Abstract
Epidermal growth factor receptor (EGFR) has attracted considerable attention as a target for cancer therapy. Wild-type (wt)EGFR is amplified/overexpressed in a number of tumor types, and several mutant forms of the coding gene have been found, with ΔEGFR, a deletion mutation lacking exons 2–7 of the external domain, being the most common and particularly associated with glioblastoma. We generated monoclonal antibodies (mAbs) against NR6ΔEGFR (mouse fibroblast line NR6 transfected with ΔEGFR). mAb 806 with selective reactivity for NR6ΔEGFR in mixed hemadsorption assays, fluorescence-activated cell sorting, Western blot, and immunohistochemistry was analyzed in detail and compared with mAbs 528 (anti-wtEGFR) and DH8.3 (anti-ΔEGFR). In xenograft tumors and molecularly pretyped glioblastomas, the reactivity pattern was as follows: 528 reactive with amplified and nonamplified wtEGFR; DH8.3 reactive with ΔEGFR; and 806 reactive with amplified/overexpressed wtEGFR (with or without ΔEGFR). In normal tissues, 528 but not DH8.3 or 806 was widely reactive with many organs, e.g., liver expressing high EGFR levels. In glioblastoma and non-CNS tumor panels, 806 was reactive with a high proportion of glioblastomas and a substantial number of epithelial cancers of lung and of head and neck. DH8.3 reactivity was restricted to ΔEGFR-positive glioblastoma. Thus, 806 represents a category of mAbs that recognizes tumors with EGFR amplification/overexpression but not normal tissues or tumors with normal EGFR levels. Our study also indicates that ΔEGFR is restricted to glioblastoma, in contrast to other reports that this mutation is found in tumors outside the brain.
Growth factor receptors are promising targets for antibody-based cancer therapies. Because of their cell-surface location, they are readily accessible, and therapeutic antibodies can exert their inhibitory effects by either interfering with cellular signaling or targeting toxic molecules or biological effectors to the tumor site (1).
The epidermal growth factor receptor (EGFR) is expressed in normal tissues and neoplastic lesions of most organs, and its expression level has been associated with biological characteristics of tumors (2). Elevated levels of EGFR have been demonstrated in many different types of cancer including glioblastoma, and EGFR overexpression seems to be associated with poor prognosis in several neoplasms (3). EGFR overexpression is often associated with gene amplification (4–8). In glioblastoma, EGFR amplification has been shown to be accompanied by gene rearrangement (9–11), frequently with deletions in the coding region. Several mutant forms have been found (12, 13), and among these the most common mutation is the Δ2-7 deletion (ΔEGFR), which lacks exons 2–7 of the external EGFR domain, resulting in the loss of an 801-bp fragment of the wild-type (wt) gene (14). Several studies have indicated that the presence of ΔEGFR enhances the tumorigenic behavior of cancer cells (15–17). ΔEGFR has only been found in neoplastic lesions and not in any normal tissue. Its tumor-restricted expression and cell-surface location make ΔEGFR a potentially ideal target for immunotherapeutic strategies.
Antibody targeting of EGFR is currently being pursued as a promising approach to treating cancers at various sites (18). Although a wide variety of antibodies to the wtEGFR are available (19, 20), only a few reagents to its mutant form have been generated (11, 21–23). Some of the currently available anti-ΔEGFR reagents are polyclonal antibodies (11, 21, 22), but these are not useful for therapeutic applications. Also, most ΔEGFR reagents have been tested on only a limited number of tissues (22, 24–27), and no comprehensive analysis of ΔEGFR expression in normal and tumor tissue has been performed.
The present study describes the generation and characterization of 806, a murine antibody raised against cells transfected with ΔEGFR. The reactivity of 806 was compared with 528, an antibody to wtEGFR (28), and DH8.3, an antibody raised against ΔEGFR (21). 806 has unique features that differentiate it from other EGFR-related mAbs, namely its ability to distinguish cells with an amplified/overexpressed EGFR phenotype from cells having wt levels of EGFR expression.
Materials and Methods
Cell Lines.
For immunization and specificity analyses, a panel of cell lines, parental or transfected with either the human wtEGFR gene or the ΔEGFR gene carrying the Δ2-7 deletion mutation, were used: murine fibroblast cell line NR6, NR6ΔEGFR, NR6wtEGFR, human glioblastoma cell line U87MG (expressing low levels of endogenous wtEGFR), U87MGwtEGFR, U87MGΔEGFR, and human squamous cell carcinoma cell line A431 (expressing high levels of wtEGFR) (29). Cell lines and transfections have been described (15).
Generation of mAbs.
The murine fibroblast line NR6ΔEGFR was used as immunogen. Mouse hybridomas were generated by immunizing BALB/c mice five times s.c. at 2- to 3-week intervals with 5 × 105 to 2 × 106 cells in Freund's adjuvant. Complete Freund's adjuvant (Difco) was used for the first injection, and incomplete Freund's adjuvant (Difco) was used for subsequent injections. Spleen cells from immunized mice were fused with mouse myeloma cell line SP2/0.
Supernatants from newly generated clones were screened in mixed hemadsorption assays for reactivity with cell lines NR6, NR6wtEGFR, and NR6ΔEGFR. Supernatants with specificity for NR6ΔEGFR were then tested on the human glioblastoma cell lines U87MG, U87wtEGFR, and U87ΔEGFR. Supernatants continuing to show selective reactivity with ΔEGFR-transfected cells were tested by fluorescence-activated cell sorting, Western blots, and immunohistochemistry.
Two additional mAbs were included in our study: 528, with specificity for wtEGFR (28), and DH8.3, generated against a synthetic peptide spanning the junctional sequence of the Δ2-7 EGFR deletion mutation (21).
Mixed Hemadsorption Assay.
