI fragment of plasmid pVDC-433. The mutations were confirmed by DNA sequencing.
Complementation Assays.
Overnight cultures of strain MG1665 (40) bearing plasmids encoding mutations in IleRS were infected with ileS∷kanr-containing P1 phage stocks and selected on LB agar plates containing 50 μg/ml ampicillin, 25 μg/ml kanamycin, and 0.02% arabinose as detailed (29). Plates were incubated at room temperature for 2 days. Individual colonies were isolated and used for protein expression.
Protein Expression and Purification.
Enzymes were expressed and purified from the ΔileS∷kanr strains harboring plasmids encoding either WT or mutant IleRS as described (29, 41). Concentrations of enzymes were determined by active-site titration (42).
Assays for Amino Acid Activation and Aminoacylation.
Aminoacylation assays were carried out essentially as described (38) with mutant or WT enzymes (7.2 nM), 20 μM [3H]Ile, and 2 mg/ml E. coli tRNA (Roche, Indianapolis, IN) at pH 7.5 and 22°C. Amino acid activation was measured by the ATP-pyrophosphate exchange assay (43) with 2 mM ATP, 2 mM Na-[32P]pyrophosphate, 2 mM Ile, and mutant or WT enzymes (250 nM) at pH 7.5 and 22°C.
Assays Related to Editing Reactions.
Assays for overall editing by measuring ATP hydrolysis were performed as described (27) by monitoring the tRNAIle-dependent hydrolysis of [γ-32P]ATP in the presence of valine and IleRS (1.0 μM) at pH 7.5 and 22°C. For posttransfer deacylation assays, the time-dependent hydrolysis of Val-tRNAIle following the addition of 12.5 nM IleRS (pH 7.5, 22°C) was monitored as described (27). Translocation was studied by measuring the reoccupation of the active site by N-methylanthraniloyl dATP (dATP†, 30 μM), after tRNAIle was added to IleRS (500 nM) complexed with an approximately stoichiometric amount of Val-AMP, by using a fluorescence assay as described (28).
Results
Rationale for Placement of Mutations.
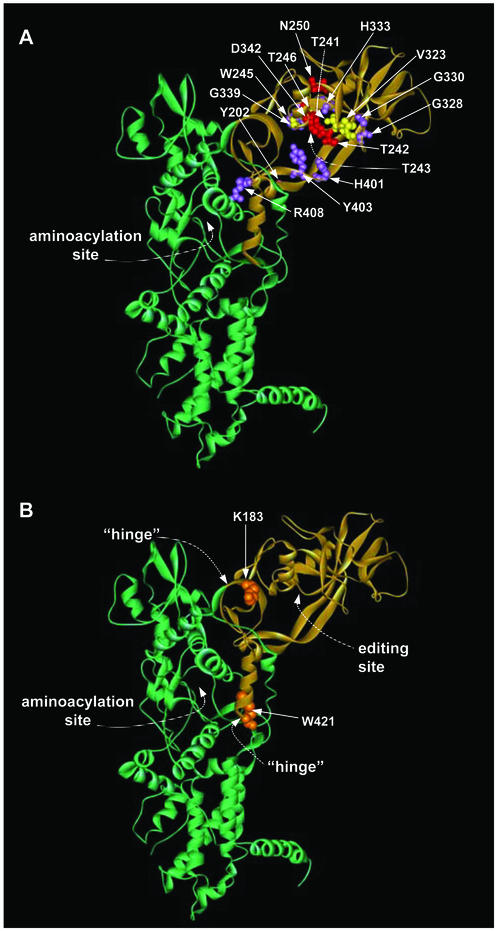
The CP1 editing domain bulges out from the body of IleRS that contains the catalytic center (Fig. 2). This domain has been cloned as an isolated unit that deacylates Val-tRNAIle but not the correctly acylated Ile-tRNAIle (13). The N and C termini of CP1 are close in space such that minimal disruption occurs with CP1 inserted into the catalytic body. The binding pocket for the valyl moiety was visualized in the structure of a cocrystal (21), and this location was consistent with mutational studies done before and after the structural determination (21, 24). Altogether, >16 mutations have been placed in CP1, and the mutant enzymes have been biochemically characterized (Fig. 2A and refs. 11–13, 21, 29, 37, and 38). These mutations have phenotypes ranging from mild to severe effects on the efficiency of deacylation of Val-tRNAIle. Other mutations affect the specificity of deacylation such as the discrimination of Val-tRNAIle versus Ile-tRNAIle (12). Most or all of them have little effect on catalysis of aminoacylation. However, none of these mutant enzymes is defective only in translocation.
