Abstract
Telomere dysfunction arising from mutations in telomerase or in telomere capping proteins leads to end-to-end chromosome fusions. Paradoxically, the Ku70/80 heterodimer, essential for nonhomologous end-joining double-strand break repair, is also found at telomeres, and in mammals it is required to prevent telomere fusion. Previously, we showed that inactivation of Ku70 in Arabidopsis results in telomere lengthening. Here, we have demonstrated that this telomere elongation is telomerase dependent. Further, we found that the terminal 3′ G overhang was significantly extended in ku70 mutants and in plants deficient in both Ku70 and the catalytic subunit of telomerase (TERT), implying that Ku is needed for proper maintenance of the telomeric C-rich strand. Consistent with inefficient C-strand maintenance, telomere shortening was accelerated in ku70 tert double mutants, and the onset of a terminal sterile phenotype was reached two to three times faster than in tert single mutants. Unexpectedly, abundant anaphase bridges were found in terminal plants harboring critically shortened telomeres, indicating that Ku is not required for the formation of end-to-end chromosome fusions in telomerase-deficient Arabidopsis. Together, these findings define Ku70 as a gene in higher eukaryotes required for maintenance of the telomeric C-rich strand and underscore the complexity and diversity of molecular interactions at telomeres.
Telomeres are nucleoprotein structures that protect chromosome ends from being recognized and processed as double-strand breaks. Telomere synthesis by telomerase provides a mechanism that assures complete replication of linear eukaryotic chromosomes and consequently maintenance of a full complement of telomeric DNA. In cells lacking telomerase, telomeric DNA tracts inexorably shorten when cells proliferate (1). Once a critical telomere length is reached, chromosome ends become uncapped and indistinguishable from DNA damage. The free ends are then subject to nucleolytic degradation and end-to-end joining reactions to create chromosome fusions (2–4).
The Ku70/80 heterodimer has a dual function in genome maintenance. As a core component of nonhomologous end-joining (NHEJ) DNA repair pathway, it binds double-strand breaks, facilitating their alignment and subsequent ligation (5, 6). The role of Ku in NHEJ has been relatively well characterized, and Ku mutants display an increased sensitivity to DNA damage (6–11). Ku has also been localized to telomeres in yeast and mammals (12, 13), but the telomere-associated phenotypes arising from a Ku deficiency vary widely among different organisms; thus, the precise function of Ku at telomeres is unclear.
Deletion of Ku in budding yeast results in shortened duplex telomere tracts with extended 3′ G overhangs (12). Mutants fail to grow at elevated temperature, presumably because of an activation of DNA checkpoints by an altered telomere structure (14–16). Shorter telomeres are also associated with Ku deletion in fission yeast and trypanosomes (17, 18). In addition, yeast strains deficient in telomerase and Ku exhibit accelerated senescence, implying a synergistic interaction.
Higher eukaryotes respond differently to a deficiency in Ku. Mouse mutants lacking Ku exhibit premature senescence and stunted growth (19, 20). At the genome level, frequent end-to-end chromosome fusions and perturbations in telomere length homeostasis are observed (21–24). We previously showed that telomeres in Arabidopsis mutants lacking Ku70 are dramatically extended, but their capping function seems to be intact, as we found no evidence for chromosome fusions (9).
To further investigate the behavior of Ku in Arabidopsis, we analyzed telomere architecture and maintenance in Ku70 mutants and in mutants deficient in both Ku70 and telomerase. Here, we provide evidence that Ku is required for proper maintenance of the telomeric C strand. We also show that Ku regulates extension of the telomeric G strand by acting as a negative regulator of telomerase. Finally, in striking contrast to what has been reported for mammals (25, 26), we demonstrate that Ku is not required for fusion of critically shortened telomeres in telomerase-deficient Arabidopsis.
Materials and Methods
Plant Material.