The hemadsorption assay, which detects surface-bound IgG by adherence of rabbit anti-mouse IgG-coated human red blood cells (blood group O) to target cells, was performed as described (30).
Western Blots.
Triton X-100 (0.3% in PBS, pH 7.5) lysates of cultured cells were resolved by SDS/PAGE 6% polyacrylamide Tris-glycine precast gels (NOVEX, San Diego) under reducing (5% β-mercaptoethanol) conditions. Proteins were blotted to poly(vinylidene difluoride) (Immobilon-P, Millipore) and incubated with hybridoma supernatant diluted 1:10 or protein A affinity-purified mAbs (5 μg/ml). Specific binding was detected by alkaline phosphatase-conjugated species-specific secondary antibody and visualized by using chemiluminescent detection (Tropix, Bedford, MA). Blocking and washing steps were carried out per manufacturer instructions.
Molecular Typing.
The gene analysis was done as described (6, 9). The presence of EGFR gene amplification and gene rearrangement was determined by PCR and hybridization of a cDNA probe to Southern blots (6). Amplification was quantitated by comparison with patient blood DNA. The consequences of rearrangements for the EGFR transcript were determined by RT-PCR.
Immunohistochemistry.
All analyses were performed on frozen tissues. Tissues were embedded in OCT compound (TissueTek, Torrance, CA) and snap-frozen in isopentane precooled in dry ice. Five-micrometer sections were applied to slides (Superfrost, Fisher) and fixed for 10 min in cold acetone (4°C). Primary antibodies were detected with a biotinylated horse anti-mouse antibody (Vector Laboratories) followed by an avidin–biotin-complex reaction (ABC Elite, Vector Laboratories). Diaminobenzidine tetrahydrochloride (BioGenex Laboratories, San Ramon, CA) was used as chromogen. As an alternative for the avidin–biotin-complex method, streptavidin–alkaline–phosphatase (SA, Roche Molecular Biochemicals) followed by new fuchsin chromogen (BioGenex Laboratories) was used. With the xenografted tumors, a preincubation with unlabelled secondary anti-mouse reagent was done in preliminary assays to block the reactivity of endogenous mouse Igs. However, this preincubation did not improve the staining, because endogenous immunoreactivity did not interfere with the interpretation of the immunohistochemical staining. The blocking step therefore was finally omitted.
The number of stained cells was estimated and graded as follows: “focal,” <5%; “+,” >5–25%; “++,” >25–50%; “+++,” >50–75%; and “++++,” >75%.
Tissues.
The reactivity of the mAbs in immunohistochemistry was assessed first on a small panel of xenografted tumors. U87MG, U87MGΔEGFR, and A431 cells were inoculated s.c. in the right thigh of nu/nu mice (5 × 105 to 2 × 106 cells per mouse) and grown until a tumor diameter of ≈10 mm was reached.
Human tissues were provided by the tissue-procurement service of Memorial Sloan–Kettering Cancer Center and the Institute of Pathology of the Karolinska Hospital (Stockholm). For the initial immunohistochemical specificity analysis, a series of 17 glioblastomas was used. These tissues were preselected on the basis of a molecular pretyping for the presence of wtEGFR, ΔEGFR, and EGFR amplification. In addition, three large frozen tissue panels were analyzed for reactivity with 806, 527, and DH8.3. These panels were (i) a set of normal tissues, (ii) a set of extracranial malignant neoplasms, and (iii) a series of 46 unselected glioblastomas. The glioblastomas were subsequently analyzed (as described above) for the presence of wtEGFR, EGFR mutations, and EGFR amplification.
Results
Selection of EGFR mAbs.
Hybridomas were established from mice immunized with NR6ΔEGFR, and three clones, 124 (IgG2a), 806 (IgG2b), and 1133 (IgG2a), were initially selected for further characterization based on high titer of supernatants with NR6ΔEGFR and background reactivity with parental NR6 and NR6wtEGFR cells in the hemadsorption assay (Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). These antibodies showed no reactivity (undiluted supernatant ≤10%) with the parental human glioblastoma cell line U87MG or U87MGwtEGFR but were strongly reactive with U87MGΔEGFR and to a lesser degree with A431. In fluorescence-activated cell-sorter analysis (data not included), 806 was unreactive with U87MG and intensively stained U87MGΔEGFR but showed some low-level reactivity with U87MGwtEGFR. In Western blot assays, 124, 806, and 1133 were tested with detergent lysates from NR6, NR6ΔEGFR, NR6wtEGFR, U87MG, U87MGΔEGFR, U87MGwtEGFR, and A431. All three mAbs showed a similar reactivity pattern, detecting both wtEGFR (170 or 160 kDa in case of A431 cells) and ΔEGFR (140 kDa), whereas DH8.3 (reference antibody for ΔEGFR) reacted only with the characteristic 140-kDa ΔEGFR band (Fig. 6, which is published as supporting information on the PNAS web site).
Tumor Xenografts.
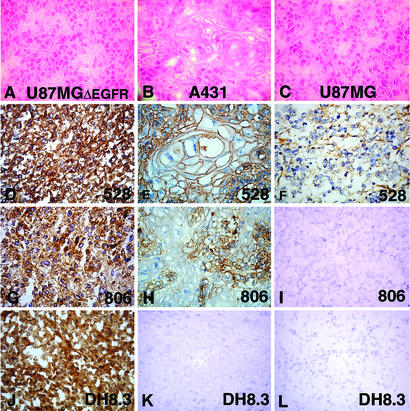
Table 1 and Fig. 1 show the results of immunohistochemical analysis with 124, 806, and 1133, as well as 528 and DH8.3, on xenografts of U87MG, U87MGΔEGFR, and A431. All mAbs showed strong staining of U87MGΔEGFR xenografts. Only 528 stained the parental U87MG xenograft. In xenografts of squamous cell carcinoma A431, 528 showed strong homogeneous staining, whereas 124, 806, and 1133 reacted primarily with basally located cells and did not react with the upper keratinizing cell layers. DH8.3 did not stain the A431 xenograft. Because of low background reactivity, 806 was selected for all subsequent analysis.