Figure 2.
Ribbon diagram of the structure of Thermus thermophilus IleRS [coordinates are from Nureki et al. (21)]. The CP1 domain is depicted in tan. (A) Sites of mutations from previous editing studies (12, 21, 29, 37, 38). Red residues symbolize mutations that had significant effects on editing, violet residues represent mutations that resulted in an alteration in the specificity of the editing reaction, and yellow side chains are those that, when mutated, resulted in a WT phenotype for editing. (B) Proposed mutation sites (shown in orange) in the IleRS hinge region. The numbering of residues is based on that of the E. coli enzyme.
With this background, we sought for a rationale to find locations in the structure that would affect translocation specifically. X-ray crystal structures of IleRS in the absence and presence of tRNAIle show that CP1 of the IleRS–tRNAIle complex is rotated ≈50o with respect to the same domain in apo-IleRS (when domains for aminoacylation are overlaid) (21, 24). As a consequence, “hinges” that connect aminoacylation and editing sites undergo a substantial conformational change (Fig. 2B). K183 and W421 are conserved residues that are at hinge points that are separated by 35 Å. We thus imagined that K183 and W421 could be critical residues for the function of hinges needed for translocation.
Aminoacylation Function Marginally Affected by Mutations of Hinge-Region Residues.
Three mutant enzymes (K183A, W421A, and K183A/W421A IleRS) were constructed and compared with each other and with the WT enzyme. The activity for adenylate synthesis (as measured by the standard isoleucine-dependent ATP-pyrophosphate exchange assay) was the same for all four enzymes at pH 7.5 and 22°C. The efficiency for catalysis of aminoacylation of tRNAIle was similar for the four proteins, with the activity varying no more than 2-fold (data not shown). Thus, K183 and W421 have little or no role in the catalytic functions associated with the aminoacylation reaction.
Overall Editing Activity Attenuated by Mutations in Hinge Region.
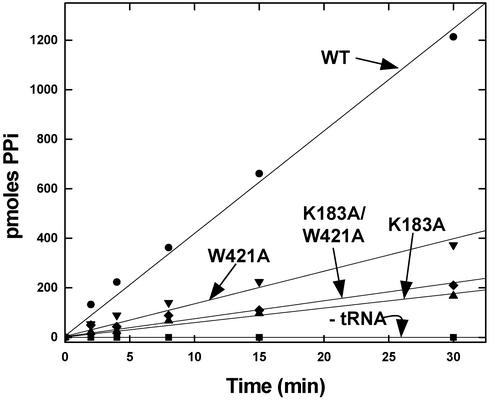
Overall editing activities for the four enzymes were tested by the standard assay for abortive ATP hydrolysis associated with the misactivation of valine. In this assay, valine is mixed with ATP, tRNAIle, and IleRS, and the consumption of ATP is monitored directly. This assay measures editing associated with both post- and pretransfer pathways. Robust activity was observed with the WT enzyme. This activity was tRNA-dependent, as expected. All three mutant enzymes were defective in editing (Fig. 3). The most significant effects were seen with K183A and K183A/W421A IleRS, with the residual activity of the double-mutant protein being about the same as that of the K183A enzyme. For these two enzymes, the ATPase activity was diminished ≈6- to 7-fold.
Figure 3.
Overall editing activities measured by tRNAIle-dependent stimulation of ATP hydrolysis [pyrophosphate (PPi)-produced] when valine, ATP, and IleRS are mixed together at pH 7.5 and 22°C.
Posttransfer Deacylation Activity Not Affected by Mutations in Hinge Region.
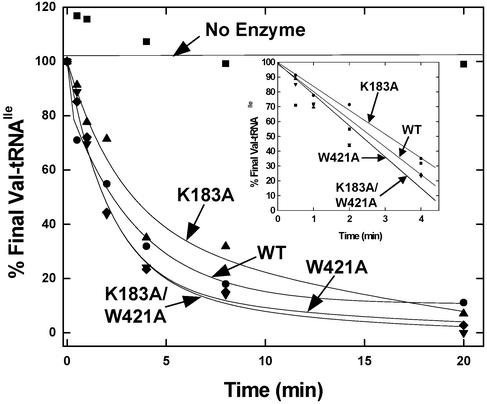
The residual editing activity of the K183A and K183A/W421A mutant enzymes represents a fraction (approximately one-sixth to one-seventh) of total editing activity. This fraction is similar to what is estimated to be the contribution of the posttransfer pathway to overall editing (10). Indeed, it seemed plausible that the reason the K183A single and the K183A/W421A double mutants had similar activities was that a limit had been reached in the reduction of overall editing. That limit would occur if only pretransfer and not pre- and posttransfer editing had been eliminated. Thus, we were especially motivated to test the effect of the mutations on posttransfer editing.