Arabidopsis thaliana plants heterozygous for a T-DNA insertion in KU70 (ecotype Ws) (9) were crossed to plants with a T-DNA insertion in TERT (ecotype Col) (27). PCR was used to determine the genotype of the F1 population (9, 27). A single F1 plant heterozygous for T-DNA insertions in both genes was used to establish two F2 ku70 tert homozygous lines (1 and 52) termed G1 (generation 1), as well as control WT (44 and 121), ku70 (87 and 90), and tert lines (119 and 120). Individual lines were propagated for successive generations by self-pollination. Plants were grown at 23°C in an environmental chamber under a 16/8-h light/dark photoperiod.
Terminal Restriction Fragment (TRF) Analysis and Cytogenetics.
DNA extraction and TRF analysis was performed as described (9, 27). The size of TRFs was determined from PhosphorImager scans by using IMAGEQUANT software (Molecular Dynamics). The average telomere size in each plant was calculated by using an algorithm that normalizes the signal intensity relative to the size of each digestion product (28). Anaphase spreads were prepared from pistils as described (29).
In-Gel Hybridization.
Genomic DNA (1 μg) was digested with HindIII and subjected to in-gel hybridization (30). Stringent hybridization was performed at 52°C for 16 h with a 32P-labeled 5′(CCCTAAA)4CCC3′ probe. The gel was washed three times in 0.25× SSC (1× SSC = 0.3 M NaCl/0.03 M sodium citrate, pH 7) at 25°C and then once at 52°C in 0.25× SSC for 1 h. Exonuclease 1 (United States Biochemical) treatment was performed as described (31).
Results and Discussion
To determine whether telomerase is responsible for telomere elongation in Ku-deficient Arabidopsis, plants lacking both KU70 and TERT, the catalytic subunit of telomerase, were analyzed. Arabidopsis heterozygous for a T-DNA insertion in TERT (27) were crossed to plants heterozygous for a T-DNA insertion in KU70 (9) (Fig. 1A). A single F1 plant heterozygous for T-DNA insertions in both genes was used to establish homozygous lines in F2 [termed G1 (generation 1)], which were propagated for several generations by self-pollination. G1 ku70 tert double mutants showed no developmental or reproductive defects.
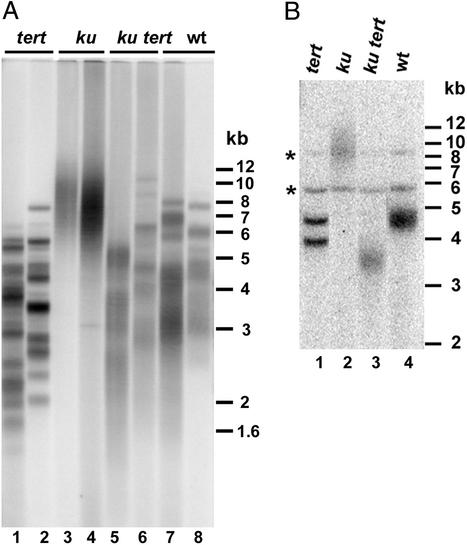
Figure 1.
Analysis of telomere length and heterogeneity in first-generation ku70 tert mutants. (A) DNA from individual G1 plants derived from tert, ku70, ku70 tert, and WT backgrounds was digested with Tru9I restriction endonuclease and subjected to TRF analysis with a radiolabeled (TTTAGGG)4 probe. (B) Telomere-length heterogeneity at 2R. DNA from individual G1 plants was digested with EcoRI and subjected to TRF analysis with a 2R probe (29). The two discrete bands in lane 1 represent signals derived from telomeres at homologous chromosomes. Asterisks show two cross-hybridizing intrachromosomal sequences.
TRF analysis of G1 mutants revealed that the telomere elongation associated with a Ku70 deficiency (Fig. 1A, lanes 3 and 4) was abolished in ku70 tert double mutants (Fig. 1A, lanes 5 and 6). These findings are consistent with recent data from mouse (25) and argue that Ku acts as a negative regulator of telomerase in higher eukaryotes.