Table 1.
Immunohistochemical analysis of U87MG, U87MGΔEGFR, and A431 xenografts
| mAb | Xenograft
|
||
|---|---|---|---|
| U87MG | U87MGΔEGFR | A431 | |
| 528 | Foc./+ | ++++ | ++++ |
| 124 | − | ++++ | ++ (predominantly basal cells) |
| 806 | − | ++++ | ++ (predominantly basal cells) |
| 1133 | − | ++++ | ++ (predominantly basal cells) |
| DH8.3 | − | ++++ | − |
Minor stromal staining is due to detection of endogenous mouse antibodies. Foc., focal.
Figure 1.
Hematoxylin/eosin (A–C) and immunohistochemical (D–O) (avidin–biotin-complex technique, diaminobenzidine tetrahydrochloride chromogen) staining of tumor xenografts U87MGΔEGFR (A, D, G, and J), squamous cell carcinoma cell line A431 (B, E, H, and K), and parental (nontransfected) U87MG cell line (C, F, I, and L) with 528 (D–F), 806 (G–I), DH8.3 (J–L). (D, G, and J) Intense staining of U87MGΔEGFR with all three antibodies. (E) Intense staining of A431 with 528. (H) Less intense staining of A431 with 806. (K) Negative staining of A431 with DH8.3. (F) Weak focal staining of parental U87MG with 528. (I and L) No staining of parental U87MG with 806 or DH8.3.
Pretyped Glioblastomas.
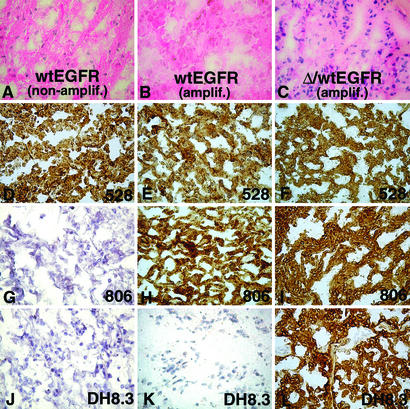
The immunohistochemical staining of glioblastomas pretyped for wtEGFR, ΔEGFR, and EGFR amplification is shown in Table 2 and Fig. 2. These tissues showed more or less extensive cracking and spacing artifacts. This is a common phenomenon with frozen glioblastoma specimens, caused by the perifocal edema typically present in this type of tumor. Whereas 528 showed intense reactivity in most tumors, DH8.3 immunostaining was present solely in tumors expressing ΔEGFR. 806 stained tumors with ΔEGFR but also tumors with EGFR amplification without ΔEGFR. No or weak 806 reactivity was present in tumors without EGFR amplification.
Table 2.
Immunoreactivity of mAbs 528, DH8.3, and 806 with glioblastomas pretyped for the presence of wtEGFR, ΔEGFR, and EGFR amplification
| No. | Frozen no. | EGFR | Amplification/ Southern blot | 528 | DH8.3 | 806 |
|---|---|---|---|---|---|---|
| 1 | 96–699 | wtEGFR | No amplification | ++ | − | − |
| 2 | 96–700 | wtEGFR | No amplification | +++ | − | + |
| 3 | 96–701 | wtEGFR | No amplification | ++ | − | − |
| 4 | 96–702 | wtEGFR | No amplification | ++ | + | − |
| 5 | 96–703 | wtEGFR | No amplification | + | − | − |
| 6 | 96–704 | wtEGFR | No amplification | ++ | + | − |
| 7 | 96–705 | wtEGFR | No amplification | ++ | − | + |
| 8 | 96–706 | wtEGFR | Amplification | +++ | − | ++ |
| 9 | 96–707 | wtEGFR | Amplification | ++ | − | +++ |
| 10 | 96–708 | wtEGFR | Amplification | +++ | − | +++ |
| 11 | 96–709 | wtEGFR | Amplification | +++ | − | +++ |
| 12 | 96–710 | wtEGFR | Amplification | +++ | − | ++ |
| 13 | 96–711 | wtEGFR + ΔEGFR | Amplification | +++ | +++ | +++ |
| 14 | 96–712 | wtEGFR + ΔEGFR | Amplification | +++ | +++ | +++ |
| 15 | 96–713 | wtEGFR + ΔEGFR | Amplification | +++ | +++ | +++ |
| 16 | 96–714 | wtEGFR + ΔEGFR | Amplification | ++ | + | ++ |
| 17 | 96–715 | wtEGFR + ΔEGFR | Amplification | +++ | +++ | +++ |
For the analysis in this table, a three-tier grading system (+–+++) was used to indicate the number of stained cells.
Figure 2.
Hematoxylin/eosin (A–C) and immunohistochemical (D–L) (avidin–biotin-complex technique, diaminobenzidine tetrahydrochloride chromogen) staining of three glioblastoma specimens pretyped for presence of wtEGFR (A, D, G, and J), amplified EGFR (B, E, H, and K), or amplified EGFR and ΔEGFR (C, F, I, and L) with 528 (D–F), 806 (G–I), or DH8.3 (J–L). 528 staining of the three glioblastomas (D–F), 806 staining of glioblastoma having EGFR amplification without ΔEGFR (H) or with ΔEGFR (I), and DH8.3 staining of glioblastoma with amplified EGFR and ΔEGFR (L) are shown.
Normal Tissue Panel.
