Abstract
The EnvZ/OmpR system in Escherichia coli, which regulates the expression of the porins OmpF and OmpC, is one of the simplest and best-characterized examples of two-component signaling. Like many other histidine kinases, EnvZ is bifunctional; it phosphorylates and dephosphorylates the response regulator OmpR. We have analyzed a mathematical model of the EnvZ-mediated cycle of OmpR phosphorylation and dephosphorylation. The model predicts that when EnvZ is much less abundant than OmpR, as is the case in E. coli, the steady-state level of phosphorylated OmpR (OmpR-P) is insensitive to variations in the concentration of EnvZ. The model also predicts that the level of OmpR-P is insensitive to variations in the concentration of OmpR when the OmpR concentration is sufficiently high. To test these predictions, we have perturbed the porin regulatory circuit in E. coli by varying the expression levels of EnvZ and OmpR. We have constructed two-color fluorescent reporter strains in which ompF and ompC transcription can be easily measured in the same culture. Using these strains we have shown that, consistent with the predictions of our model, the transcription of ompC and ompF is indeed robust or insensitive to a wide range of expression levels of both EnvZ and OmpR.
Among prokaryotes, a remarkable number of cellular functions are controlled by two-component regulatory systems. Dedicated circuits transduce and interpret specific signals such as pH, temperature, osmolarity, light, nutrients, ions, pheromones, and toxins to regulate a wide range of processes including motility, virulence, metabolism, the cell cycle, developmental switches, antibiotic resistance, and stress responses (for reviews see for example refs. 1–4). Most of the detailed features of these circuits are quite varied and depend on the particular signals that are detected and the responses that are effected. However, all two-component systems transmit information via phosphorylation of a histidine on one protein (a histidine kinase) followed by transfer of phosphate to an aspartic acid, which is usually on a distinct protein (a response regulator). A notable feature of many of these systems is that the histidine kinase is bifunctional: it phosphorylates and dephosphorylates its cognate response regulator. This feature raises the interesting question of what is gained by implementing a regulatory circuit in this manner. Would a circuit in which, for example, phosphorylation is controlled by the histidine kinase and dephosphorylation is caused by spontaneous hydrolysis or a separate phosphatase have important differences from a circuit in which both reactions are mediated by the same enzyme?
A particularly well-characterized example of a bifunctional histidine kinase is EnvZ. In Escherichia coli, EnvZ responds to changes in the extracellular osmolarity of inner-membrane impermeable compounds and controls the phosphorylation of the response regulator OmpR. OmpR, in turn, regulates the transcription of a number of genes, the best characterized of which are ompF and ompC. These two genes, which code for porins in the outer membrane, are reciprocally regulated: OmpF is expressed primarily at low osmolarity and OmpC is expressed primarily at high osmolarity. Based on a variety of in vitro and genetic data, a model of this system has emerged in which EnvZ functions as an autokinase, an OmpR phosphotransferase, and as a phospho-OmpR (OmpR-P) phosphatase (reviewed in refs. 2, 3, 5, and 6). It is not yet understood how OmpR regulates ompF and ompC expression (7–11), although it appears that at low concentrations of OmpR-P, ompF is activated, whereas at high concentrations of OmpR-P, ompF is repressed and ompC is activated (12–14).
Previously, it was suggested on the basis of a simplified model for the EnvZ-mediated cycle of phosphorylation and dephosphorylation of OmpR that the output of the circuit (the concentration of OmpR-P) should be independent of the total concentration of EnvZ and OmpR in the cell (13, 15). The notion that the complex networks of interacting molecules within cells should be robust (i.e., insensitive) to variations in many of the parameters that characterize the constituent reactions has appeared in various contexts in cell biology (e.g., refs. 16–21). Robustness has been particularly well studied in development (19, 22) and bacterial chemotaxis (18, 23, 24). Although the extent to which robustness is a general feature of biochemical networks remains an open question, it is difficult to imagine that the integrated behavior of all of the pathways in a cell could function effectively if they required precise adjustment of their underlying components. The model of the EnvZ/OmpR system in refs. 13 and 15 represents a particularly simple mechanism for achieving robust behavior. The essential assumptions of the model are (i) the phosphorylation and dephosphorylation reactions are mediated by the same enzyme and (ii) the two reactions are independent processes. Recent experimental work, however, has revealed that the domains in EnvZ involved in the autokinase, phosphotransfer, and phosphatase reactions overlap and interact (25–29), making it unlikely that the second assumption, that the reactions function independently, applies to the EnvZ/OmpR system. We describe here the analysis of a more realistic model of the EnvZ/OmpR circuit that is consistent with the interdependence of the various enzymatic activities of EnvZ. We also present the results from experiments in which we have explored the extent to which the system is robust. We have perturbed the porin regulatory circuit by varying the expression levels of EnvZ and OmpR and measured the effect on ompF and ompC transcription. The results are consistent with the predictions from our model and more generally provide an important constraint on potential models of the EnvZ/OmpR system.