TRF analysis performed on DNA from individual plants uncovered remarkably different profiles in the mutants. Because each plant is composed of a population of cells with different proliferation histories, TRF heterogeneity reflects the kinetics of the telomere tract at individual telomeres throughout plant growth and development. The exceptionally sharp bands in tert mutants (Fig. 1A, lanes 1 and 2) correspond to the 20 unique chromosome ends (27). They not only indicate that telomerase is the major source of telomere length heterogeneity in WT, they also indicate that the rate of telomere shortening caused by the end replication problem is slow (27, 29). Although telomerase inactivation suppressed telomere extension in ku70 mutants, TRFs in the double mutants were more heterogeneous than in tert plants (Fig. 1A, lanes 1, 2, 5, and 6) and closely resembled WT (Fig. 1A, lanes 7 and 8).
To further examine telomere length heterogeneity, we analyzed the TRF associated with a single chromosome end by using a probe specific for the right arm of chromosome two (2R). In WT, telomere length heterogeneity for 2R ranges from 500 to 700 bp, but in tert mutants heterogeneity spanned <200 bp (Fig. 1B, lanes 1 and 4; data not shown). Inactivation of Ku in a tert background restored TRF heterogeneity to WT (Fig. 1B, lane 3), arguing that Ku regulates telomere-associated activities besides telomerase.
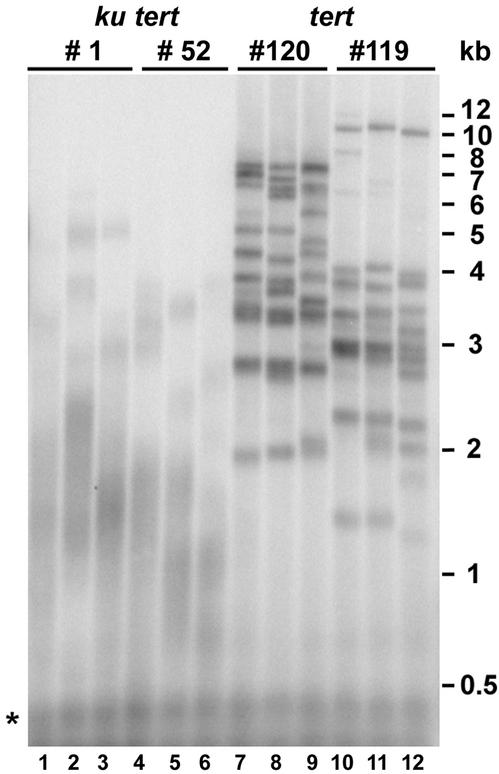
One explanation for increased TRF heterogeneity in ku70 tert mutants is accelerated telomere shortening during plant development. Supporting this idea, the 2R telomere was shorter in G1 ku70 tert mutants than in G1 tert plants (Fig. 1B, lanes 1 and 3). To ascertain whether all telomeres shortened faster in the absence of Ku70, we compared the average telomere length for tert and ku70 tert plants in four independent lines after three generations of a telomerase deficiency (Fig. 2). The average telomere length in WT was 4.87 ± 0.44 kb (n = 6) (data not shown), and the lengths in G3 tert lines 119 and 120 were 3.06 ± 0.18 kb (n = 5) and 3.92 ± 0.15 kb (n = 5), respectively (Fig. 2). By contrast, the average telomere length in G3 ku70 tert double mutants was 1.59 ± 0.21 kb (n = 7) for line 1 and only 1.15 ± 0.14 kb (n = 5) for line 52 (Fig. 2; data not shown). Hence, telomeres shortened approximately two to three times faster in telomerase-deficient cells that lack Ku.
Figure 2.
TRF analysis of individual plants from G3 ku70 tert and tert mutant populations. Three G3 plants in each line were analyzed. Asterisk denotes signals from interstitial telomeric sequences.