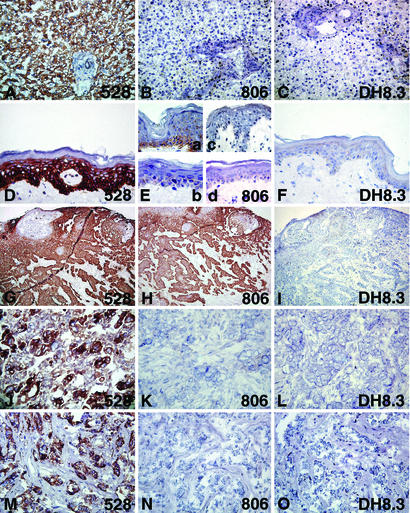
The results of immunohistochemical staining of normal tissues are shown in Fig. 3 and Table 6 (which is published as supporting information on the PNAS web site). 528 showed immunoreactivity in all normal tissues tested. The 528-reactivity pattern is not described in detail here, because the distribution of EGFR in normal tissues has been reported in detail (31, 32). No normal tissue showed significant 806 staining. Some inconsistent focal staining was present in basal cells of one of five samples of normal skin and in the squamous epithelium in one of five samples of tonsil mucosa. This staining was not present in the other four samples and was variable within the single positive tissues. In placenta, occasional staining of the trophoblast epithelium was observed. This low-level reactivity of 806 with skin and placenta might be expected, because both keratinocytes and trophoblastic epithelia are known to express high levels of EGFR (33). DH8.3 showed virtually no staining of normal tissues with the exception of faint focal stromal staining in several tissues, which was considered nonspecific. Liver, an organ with high expression of EGFR, was negative for 806 and DH8.3.
Figure 3.
Immunohistochemical staining of normal tissues (A–F) and non-CNS tumors (G–O) with 528 (A, D, G, J, and M), 806 (B, E, H, K, and N), or DH8.3 (C, F, I, L, and O). Intense immunoreactivity of 528 with normal liver (A) and skin (D) is shown. (B) No 806 staining of liver. (E) Patchy focal staining of basal cells in epidermis of one skin (specimens from four individuals shown). No DH8.3 staining of liver (C) or skin (F). Squamous cell carcinoma of the head and neck (G–I) with intense immunoreactivity of 528 (G) and with 806 (H) but no staining with DH8.3 (I); serous-papillary ovarian carcinoma (J–L), staining with 528 (J) but not with 806 (K) or DH8.3 (L); invasive ductal carcinoma of the breast (M–O) with intense 528 staining (M) but no staining with 806 (N) or DH8.3 (O).
Tumor Panel.
Table 3 and Fig. 3 G–O show the results of immunostaining of the tumor panel. 528 gave intense and often homogeneous staining in a wide variety of different tumor types. Immunoreactivity was present in the tumoral as well as the stromal components of epithelial neoplasms and sarcomas. Only melanomas and seminomas revealed a generally weak staining pattern. The reactivity pattern of 528 in tumors is not specified further, because it has been analyzed in several studies (34–37).
Table 3.
Immunohistochemical analysis of human tumors with 528, DH8.3, and 806
| Tumor | 528 | DH8.3 | 806 |
|---|---|---|---|
| Melanoma metastases | 7/10 | 0/10 | 0/10 |
| Urinary bladder carcinoma | 15/16 | 0/16 | 6/16 |
| Breast carcinoma | 8/10 | 0/10 | 0/10 |
| Metastatic head and neck cancer (sqcc) | 10/10 | 0/10 | 10/10 |
| Esophagus cancer (sqcc) | 9/10 | 0/10 | 7/10 |
| Non-small-cell lung cancer | 63/65 | 0/65 | 23/65 |
| Sqcc | 25/26 | 17/26 | |
| Adenocarcinoma | 31/32 | 4/32 | |
| Large cell carcinoma | 7/7 | 2/7 | |
| Leiomyosarcoma | 5/5 | 0/5 | 0/5 |
| Liposarcoma | 4/5 | 0/5 | 0/5 |
| Synovial sarcoma | 5/5 | 0/5 | 0/5 |
| Malignant fibrous histiocytoma | 5/5 | 0/5 | 1/5 |
| Colonic carcinoma | 10/10 | 0/10 | 0/10 |
| Seminoma | 7/10 | 0/10 | 0/10 |
| Ovarian carcinoma (serous-papillary, endometroid) | 12/15 | 0/15 | 0/15 |
sqcc, squamous cell carcinoma.
806 showed positive staining in carcinomas of head and neck, esophagus, lung, and urinary bladder. These 806-positive tumors were mostly squamous cell carcinomas or tumors with varying degrees of squamous cell differentiation. In non-small-cell lung carcinomas, 17 of 26 squamous cell carcinomas were 806-positive, whereas large-cell carcinomas and adenocarcinomas were less immunoreactive. Of the 17 positive cases, 11 showed homogeneous 806 staining, i.e., immunoreactivity in >50% of the tumor cells. In metastatic head and neck squamous cell carcinoma, 10 of 10 cases were positive for 806, with 4 cases showing homogeneous staining. In esophageal squamous cell carcinoma, 7 of 10 were positive, and 5 of the 7 positive cases showed homogeneous staining. In tumors of the urinary bladder, those with squamous differentiation and occasionally those with urothelial differentiation were reactive with 806. Little 806 staining was observed in other tumor types.
mAb DH8.3 showed no immunoreactivity in this tumor panel, indicating no or little expression of ΔEGFR in extracranial neoplasms. For instance, cancers of breast and ovary, tumors reported to have ΔEGFR (24, 26), were negative. There was some inconsistent focal endothelial staining of blood vessels and a varying weak diffuse staining of connective tissue, which was observed previously in normal tissues. The latter staining was restricted to small areas and also strongly depended on the concentration of antibody and was finally regarded as nonspecific background reactivity. Importantly, no convincing reactivity in tumor cells was seen.