For this assay, Val-tRNAIle was mixed separately with all four enzymes, and the time-dependent clearance of the valyl moiety was investigated. The activities of the four enzymes were the same within experimental error (Fig. 4). Thus, the K183A and W421A substitutions do not affect catalytic residues needed for the chemical step of editing. (This result is consistent with the failure to detect accumulation of Val-tRNAIle with either W421A or K183A/W421A.) This result suggested that the defect in overall editing caused by these mutations was because pretransfer editing was affected in some way. Because K183 and W421 are distant from each active site and have no apparent role in catalysis, it seemed most likely that these hinge-region residues were important for the translocation step associated with movement of the misactivated adenylate from the site for aminoacylation to the center for editing.
Figure 4.
Posttransfer editing activities of WT and mutant enzymes (12.5 nM) at pH 7.5 and 22°C. The clearance of Val-tRNAIle is plotted as a function of time. (Inset) Initial 4 min.
Translocation Specifically Disrupted by Mutations in Hinge-Region Residues.
An assay for translocation based on fluorescence energy transfer has been developed (28). In this assay, use is made of the fluorescent nucleotide dATP†. Although this nucleotide binds to the catalytic site for aminoacylation with a Kd of ≈2 μM, it is a poor substrate for the enzyme. The excitation maximum of dATP† is at ≈350 nm, which is near the peak of the emission spectrum for the indole group of tryptophan. The emission spectrum of dATP† peaks at ≈440 nm. Thus, when dATP† is bound at the active site for aminoacylation, excitation at 290 nm of any tryptophan near the active-site pocket leads to emission at 440 nm via energy transfer. When dATP† is absent from the active site, no emission at 440 nm is seen.
In a single turnover reaction for clearance of the misactivated adenylate, the energy transfer-dependent emission of dATP† at 440 nm can be used to study translocation (Fig. 5). When the valyl moiety is translocated (either in the form of the adenylate (Val-AMP) or as Val-tRNAIle from the site for aminoacylation to the CP1 editing domain, the presence of excess dATP† ensures instantaneous binding of the nucleotide analog and the appearance of emission at 440 nm. The rate of appearance of this emission is a direct measure of translocation. Thus, the enzyme is first complexed with the tightly bound Val-AMP (Kd of ≈10 nM), and translocation is initiated by the addition of tRNAIle. By reducing the amount of tRNAIle, the apparent rate of translocation can be slowed enough to be measured by conventional methods (28).
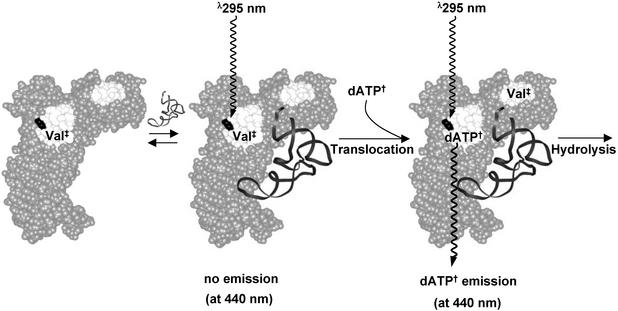
Figure 5.
Schematic illustration of the energy transfer-dependent emission of dATP† associated with the translocation step. Val‡ represents Val-AMP or Val-tRNAIle. Binding of tRNAIle triggers translocation of Val-AMP or Val-tRNAIle from the active site to the editing site with subsequent occupation of dATP† in the vacated active site. Energy transfer-dependent emission (350 nm) from an excited Trp (shown in black) is transferred to dATP†, resulting in emission at 440 nm.