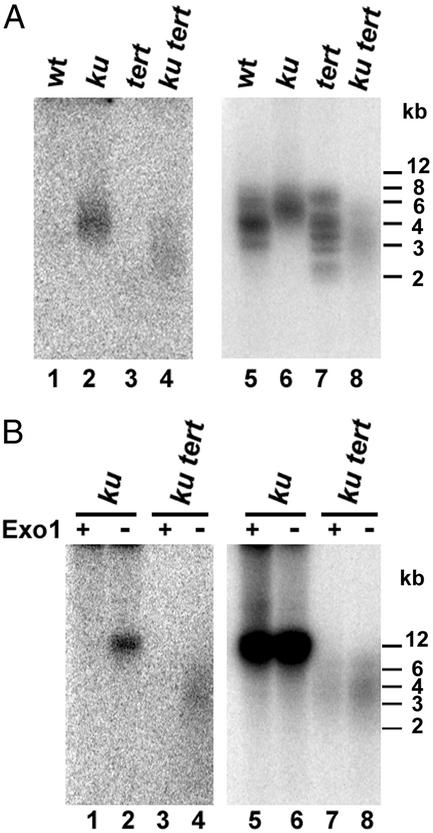
The rapid loss of telomeric DNA in ku70 tert mutants may reflect an inability to properly maintain the telomeric C-rich strand, a defect that could give rise to extended G overhangs. G overhangs on WT Arabidopsis telomeres are ≈20–30 nt (31). To determine whether G overhangs were longer in the mutants, we used in-gel hybridization (30). Under standard 37°C hybridization conditions (30), a signal was detected for WT, tert, ku70, and ku70 tert mutants (data not shown). However, when a more a stringent hybridization protocol was used (see Materials and Methods), G-overhang signals were observed only in ku70 and ku70 tert mutants (Fig. 3A, lanes 2 and 4). G-overhang elongation was not caused by the action of telomerase, as extended G overhangs were present in ku70 tert plants. Instead, the defect seems to be in C-strand maintenance. Furthermore, because the rate of telomere shortening is proportional to the size of G overhangs (32), results from the G-overhang assays are consistent with an accelerated rate of telomere loss in ku70 tert mutants.
Figure 3.
G-overhang analysis. (A) HindIII restriction fragments from WT, G1 ku70, and G2 tert and ku70 tert plants were hybridized with (CCCTAAA)4CCC probe under nondenaturing conditions (Left). After PhosphorImager analysis, the DNA in the gel was denatured by treatment with 0.5 M NaOH/0.15 M NaCl, neutralized in 0.5 M Tris-Cl/0.15 M NaCl solution (pH 7), and rehybridized with the telomeric probe (Right). (B) DNA from G2 ku70 and G2 ku70 tert plants was treated with exonuclease 1 (Exo 1) before restriction digest and then subjected to sequential in-gel hybridization under nondenaturing (Left) and denaturing (Right) conditions. The sensitivity of the signal to exonuclease 1 confirmed that it was derived from single-stranded DNA at the chromosome terminus.
Our data define KU70 as a gene in higher eukaryotes required for C-strand maintenance. Replication of the C strand is thought to occur by the conventional lagging strand machinery via a mechanism physically coupled to G-strand synthesis by telomerase (33–36). Two models can be proposed for the perturbation of telomere architecture in ku70 mutants. First, Ku may control exonuclease activities required for postreplicative telomere processing. In yeast and Tetrahymena, the C strand undergoes additional nucleolytic processing to generate a G overhang on both the lagging and leading strands (37, 38). Recent studies indicate that exonuclease 1 is responsible for creating extended G overhangs in yku70-deficient yeast (16). In addition, DNA-PK, another component of the NHEJ pathway, has been implicated in postreplicative processing of mammalian telomeres (39). Alternatively, Ku may facilitate coupling of C- and G-strand DNA replication by helping to position the primosome at the telomere for synthesis of the terminal Okazaki fragment (15). Telomere structure and length regulation in Arabidopsis ku70 mutants strongly resemble yeast pol1 (polymerase α) mutants, which are characterized by telomerase-dependent telomere lengthening and extended G overhangs (40).