Unselected Glioblastomas.
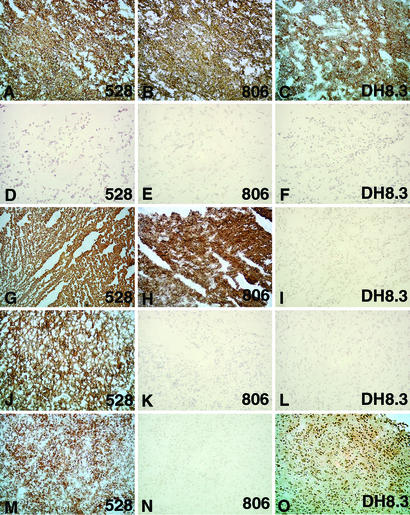
Tables 4 and 5 summarize the results of immunostaining of 46 unselected glioblastomas. 528 showed strong reactivity with all but two tumors, and in most cases there was a predominantly homogeneous staining (Fig. 4). 806 was reactive with 27 of 46 (58.7%) glioblastomas, with 22 of the positive cases showing staining of >50% of the tumor cells (+++–++++ corresponding to our grading system). DH8.3 stained 15 of 46 (32.6%) glioblastomas, 9 of which showed immunostaining in >50% (+++–++++) of the tumor cells (Fig. 4).
Table 4.
Immunohistochemical analysis of 46 unselected glioblastomas with 528, 806, and DH8.3 and correlation with EGFR amplification and ΔEGFR
| No. | 528 | DH8.3 | 806 | EGFR amplification* | EGFR status |
|---|---|---|---|---|---|
| 1 | ++++ | ++ | ++++ | A | ΔEGFR |
| 2 | ++++ | ++++ | ++++ | N | wt |
| 3 | ++++ | Neg. | ++++ | N | A2 |
| 4 | ++++ | Neg. | ++++ | N | wt |
| 5 | ++++ | ++++ | ++++ | N | ΔEGFR |
| 6 | ++++ | Neg. | ++++ | A | wt |
| 7 | ++++ | ++++ | ++++ | N | ΔEGFR |
| 8 | ++++ | ++++ | ++++ | A | ΔEGFR |
| 9 | ++++ | Neg. | ++++ | A | wt |
| 10 | ++++ | Neg. | Neg. | N | wt |
| 11 | ++ | ++ | ++ | A | ΔEGFR |
| 12 | ++++ | Neg. | ++ | A | ΔEGFR |
| 13 | ++++ | Neg. | ++++ | N | wt |
| 14 | ++ | Neg. | Neg. | ND | ND |
| 15 | ++ | Neg. | ++ | N | wt |
| 16 | + | Neg. | Neg. | N | ND |
| 17 | ++++ | Neg. | Neg. | N | wt |
| 18 | ++++ | Neg. | ++++ | A | ΔEGFR |
| 19 | ++++ | Neg. | ++++ | N | wt |
| 20 | ++++ | Neg. | Neg. | N | wt |
| 21 | ++++ | Neg. | ++++ | N | wt |
| 22 | +++ | Neg. | Neg. | N | wt |
| 23 | ++++ | ++ | ++++ | N | ΔEGFR |
| 24 | ++++ | Neg. | ++++ | A | wt |
| 25 | ++++ | Neg. | Neg. | N | wt |
| 26 | ++++ | +++ | ++++ | A | ΔEGFR |
| 27 | Neg. | Neg. | Neg. | N | wt |
| 28 | +++ | Neg. | Neg. | N | wt |
| 29 | Neg. | Neg. | Neg. | N | wt |
| 30 | ++++ | Neg. | ++++ | N | wt |
| 31 | ++++ | Neg. | Neg. | N | ND |
| 32 | ++ | ++ | +++ | N | ΔEGFR |
| 33 | +++ | ++++ | ++++ | A | ΔEGFR |
| 34 | ++++ | ++++ | +++ | N | wt |
| 35 | ++++ | ++++ | Neg. | A | ΔEGFR |
| 36 | +++ | +++ | ++ | A | ΔEGFR |
| 37 | ++++ | + | + | A | ΔEGFR |
| 38 | ++++ | Neg. | Neg. | N | wt |
| 39 | ++ | Neg. | Neg. | N | ΔEGFR |
| 40 | ++++ | + | ++++ | A | wt |
| 41 | ++ | Neg. | Neg. | N | wt |
| 42 | ++++ | Neg. | ++++ | A | wt |
| 43 | ++++ | Neg. | Neg. | ND | ND |
| 44 | ++++ | Neg. | Neg. | N | wt |
| 45 | ++++ | Neg. | Neg. | N | wt |
| 46 | ++++ | Neg. | Neg. | N | ND |
N, not amplified; A, amplified; ND, not done.
Table 5.
Summary of Table 4
| EGFR amplification | ΔEGFR | No. of cases | 528- positive | 806- positive | DH8.3- positive |
|---|---|---|---|---|---|
| Not present | Not present | 20 | 18 | 8 | 2 |
| Present | Present | 10 | 10 | 9 | 8 |
| Not present | Present | 5 | 5 | 4 | 4 |
| Present | Not present | 5 | 5 | 5 | 1 |
| ND | ND | 2 | 2 | 0 | 0 |
| Not present | ND | 3 | 3 | 0 | 0 |
| Not present | Other mutation suspected | 1 | 1 | 1 | 0 |
| Total | 46 | 44 | 27 | 15 |
Figure 4.