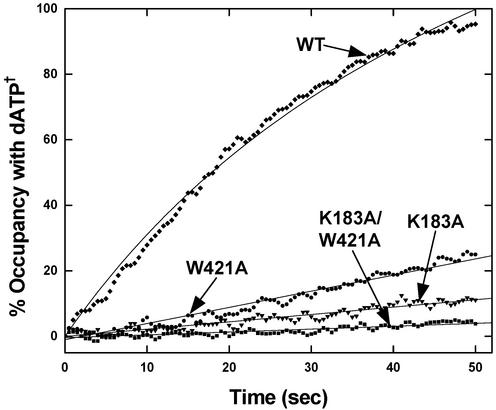
With this assay, K183A and W421A substitutions substantially reduced translocation. The double K183A/W421A replacement further reduced the rate of translocation (Fig. 6). Unlike the assay for overall editing in which the K183A and K183A/W421A mutations had the same effect (within experimental error), the double-mutant protein was more defective than the K183A mutant in the translocation assay. Indeed, K183A/W421A IleRS is completely inactive for translocation. Because translocation is required for clearance of misactivated Val-AMP, the residual overall editing activity observed for the K183A/W421A enzyme is most likely due to an “in trans” posttransfer pathway. Unlike pretransfer editing, the posttransfer pathway can operate without translocation by dissociation of the mischarged Val-tRNAIle and rebinding with the valyl moiety of Val-tRNAIle placed in the CP1 editing domain.
Figure 6.
Activities for translocation of misactivated valine by mutant and WT enzymes at pH 7.5 and 22°C. Reactions were initiated after the addition of substoichiometric tRNAIle (≈0.5 equivalents) to a near stoichiometric Val-AMP–IleRS complex (0.8:1.0). Translocation of Val-AMP/Val-tRNAIle to the editing site results in the rapid binding of excess dATP† (30 μM) to the active site.
Discussion
Previous work established that under similar conditions to those used here the translocation rate of misactivated Val-AMP is ≈1.4 sec−1 (28). This rate is the same within experimental error as that measured for the maximal rate of ATP hydrolysis when valine, ATP, and tRNAIle are mixed together with IleRS (10, 28). Thus, translocation is rate-limiting for the overall editing reaction. However, as discussed above, elimination of translocation does not preclude the editing reaction, because mischarged Val-tRNAIle can dissociate and rebind to the enzyme with the valyl moiety placed in the editing pocket. Indeed, the CP1 domain in isolation catalyzes deacylation of Val-tRNAIle (13, 29). From an evolutionary perspective, a plausible scenario is one where the CP1 progenitor was separate from the “primordial” IleRS and cleared all errors of aminoacylation by acting in trans on mischarged tRNAIle. At some point the CP1 domain was inserted into the body of the enzyme, which in turn led to the ability to translocate Val-AMP in cis directly from the active site to the editing pocket in CP1. In searching sequence databases, no example of a “free-standing” CP1-like domain has been identified. Moreover, the CP1 domain itself is an integral part of the isoleucyl-, valyl-, and leucyl-tRNA synthetases throughout all three kingdoms and is found in the most basal organisms such as Aquifex aeolicus (44). Thus, the incorporation of CP1 into these enzymes may have occurred at the time of the last common ancestor.
Critical to understanding translocation of the misactivated aminoacyl group (in the form of aminoacyl adenylate or aminoacyl tRNA) is the requirement for a specific tRNA. For example, tRNAVal binds to IleRS and competitively displaces tRNAIle. But when tRNAVal is mixed with valine, ATP, and IleRS, ATPase activity is not stimulated, that is, there is no editing reaction (25). Two factors are thought to account for this behavior. First, an initial priming step requires formation of charged tRNA, and tRNAVal is not acylated by IleRS even though it can bind (25). Second, despite an alteration to the anticodon of tRNAVal so as make it a substrate for IleRS, the editing response is not triggered. Three tRNAIle-specific nucleotides in the dihydrouridine loop are also required and when they are introduced into the body of the anticodon-modified tRNAVal, the latter then is activated for editing (25). These nucleotides are located at the outside corner, or elbow, of the L-shaped tRNA structure where they play a role in facilitating translocation (27), presumably through a conformational change that also involves the hinge region of IleRS. In the cocrystal of IleRS and tRNAIle (24), and of the homologous Val-tRNA synthetase with tRNAVal (45), residues in the hinge region of IleRS make no contact with the bound nucleic acid. Possibly, the outside corner of tRNAIle is the counterpart of the hinge associated with IleRS, that is, a region of structure requiring a specific sequence for making a conformational change associated with the translocation event.
The presumptive enzyme and tRNA hinges may act in concert during translocation. With this scenario it is conceivable that the mutations such as K183A and W421A investigated here interfere with a coordinated conformational change in tRNAIle during translocation. If so, then it may be possible to compensate for a K183A mutation, with, for example, a substitution in tRNAIle. The success of an experiment such as this would support the idea that conformational events in IleRS and tRNAIle are critically linked during translocation.