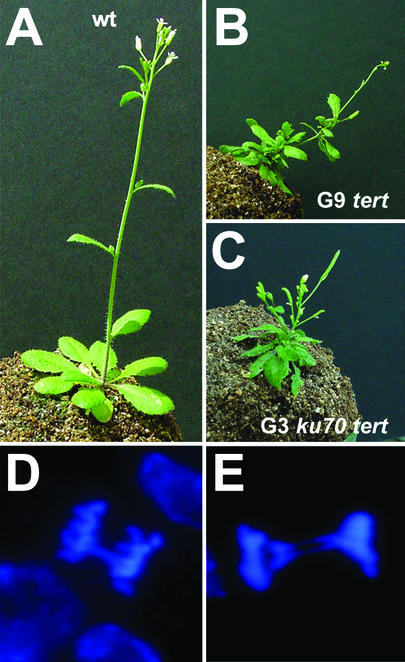
The accelerated loss of telomeric DNA in ku70 tert mutants is predicted to advance the onset of telomere dysfunction and the accompanying proliferation defects. A telomerase deficiency in Arabidopsis results in mild growth and developmental defects (type I phenotype) beginning in G6 that worsen in successive generations. By G10, all plants in the population arrest at a terminal, infertile vegetative state (terminal phenotype; compare Fig. 4 A and B) (29). In the population of G3 ku70 tert mutants from line 52, 19 of 42 plants displayed the type I phenotype (data not shown) and 8 of 42 of the terminal phenotype (Fig. 4C). In addition, ku70 tert mutants in line 1 began to exhibit proliferation defects in G4, consistent with the slightly longer telomeres in this line (Fig. 2; data not shown). In contrast, all of the tert and ku single mutants appeared WT through G4 (data not shown). These data indicate that Ku and telomerase act synergistically in Arabidopsis to maintain telomere tracts.
Figure 4.
Developmental arrest and chromosome fusions in third-generation ku70 tert mutants. (A–C) Developmental defects associated with telomere dysfunction. The WT control is shown in A. A terminal plant displaying severe growth and developmental defects from line 52 in G3 (C) exhibits a phenotype indistinguishable from a G9 tert mutant (line 69) (B) (29). (D and E) Cytogenetic analysis ku70 tert double mutants. A high incidence of chromosome fusions was observed in plants exhibiting the terminal phenotype from G3 line 52 mutants. Examples of anaphases with one (D) or two (E) bridges are shown.
In fission yeast chromosome fusions in taz1 mutants depend on the NHEJ pathway (41), although chromosome circularization occurs in telomerase-negative cells in the absence of Lig4/Ku70 (17). In contrast, Ku is required for end-to-end fusions in mammalian cells with critically shortened telomeres (25). To uncover the basis for the cell-proliferation defect in Arabidopsis, cytogenetic analysis was performed. As expected, no cytogenetic abnormalities were detected in the third generation of ku70 or tert single mutants (Table 1). However, chromosome bridges were observed in 36.4% of the anaphases examined from G3 ku70 tert mutants from line 52 that displayed the terminal phenotype (Fig. 4 D and E; Table 1). A similar frequency of anaphase bridges (44%) was associated with terminal plants from late-generation tert mutants (29). These chromosomal defects do not arise from a Ku70 deficiency in a telomerase-negative background, as only a single fusion event was detected among 262 anaphases from G2 ku70 tert mutants (Table 1). Instead, the onset of anaphase bridges correlates with continued telomere shortening. Hence, critically shortened telomeres seem to trigger a Ku-independent mechanism of chromosome fusion. Sequence analysis will be necessary to determine whether the fusion junctions in telomerase-deficient plants differ in the absence of Ku.
Table 1.
Frequency of chromosome fusions in ku70 tert mutants
| Genotype | Phenotype* | Anaphases with bridges, % | Bridges per anaphase | No. of anaphases scored |
|---|---|---|---|---|
| WT | WT | 0 | 0 | 257 |
| G3ku70 | WT-like | 0 | 0 | 241 |
| G3tert | WT-like | 0 | 0 | 315 |
| G2ku70 tert (52) | WT-like | 0.4 | 0.004 | 262 |
| G3ku70 tert (52) | I | 13.8 | 0.209 | 363 |
| G3ku70 tert (52) | T | 36.4 | 0.586 | 198 |
WT-like plants exhibit no obvious defects in growth or development. Phenotype I is characterized by mild defects in leaf morphology and reduced fertility. Plants displaying the terminal phenotype (T) display severe defects in leaf development and stunted growth, and are unable to produce fertile flowers (29).