Immunohistochemical staining of glioblastomas with 528 (A, D, G, J, and M), 806 (B, E, H, K, and N), and DH8.3 (C, F, I, L, and O). (A–C) Case 26, amplified EGFR/ΔEGFR: 528- (A), 806- (B), and DH8.3-positive (C). (D–F) Case 27, no EGFR amplification/no ΔEGFR: 528- (D), 806- (E), and DH8.3-negative (F). (G–I) Case 6, amplified EGFR/no ΔEGFR: 528-positive (G), 806-positive (H), and DH8.3-negative (I). (J–L) Case 44, no EGFR amplification/no ΔEGFR: 528-positive (I), 806-negative (K), and DH8.3-negative (L). (M–O) Case 35, amplified EGFR/ΔEGFR: 528-positive (M), 806-negative (N), and DH8.3-positive (O).
We next compared 806 and DH8.3 reactivity with the presence or absence of EGFR amplification and ΔEGFR. In the 44 cases tested for EGFR amplification, 30 cases cotyped with 806, i.e., 16 806-negative cases had no EGFR amplification, and 14 806-positive cases had EGFR amplification. However, 13 cases with 806 reactivity were negative for EGFR amplification, whereas 1 EGFR-amplified case was 806-negative (case 35).
In the 41 cases typed for ΔEGFR by RT-PCR, 34 cases cotyped with DH8.3: 12 cases were positive in RT-PCR and immunohistochemistry, and 22 cases were negative in both assays. Six cases showed discrepant results. Three of these (cases 2, 34, and 40) were DH8.3-positive/ΔEGFR-negative, and three others (cases 12, 18, and 39) were DH8.3-negative/ΔEGFR-positive. Case 3 (DH8.3-negative) revealed a mutation in the EGFR gene, but this was distinct from the classical Δ2-7 deletion. As expected, 806 stained all DH8.3-positive tumors with the exception of one (case 35) (Fig. 4).
Discussion
Antibody-based immunotherapy of tumors has experienced renewed interest based on the success of targeting the CD20 antigen of B cell lymphomas (38) and two members of the EGFR family, namely EGFR and HER-2neu (39, 40). Several clinical studies using EGFR antibodies have been conducted and reveal promising results, especially when combined with additional treatment modalities such as radiation or chemotherapy (39). However, EGFR is a normal constituent of many cell types, and organs such as liver and skin express high levels of EGFR, factors that complicate clinical use of antibodies to the wt receptor (2, 31, 41). In fact, skin toxicity is commonly seen in patients treated with anti-EGFR antibodies (42). For this reason, antibodies that target mutant EGFR, such as ΔEGFR, or otherwise altered forms of EGFR would be highly desirable.
The present study was prompted by the availability of a panel of mouse (NR6 fibroblast) and human (U87MG glioblastoma) cell lines transfected with wtEGFR or ΔEGFR. The intention was to develop antibodies with specificity for ΔEGFR, using the ΔEGFR-transfected NR6 cell line as immunogen, and screening for antibodies that have selective cell-surface reactivity with ΔEGFR-transfected NR6 and U87MG cells. We considered this an attractive alternative strategy to the one used to develop previous antibodies to ΔEGFR, i.e., immunization with the peptide spanning the EGFR deletion mutation (21, 22). To our surprise, the antibodies we isolated, such as 806, had specificity for cells with amplified EGFR expression regardless of the presence or absence of ΔEGFR and did not react with cells expressing normal levels of EGFR. This reactivity pattern distinguishes 806 from antibodies to wtEGFR such as 225 and 528 (34, 41, 43, 44) and antibodies to ΔEGFR such as DH8.3 (21). In normal tissues, 528 stained a broad range of cell types in virtually every organ in the body, whereas DH8.3 showed no reactivity with any normal cell type. The normal tissue reactivity of 806 was also extremely limited, with the only exception being the occasional staining of basal cells in skin epithelium (a cell type known to have high levels of EGFR expression) in one of five specimens tested and the corresponding cell type in the mucosal epithelium of the tonsil in one of five specimens. Most importantly, 806 showed no reactivity with normal liver.
The reactivity of 528, DH8.3, and 806 with the tumor panel also revealed clear distinctions among these antibodies. As with normal tissues, 528 reacted broadly with different tumor types as well as the normal tissue constituents of tumors. In contrast, DH8.3 reactivity was restricted to a subset of glioblastoma, and in 34 of 41 glioblastomas tested for both DH8.3 and ΔEGFR, there was a clear correlation between immunostaining and presence of the mutation. In contrast to reports that non-small-cell lung cancer (26), breast cancer (24, 27), and ovarian cancer (24) show a high frequency of ΔEGFR as detected by mAbs or polyclonal antibodies, we found no evidence for DH8.3 immunoreactivity with tumors outside the CNS. The serologic reagents used in these other studies have not been subjected to a comprehensive specificity analysis, and the discrepancies between our results and the previous reports may have to do with undefined cross-reactivities. However, a recent study reporting a high frequency of ΔEGFR in invasive breast cancer indicates that the issue of ΔEGFR in non-CNS awaits further study (27).
In relation to DH8.3, 806 reacts with a higher percentage of glioblastomas than DH8.3 (58.7% vs. 32.6%) and also with tumor types known to have EGFR overexpression, e.g. cancers of the head and neck, lung, and bladder. Although 806 reactivity clearly correlated with EGFR amplification in glioblastoma, e.g. 14 of 15 glioblastomas with EGFR amplification were 806-positive, there were 13 glioblastomas that were 806-positive without having amplified EGFR expression. Because overexpression of EGFR in the absence of gene amplification occurs in a variety of tumors, e.g. breast, colorectum, and bladder (2, 3, 6), it is likely that 806-positive/EGFR amplification-negative glioblastomas have an EGFR overexpression phenotype. However, because many tumor types with reported EGFR overexpression, e.g. breast, showed little or no 806 staining, it seems likely that only a subset of EGFR overexpressing tumors, perhaps those exceeding a certain expression level, shows 806 reactivity.