Materials and Methods
Media, Chemicals, and Other Reagents.
Low osmolarity cultures were grown in medium A (30) (for isolation of cell envelopes) or minimal A medium (31) with 0.2% glycerol (for fluorescence measurements). Osmolarity was varied by supplementing with varying concentrations of sucrose. Plasmids were maintained with 50 μg/ml ampicillin. The lac promoter was induced with isopropyl-β-d-thiogalactoside (IPTG).
Construction of Fluorescent Reporter Strains.
Plasmids containing the genes for cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) were gifts from M. Elowitz (The Rockefeller University, New York). The strain MDG131, which contains operon fusions of yfp to ompF and cfp to ompC [MC4100 Φ(ompF+-yfp+) Φ (ompC+-cfp+)], was constructed in two steps. A cassette consisting of the last ≈1 kb of ompF (including the translation stop codon but preceding the ompF terminator), followed by a promoterless yfp with a ribosome binding site, followed by ≈1 kb of the DNA sequence downstream of ompF in the E. coli K-12 genome (including the ompF transcription terminator), was assembled and cloned into the suicide vector pCVD442 (32) to give pMG37. This plasmid was introduced into MC4100 [F− araD139 Δ(argF-lac)169 λ− flhD5301 fruA25 relA1 rpsL150 rbsR22 deoC1] (33). Cells were selected for ampicillin resistance and then counterselected for sucrose resistance as in ref. 32. Colonies were screened by PCR using primers that annealed to sites on the chromosome outside of the regions of flanking homology used in the construction of pMG37. This process resulted in a strain containing a transcriptional fusion of yfp to ompF. A second cassette was assembled that contained the last ≈1 kb of ompC (including the translation stop but not including the transcription terminator), followed by a promoterless cfp with a ribosome binding site, followed by ≈1 kb of the DNA sequence downstream of ompC (including the ompC transcription terminator). This cassette was cloned into pCVD442 to give pMG35. This plasmid was introduced into the above ompF-yfp transcriptional fusion strain and again cells were selected for ampicillin resistance, counterselected for sucrose resistance, and screened by PCR. This process resulted in MDG131. MDG133 (MDG131 ompR101) and MDG135 (MDG131 envZ∷kan) were constructed by P1 transduction from FR195 (13) and WH57 (25), respectively.
Analysis of Cell Envelopes.
To assay the levels of outer membrane proteins, cultures were grown in medium A (with or without 15% sucrose) to an OD600 of ≈0.7. Cell envelopes were isolated as in ref. 34, separated by SDS/PAGE on 12% polyacrylamide gels containing 6 M urea, and stained with Coomassie brilliant blue R-250.
Expression Plasmids.
To control EnvZ levels we used the plasmid pEnvZ (25). For OmpR expression, we constructed the plasmid pEB15, which consists of the ompR coding sequence (without the upstream regulatory regions) from pFR29 (13) cloned downstream of the lac promoter in the low-copy vector pWSK29 (35). The plasmid also contains lacIq. The control plasmid pEB5 was constructed by deleting the envZ gene from pEnvZ. Similarly, the control plasmid pEB16 consists of pEB15 with ompR deleted. More detailed descriptions of the plasmid constructions are provided in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Fluorescence Measurements.
Two-milliliter cultures were grown at 37° with aeration to saturation in minimal media (supplemented with ampicillin, IPTG, and sucrose when appropriate). The saturated cultures were then diluted with sufficient prewarmed media such that at least 10 doublings would occur before the cultures reached an OD600 of ≈0.2. When the cultures reached this optical density they were rapidly cooled by stirring the tubes in an ice-water slurry, and chloramphenicol was added to a concentration of 75 μg/ml to inhibit protein synthesis. Two-milliliter samples of the cultures were warmed to room temperature and used for fluorescence measurements. The remainder of the cultures was aliquoted and centrifuged, and the pellets were stored at −80° until they were needed for protein gels.
All fluorescence measurements were performed by using an Aminco Bowman SLM 8100 spectro-fluorometer (Jobin Yvon, Edison, NJ). YFP fluorescence was measured by using an excitation wavelength of 505 nm and an emission wavelength of 527 nm, and CFP fluorescence was measured by using an excitation wavelength of 434 nm and an emission wavelength of 477 nm.
Protein Quantification of EnvZ and OmpR.