Whereas many of the structural components of the telomere complex are conserved, Ku is an example of an essential chromosome-capping protein whose precise function varies among different kingdoms. Even among higher eukaryotes, the role of Ku (and presumably NHEJ) in facilitating end-to-end chromosome fusions is puzzling. NHEJ is required for fusing dysfunctional mammalian telomeres (25, 26), but this does not seem to be the case for Arabidopsis (this study). Clues to the function of Ku may come from understanding its role in maintaining proper telomere architecture. Intriguingly, Ku acts as a positive regulator of telomerase in yeast (42, 43), but in plants and mammals it behaves as a negative regulator (this study) (25). The degree of telomerase deregulation in higher eukaryotes varies markedly, however. In mammals, telomere length increases only slightly in a Ku-deficient background (24, 25), but in Arabidopsis telomeres expand to more than twice their normal size (9, 11). It is possible that telomerase activity levels are more limiting in mammals than in plants (44). Alternatively, telomere elongation in Arabidopsis may reflect greater access of telomerase afforded by the extended G overhangs generated in Ku-deficient mutants. Consistent with this model, extension of G overhangs is not observed in the Ku-deficient mouse (24, 25). The current data illustrate the importance of gathering information from diverse model systems to elucidate complex molecular interactions.
Acknowledgments
We thank Naduparambil Jacob and Chao Wei for helpful advice on in-gel hybridization and Titia de Lange for communicating results before publication. We are also indebted to Carolyn Price, Tom McKnight, and Jeff Kapler for insightful comments on the manuscript. This work was supported by National Science Foundation Grant MCB9982499 and National Institutes of Health Grant GM65383 (to D.E.S.).
Abbreviations
- NHEJ
nonhomologous end-joining
- TRF
terminal restriction fragment
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Harrington L, Robinson M O. Oncogene. 2002;21:592–597. doi: 10.1038/sj.onc.1205084. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E H. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 3.Cervantes R B, Lundblad V. Curr Opin Cell Biol. 2002;14:351–356. doi: 10.1016/s0955-0674(02)00325-3. [DOI] [PubMed] [Google Scholar]
- 4.McKnight T D, Riha K, Shippen D E. Plant Mol Biol. 2002;48:331–337. doi: 10.1023/a:1014091032750. [DOI] [PubMed] [Google Scholar]
- 5.Critchlow S E, Jackson S P. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 6.Featherstone C, Jackson S P. Mutat Res. 1999;434:3–15. doi: 10.1016/s0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- 7.Mages G J, Feldmann H M, Winnacker E L. J Biol Chem. 1996;271:7910–7915. doi: 10.1074/jbc.271.14.7910. [DOI] [PubMed] [Google Scholar]
- 8.Manolis K G, Nimmo E R, Hartsuiker E, Carr A M, Jeggo P A, Allshire R C. EMBO J. 2001;20:210–221. doi: 10.1093/emboj/20.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riha K, Watson J M, Parkey J, Shippen D E. EMBO J. 2002;21:2819–2826. doi: 10.1093/emboj/21.11.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.West C E, Waterworth W M, Story G W, Sunderland P A, Jiang Q, Bray C M. Plant J. 2002;31:517–528. doi: 10.1046/j.1365-313x.2002.01370.x. [DOI] [PubMed] [Google Scholar]
- 11.Bundock P, van Attikum H, Hooykaas P. Nucleic Acids Res. 2002;30:3395–3400. doi: 10.1093/nar/gkf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gravel S, Larrivee M, Labrecque P, Wellinger R J. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 13.Hsu H L, Gilley D, Blackburn E H, Chen D J. Proc Natl Acad Sci USA. 1999;96:12454–12458. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teo S H, Jackson S P. EMBO Rep. 2001;2:197–202. doi: 10.1093/embo-reports/kve038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gravel S, Wellinger R J. Mol Cell Biol. 2002;22:2182–2193. doi: 10.1128/MCB.22.7.2182-2193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maringele L, Lydall D. Genes Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumann P, Cech T R. Mol Biol Cell. 2000;11:3265–3275. doi: 10.1091/mbc.11.10.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conway C, McCulloch R, Ginger M L, Robinson N P, Browitt A, Barry J D. J Biol Chem. 2002;277:21269–21277. doi: 10.1074/jbc.M200550200. [DOI] [PubMed] [Google Scholar]