The epitope recognized by 806 is currently unknown. 806 does not react with the junction peptide characteristic of the ΔEGFR deletion mutation, clearly distinguishing 806 from antibodies detecting the ΔEGFR mutation (unpublished data). The preferential reactivity of 806 with cells having high EGFR expression could be explained on the basis of affinity, i.e., 806 has a lower affinity than 528 for wtEGFR. Scatchard analysis of 806 and titration experiments with 806 and 528 do not lend support to this idea (45–47). Our current hypothesis is that the EGFR expressed by cells with amplification or overexpression has an altered configuration or composition, e.g., aberrant glycosylation, and that 806 recognizes this altered state independent of the ΔEGFR mutation. We have found recently that only a fraction of the EGFRs in overexpressing cells seems to have the 806 epitope; only 10% of the EGFRs from the A431 cell lines is immunoprecipitated by 806 (45–47).
In summary, 806 represents a way to target cancer cells with EGFR amplification/overexpression. Because of the higher frequency of glioblastoma with 806 reactivity than with antibodies detecting ΔEGFR, and the substantial number of head and neck, lung, and bladder cancers that show 806 reactivity, therapeutic strategies based on 806-targeting offer attractive opportunities. The antitumor activity of 806 in mouse systems, including a model of human brain cancer (47), provides encouragement for the clinical application of 806.
Supplementary Material
Acknowledgments
We dedicate this article to our coauthor Dr. Elisabeth Stockert, who died shortly before its publication. Her work was essential for the realization of this project.
Abbreviations
- EGFR
epidermal growth factor receptor
- wt
wild type
References
- 1.Mach J P. In: Oxford Textbook of Oncology. Peckham M, Pinedo H M, Veronesi U, editors. Vol. 1. Oxford: Oxford Univ. Press; 1995. pp. 81–103. [Google Scholar]
- 2.Prenzel N, Fischer O M, Streit S, Hart S, Ullrich A. Endocr Relat Cancer. 2001;8:11–31. doi: 10.1677/erc.0.0080011. [DOI] [PubMed] [Google Scholar]
- 3.Nicholson R I, Gee J M, Harper M E. Eur J Cancer. 2001;37, Suppl. 4:S9–S15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 4.Libermann T A, Nusbaum H R, Razon N, Kris R, Lax I, Soreq H, Whittle N, Waterfield M D, Ullrich A, Schlessinger J. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 5.Wong A J, Bigner S H, Bigner D D, Kinzler K W, Hamilton S R, Vogelstein B. Proc Natl Acad Sci USA. 1987;84:6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ekstrand A J, James C D, Cavenee W K, Seliger B, Pettersson R F, Collins V P. Cancer Res. 1991;51:2164–2172. [PubMed] [Google Scholar]
- 7.Collins V P. Semin Cancer Biol. 1993;4:27–32. [PubMed] [Google Scholar]
- 8.Malden L T, Novak U, Kaye A H, Burgess A W. Cancer Res. 1988;48:2711–2714. [PubMed] [Google Scholar]
- 9.Sugawa N, Ekstrand A J, James C D, Collins V P. Proc Natl Acad Sci USA. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamazaki H, Fukui Y, Ueyama Y, Tamaoki N, Kawamoto T, Taniguchi S, Shibuya M. Mol Cell Biol. 1988;8:1816–1820. doi: 10.1128/mcb.8.4.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humphrey P A, Wong A J, Vogelstein B, Zalutsky M R, Fuller G N, Archer G E, Friedman H S, Kwatra M M, Bigner S H, Bigner D D. Proc Natl Acad Sci USA. 1990;87:4207–4211. doi: 10.1073/pnas.87.11.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ekstrand A J, Sugawa N, James C D, Collins V P. Proc Natl Acad Sci USA. 1992;89:4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humphrey P A, Gangarosa L M, Wong A J, Archer G E, Lund-Johansen M, Bjerkvig R, Laerum O D, Friedman H S, Bigner D D. Biochem Biophys Res Commun. 1991;178:1413–1420. doi: 10.1016/0006-291x(91)91051-d. [DOI] [PubMed] [Google Scholar]
- 14.Frederick L, Wang X Y, Eley G, James C D. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- 15.Nishikawa R, Ji X D, Harmon R C, Lazar C S, Gill G N, Cavenee W K, Huang H J. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagane M, Coufal F, Lin H, Bogler O, Cavenee W K, Huang H J. Cancer Res. 1996;56:5079–5086. [PubMed] [Google Scholar]
- 17.Huang H S, Nagane M, Klingbeil C K, Lin H, Nishikawa R, Ji X D, Huang C M, Gill G N, Wiley H S, Cavenee W K. J Biol Chem. 1997;272:2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 18.Mendelsohn J, Baselga J. Oncogene. 2000;19:6550–6565. doi: 10.1038/sj.onc.1204082. [DOI] [PubMed] [Google Scholar]