As a straightforward possibility, the K183A and W421A mutations studied here may interfere with the priming stem that is required to establish translocation of the adenylate. For the priming step, mischarged Val-tRNAIle is proposed to switch its 3′ end between two defined conformational states (24). One state of Val-tRNAIle is analogous to that seen in the cocrystal structure of Gln-tRNA synthetase with tRNAGln, where the 3′ end of tRNAGln is bent back into the aminoacylation active site through a hairpin-like structure. After straightening the 3′ end of Val-tRNAIle into a conventional form, the editing pocket can be reached by the valyl moiety. Plausibly, the nucleotides at the outside corner of tRNAIle could be sensitive to this conformational switch at the 3′ end. At the same time, the hinge region could allow (or be sensitive to) rotation of the CP1 domain relative to the active site and could be coupled to the conformational switch in the tRNA structure. Possibly, these coupled events are rate-limiting for translocation and therefore for overall editing.
Acknowledgments
We thank Professor Tamara Hendrickson for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant GM 15539 and a fellowship from the National Foundation for Cancer Research. K.B. is a National Institutes of Health postdoctoral fellow. A.C.B. was supported by Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation Fellowship Grant DRG-1637.
Abbreviations
- CP1
connective polypeptide 1
- IleRS
isoleucyl-tRNA synthetase
- dATP†
N-methylanthraniloyl dATP
References
- 1.Döring V, Mootz H D, Nangle L A, Hendrickson T L, de Crécy-Lagard V, Schimmel P, Marliere P. Science. 2001;292:501–504. doi: 10.1126/science.1057718. [DOI] [PubMed] [Google Scholar]
- 2.Nangle L A, de Crécy-Lagard V, Döring V, Schimmel P. J Biol Chem. 2003;277:45729–45733. doi: 10.1074/jbc.M208093200. [DOI] [PubMed] [Google Scholar]
- 3. Beebe, K., Ribas de Pouplana, L. & Schimmel, P. (2002) EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 4.Lapointe J, Giegé R. In: Translation in Eukaryotes. Trachsel H, editor. Boca Raton, FL: CRC; 1991. pp. 35–69. [Google Scholar]
- 5.Moras D. Trends Biochem Sci. 1992;17:159–164. doi: 10.1016/0968-0004(92)90326-5. [DOI] [PubMed] [Google Scholar]
- 6.Carter C W., Jr Annu Rev Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 7.Giegé R, Puglisi J D, Florentz C. Prog Nucleic Acid Res Mol Biol. 1993;45:129–206. doi: 10.1016/s0079-6603(08)60869-7. [DOI] [PubMed] [Google Scholar]
- 8.Ibba M, Söll D. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 9.Pauling L. Festschrift Prof. Dr. Arthur Stoll. Basel: Birkhauser; 1957. pp. 597–602. [Google Scholar]
- 10.Fersht A R. Biochemistry. 1977;16:1025–1030. doi: 10.1021/bi00624a034. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt E, Schimmel P. Science. 1994;264:265–267. doi: 10.1126/science.8146659. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt E, Schimmel P. Biochemistry. 1995;34:11204–11210. doi: 10.1021/bi00035a028. [DOI] [PubMed] [Google Scholar]
- 13.Lin L, Hale S P, Schimmel P. Nature. 1996;384:33–34. doi: 10.1038/384033b0. [DOI] [PubMed] [Google Scholar]
- 14.Beuning P J, Musier-Forsyth K. Proc Natl Acad Sci USA. 2000;97:8916–8920. doi: 10.1073/pnas.97.16.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mursinna R S, Lincecum T L, Martinis S A. Biochemistry. 2001;40:5376–5381. doi: 10.1021/bi002915w. [DOI] [PubMed] [Google Scholar]
- 16.Webster T, Tsai H, Kula M, Mackie G A, Schimmel P. Science. 1984;226:1315–1317. doi: 10.1126/science.6390679. [DOI] [PubMed] [Google Scholar]