- 19.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig M C, Li G C. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 20.Vogel H, Lim D S, Karsenty G, Finegold M, Hasty P. Proc Natl Acad Sci USA. 1999;96:10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu H L, Gilley D, Galande S A, Hande M P, Allen B, Kim S H, Li G C, Campisi J, Kohwi-Shigematsu T, Chen D J. Genes Dev. 2000;14:2807–2812. doi: 10.1101/gad.844000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey S M, Meyne J, Chen D J, Kurimasa A, Li G C, Lehnert B E, Goodwin E H. Proc Natl Acad Sci USA. 1999;96:14899–14904. doi: 10.1073/pnas.96.26.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.d'Adda di Fagagna F, Hande M P, Tong W M, Roth D, Lansdorp P M, Wang Z Q, Jackson S P. Curr Biol. 2001;11:1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- 24.Samper E, Goytisolo F A, Slijepcevic P, van Buul P P, Blasco M A. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Espejel S, Franco S, Rodriguez-Perales S, Bouffler S D, Cigudosa J C, Blasco M A. EMBO J. 2002;21:2207–2219. doi: 10.1093/emboj/21.9.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. Curr Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- 27.Fitzgerald M S, Riha K, Gao F, Ren S, McKnight T D, Shippen D E. Proc Natl Acad Sci USA. 1999;96:14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oexle K. J Theor Biol. 1998;190:369–377. doi: 10.1006/jtbi.1997.0559. [DOI] [PubMed] [Google Scholar]
- 29.Riha K, McKnight T D, Griffing L R, Shippen D E. Science. 2001;291:1797–1800. doi: 10.1126/science.1057110. [DOI] [PubMed] [Google Scholar]
- 30.Dionne I, Wellinger R J. Proc Natl Acad Sci USA. 1996;93:13902–13907. doi: 10.1073/pnas.93.24.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riha K, McKnight T D, Fajkus J, Vyskot B, Shippen D E. Plant J. 2000;23:633–641. doi: 10.1046/j.1365-313x.2000.00831.x. [DOI] [PubMed] [Google Scholar]
- 32.Huffman K E, Levene S D, Tesmer V M, Shay J W, Wright W E. J Biol Chem. 2000;275:19719–19722. doi: 10.1074/jbc.M002843200. [DOI] [PubMed] [Google Scholar]
- 33.Fan X, Price C M. Mol Biol Cell. 1997;8:2145–2155. doi: 10.1091/mbc.8.11.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diede S J, Gottschling D E. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- 35.Qi H, Zakian V A. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 36.Ray S, Karamysheva Z, Wang L, Shippen D E, Price C M. Mol Cell Biol. 2002;22:5859–5868. doi: 10.1128/MCB.22.16.5859-5868.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wellinger R J, Ethier K, Labrecque P, Zakian V A. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- 38.Jacob N K, Skopp R, Price C M. EMBO J. 2001;20:4299–4308. doi: 10.1093/emboj/20.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey S M, Cornforth M N, Kurimasa A, Chen D J, Goodwin E H. Science. 2001;293:2462–2465. doi: 10.1126/science.1062560. [DOI] [PubMed] [Google Scholar]
- 40.Adams Martin A, Dionne I, Wellinger R J, Holm C. Mol Cell Biol. 2000;20:786–796. doi: 10.1128/mcb.20.3.786-796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferreira M G, Cooper J P. Mol Cell. 2001;7:55–63. doi: 10.1016/s1097-2765(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 42.Peterson S E, Stellwagen A E, Diede S J, Singer M S, Haimberger Z W, Johnson C O, Tzoneva M, Gottschling D E. Nat Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- 43.Grandin N, Damon C, Charbonneau M. Mol Cell Biol. 2000;20:8397–8408. doi: 10.1128/mcb.20.22.8397-8408.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Snow B E, Hande M P, Yeung D, Erdmann N J, Wakeham A, Itie A, Siderovski D P, Lansdorp P M, Robinson M O, Harrington L. Curr Biol. 2000;10:1459–1462. doi: 10.1016/s0960-9822(00)00805-8. [DOI] [PubMed] [Google Scholar]