- 19.Kawamoto T, Sato J D, Le A, Polikoff J, Sato G H, Mendelsohn J. Proc Natl Acad Sci USA. 1983;80:1337–1341. doi: 10.1073/pnas.80.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waterfield M D, Mayes E L, Stroobant P, Bennet P L, Young S, Goodfellow P N, Banting G S, Ozanne B. J Cell Biochem. 1982;20:149–161. doi: 10.1002/jcb.240200207. [DOI] [PubMed] [Google Scholar]
- 21.Hills D, Rowlinson-Busza G, Gullick W J. Int J Cancer. 1995;63:537–543. doi: 10.1002/ijc.2910630414. [DOI] [PubMed] [Google Scholar]
- 22.Wikstrand C J, Hale L P, Batra S K, Hill M L, Humphrey P A, Kurpad S N, McLendon R E, Moscatello D, Pegram C N, Reist C J, et al. Cancer Res. 1995;55:3140–3148. [PubMed] [Google Scholar]
- 23.Okamoto S, Yoshikawa K, Obata Y, Shibuya M, Aoki S, Yoshida J, Takahashi T. Br J Cancer. 1996;73:1366–1372. doi: 10.1038/bjc.1996.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moscatello D K, Holgado-Madruga M, Godwin A K, Ramirez G, Gunn G, Zoltick P W, Biegel J A, Hayes R L, Wong A J. Cancer Res. 1995;55:5536–5539. [PubMed] [Google Scholar]
- 25.Olapade-Olaopa E O, Moscatello D K, MacKay E H, Horsburgh T, Sandhu D P, Terry T R, Wong A J, Habib F K. Br J Cancer. 2000;82:186–194. doi: 10.1054/bjoc.1999.0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia de Palazzo I E, Adams G P, Sundareshan P, Wong A J, Testa J R, Bigner D D, Weiner L M. Cancer Res. 1993;53:3217–3220. [PubMed] [Google Scholar]
- 27.Ge H, Gong X, Tang C K. Int J Cancer. 2002;98:357–361. doi: 10.1002/ijc.10224. [DOI] [PubMed] [Google Scholar]
- 28.Sato J D, Kawamoto T, Le A D, Mendelsohn J, Polikoff J, Sato G H. Mol Biol Med. 1983;1:511–529. [PubMed] [Google Scholar]
- 29.Lin C R, Chen W S, Kruiger W, Stolarsky L S, Weber W, Evans R M, Verma I M, Gill G N, Rosenfeld M G. Science. 1984;224:843–848. doi: 10.1126/science.6326261. [DOI] [PubMed] [Google Scholar]
- 30.Pfreundschuh M, Shiku H, Takahashi T, Ueda R, Ransohoff J, Oettgen H F, Old L J. Proc Natl Acad Sci USA. 1978;75:5122–5126. doi: 10.1073/pnas.75.10.5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damjanov I, Mildner B, Knowles B B. Lab Invest. 1986;55:588–592. [PubMed] [Google Scholar]
- 32.Sobol R E, Astarita R W, Hofeditz C, Masui H, Fairshter R, Royston I, Mendelsohn J. J Natl Cancer Inst. 1987;79:403–407. [PubMed] [Google Scholar]
- 33.Wang D P, Konishi I, Koshiyama M, Nanbu Y, Iwai T, Nonogaki H, Mori T, Fujii S. Virchows Arch A Pathol Anat Histopathol. 1992;421:393–400. doi: 10.1007/BF01606911. [DOI] [PubMed] [Google Scholar]
- 34.Zarcone R, Bellini P, Cardone G, Vicinanza G, Cardone A. Clin Exp Obstet Gynecol. 1995;22:298–300. [PubMed] [Google Scholar]
- 35.Kikkawa K, Kato M, Saitoh Y. Nippon Geka Gakkai Zasshi. 1993;94:1231–1238. [PubMed] [Google Scholar]
- 36.Martinazzi M, Crivelli F, Zampatti C, Martinazzi S. J Clin Pathol. 1993;46:1009–1010. doi: 10.1136/jcp.46.11.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugimura M, Kobayashi H, Kanayama N, Terao T. Nippon Sanka Fujinka Gakkai Zasshi. 1992;44:689–694. [PubMed] [Google Scholar]
- 38.Kaminski M S, Zasadny K R, Francis I R, Fenner M C, Ross C W, Milik A W, Estes J, Tuck M, Regan D, Fisher S, Glenn S D, Wahl R L. J Clin Oncol. 1996;14:1974–1981. doi: 10.1200/JCO.1996.14.7.1974. [DOI] [PubMed] [Google Scholar]
- 39.Baselga J. J Clin Oncol. 2001;19:41S–44S. [PubMed] [Google Scholar]
- 40.Goldenberg D M. J Nucl Med. 2002;43:693–713. [PubMed] [Google Scholar]
- 41.Divgi C R, Welt S, Kris M, Real F X, Yeh S D, Gralla R, Merchant B, Schweighart S, Unger M, Larson S M, et al. J Natl Cancer Inst. 1991;83:97–104. doi: 10.1093/jnci/83.2.97. [DOI] [PubMed] [Google Scholar]
- 42.Busam K J, Capodieci P, Motzer R, Kiehn T, Phelan D, Halpern A C. Br J Dermatol. 2001;144:1169–1176. doi: 10.1046/j.1365-2133.2001.04226.x. [DOI] [PubMed] [Google Scholar]
- 43.Kawagoe K, Akiyama J, Kawamoto T, Morishita Y, Mori S. Placenta. 1990;11:7–15. doi: 10.1016/s0143-4004(05)80438-2. [DOI] [PubMed] [Google Scholar]
- 44.Messing E M, Hanson P, Ulrich P, Erturk E. J Urol. 1987;138:1329–1335. doi: 10.1016/s0022-5347(17)43593-2. [DOI] [PubMed] [Google Scholar]
- 45.Johns T G, Stockert E, Ritter G, Jungbluth A A, Huang H J, Cavenee W K, Smyth F E, Hall C M, Watson N, Nice E C, et al. Int J Cancer. 2002;98:398–408. doi: 10.1002/ijc.10189. [DOI] [PubMed] [Google Scholar]
- 46.Luwor R B, Johns T G, Murone C, Huang H J, Cavenee W K, Ritter G, Old L J, Burgess A W, Scott A M. Cancer Res. 2001;61:5355–5361. [PubMed] [Google Scholar]
- 47.Mishima K, Johns T G, Luwor R B, Scott A M, Stockert E, Jungbluth A A, Ji X D, Suvarna P, Voland J R, Old L J, Huang H J, Cavenee W K. Cancer Res. 2001;61:5349–5354. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.