- 17.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 18.Steitz T A. Curr Opin Struct Biol. 1991;1:139–143. [Google Scholar]
- 19.Cavarelli J, Moras D. FASEB J. 1993;7:79–86. doi: 10.1096/fasebj.7.1.8422978. [DOI] [PubMed] [Google Scholar]
- 20.Cusack S. Nat Struct Biol. 1995;2:824–831. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- 21.Nureki O, Vassylyev D G, Tateno M, Shimada A, Nakama T, Fukai S, Konno M, Hendrickson T L, Schimmel P, Yokoyama S. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 22.Hale S P, Schimmel P. Tetrahedron. 1997;53:11985–11994. [Google Scholar]
- 23.Starzyk R M, Webster T A, Schimmel P. Science. 1987;237:1614–1618. doi: 10.1126/science.3306924. [DOI] [PubMed] [Google Scholar]
- 24.Silvian L F, Wang J, Steitz T A. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]
- 25.Hale S P, Auld D S, Schmidt E, Schimmel P. Science. 1997;276:1250–1252. doi: 10.1126/science.276.5316.1250. [DOI] [PubMed] [Google Scholar]
- 26.Baldwin A N, Berg P. J Biol Chem. 1966;241:839–845. [PubMed] [Google Scholar]
- 27.Farrow M A, Nordin B E, Schimmel P. Biochemistry. 1999;38:16898–16903. doi: 10.1021/bi9920782. [DOI] [PubMed] [Google Scholar]
- 28.Nomanbhoy T K, Hendrickson T L, Schimmel P. Mol Cell. 1999;4:519–528. doi: 10.1016/s1097-2765(00)80203-8. [DOI] [PubMed] [Google Scholar]
- 29.Bishop A C, Nomanbhoy T K, Schimmel P. Proc Natl Acad Sci USA. 2002;99:585–590. doi: 10.1073/pnas.012611299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fersht A R, Kaethner M M. Biochemistry. 1976;15:3342–3346. doi: 10.1021/bi00660a026. [DOI] [PubMed] [Google Scholar]
- 31.Hale S P, Schimmel P. Proc Natl Acad Sci USA. 1996;93:2755–2758. doi: 10.1073/pnas.93.7.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eldred E W, Schimmel P R. J Biol Chem. 1972;247:2961–2964. [PubMed] [Google Scholar]
- 33.Dock-Bregeon A, Sankaranarayanan R, Romby P, Caillet J, Springer M, Rees B, Francklyn C S, Ehresmann C, Moras D. Cell. 2000;103:877–884. doi: 10.1016/s0092-8674(00)00191-4. [DOI] [PubMed] [Google Scholar]
- 34.Yarus M. Proc Natl Acad Sci USA. 1972;69:1915–1919. doi: 10.1073/pnas.69.7.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J F, Guo N N, Li T, Wang E D, Wang Y L. Biochemistry. 2000;39:6726–6731. doi: 10.1021/bi000108r. [DOI] [PubMed] [Google Scholar]
- 36.Wong F C, Beuning P J, Nagan M, Shiba K, Musier Forsyth K. Biochemistry. 2002;41:7108–7115. doi: 10.1021/bi012178j. [DOI] [PubMed] [Google Scholar]
- 37.Hendrickson T L, Nomanbhoy T K, de Crécy-Lagard V, Fukai S, Nureki O, Yokoyama S, Schimmel P. Mol Cell. 2002;9:353–362. doi: 10.1016/s1097-2765(02)00449-5. [DOI] [PubMed] [Google Scholar]
- 38.Hendrickson T L, Nomanbhoy T K, Schimmel P. Biochemistry. 2000;39:8180–8186. doi: 10.1021/bi0004798. [DOI] [PubMed] [Google Scholar]
- 39.Glasfeld E, Schimmel P. Biochemistry. 1996;35:4139–4145. doi: 10.1021/bi9527810. [DOI] [PubMed] [Google Scholar]
- 40.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 41.Shepard A, Shiba K, Schimmel P. Proc Natl Acad Sci USA. 1992;89:9964–9968. doi: 10.1073/pnas.89.20.9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fersht A R, Ashford J S, Bruton C J, Jakes R, Koch G L E, Hartley B S. Biochemistry. 1975;14:1–4. doi: 10.1021/bi00672a001. [DOI] [PubMed] [Google Scholar]
- 43.Calendar R, Berg P. Biochemistry. 1966;5:1690–1695. doi: 10.1021/bi00869a034. [DOI] [PubMed] [Google Scholar]
- 44.Ribas de Pouplana L, Schimmel P. Cold Spring Harbor Symp Quant Biol. 2001;LXVI:131–166. doi: 10.1101/sqb.2001.66.161. [DOI] [PubMed] [Google Scholar]
- 45.Fukai S, Nureki O, Sekine S, Shimada A, Tao J, Vassylyev D G, Yokoyama S. Cell. 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]