The pellets from either 1- or 2-ml aliquots of cultures expressing various levels of EnvZ and OmpR were analyzed on 12% SDS/PAGE gels. Standard buffers and conditions were used for electrophoresis and Western blots (36). Anti-EnvZ and anti-OmpR antibodies were gifts from T. J. Silhavy, Princeton University (Princeton). Alkaline phosphatase-conjugated goat anti-rabbit whole molecule IgG (Sigma) was used for the secondary antibody. Bands were visualized with a Nitro blue tetrazolium, 5-bromo-4-chloro-3-indolyl phosphate assay (36). Digitized images were acquired with a Kodak EDAS290 gel documentation system with the yellow filter removed and with the membranes back-illuminated by using a standard fluorescent light box. Images were analyzed by using the program scion image (Scion, Frederick, MD).
Results
A Model of the EnvZ/OmpR Circuit Predicts Robust Output.
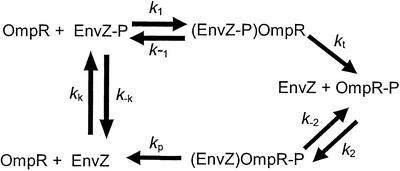
We have constructed a model of the EnvZ/OmpR circuit that incorporates the autokinase, phosphotransfer, and phosphatase activities of EnvZ (Fig. 1). Enzyme–substrate complexes for both the phosphotransfer and dephosphorylation steps, [(EnvZ-P)OmpR] and [(EnvZ)OmpR-P], have been included to allow for the possibility of enzyme saturation. The model depends on 10 parameters: eight rate constants, the total concentration of OmpR ([OmpR]T), and the total concentration of EnvZ ([EnvZ]T) (Fig. 1). The concentration of ATP is assumed constant and is absorbed into the rate constant kk. The system is characterized by six first-order ordinary differential equations, two of which can be eliminated after imposing the constraints that [OmpR]T and [EnvZ]T are both constant. Note that an important feature of this model, which distinguishes it from the model in refs. 13 and 15, is that at any given moment the total amount of EnvZ is divided among the various forms [EnvZ], [EnvZ-P], [(EnvZ-P)OmpR], and [(EnvZ)OmpR-P] and that these different forms can participate only in specific reactions. For example, the model assumes that EnvZ-P cannot dephosphorylate OmpR-P.
Figure 1.
A model of the EnvZ/OmpR two-component circuit. The kinetics are described by four differential equations together with the constraints that the total amount of EnvZ and OmpR are constant. The concentration of ATP is assumed constant and is absorbed into the rate constant kk. The explicit form of the equations is given in Supporting Text.
To find the steady-state behavior, we set the time derivatives in the kinetic equations equal to zero, which gives six algebraic equations for the six concentrations in the model. These equations reduce to a cubic equation for [OmpR-P], the solution of which is a complicated function of all 10 parameters, including in particular [EnvZ]T. We thus find that in this model, in contrast to a model in which the phosphorylation and dephosphorylation activities of EnvZ function independently (13, 15), the output of the circuit ([OmpR-P]) depends on the total amount of EnvZ. The EnvZ/OmpR circuit in E. coli, however, has an interesting operating point: the amount of EnvZ in the cell is much less than the amount of OmpR (37, 38). When we take this limit in our model, i.e., [OmpR]T ≫ [EnvZ]T, it is straightforward to derive for the steady-state concentration of [OmpR-P] (see Supporting Text for more details):
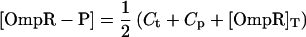 |
1 |
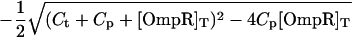 |
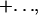 |
where the constants Ct and Cp are given by
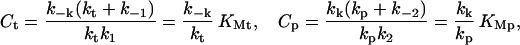 |
and … denotes terms that are small when [EnvZ]T/[OmpR]T is small. KMt and KMp are the Michaelis constants for the phosphotransfer and phosphatase reactions, respectively. Thus, to leading order in the limit [OmpR]T ≫ [EnvZ]T, the circuit output, [OmpR-P], is independent of [EnvZ]T. Within the context of this model, the fact that the histidine kinase EnvZ is in much lower abundance compared with the response regulator OmpR makes the circuit output robust with respect to variations in [EnvZ]T.
If we assume that [OmpR]T is not only much greater than [EnvZ]T but also much greater than the concentrations Ct and Cp, Eq. 1 simplifies to (see Supporting Text for more details as well as a simple alternative derivation):
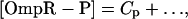 |
2 |
where … denotes terms that are small when [EnvZ]T/[OmpR]T, Cp/[OmpR]T, and Ct/[OmpR]T, are small. Thus, in this limit the circuit output is robust with respect to variations in [OmpR]T as well. Interestingly, the model predicts that in this limit [OmpR-P] is also independent of the phophotransfer rate constants kt, k1, and k−1.
To summarize the results of our analysis, for the model depicted in Fig. 1 the steady-state level of OmpR-P in general depends on the concentrations of EnvZ and OmpR. However, when the amount of EnvZ in the cell is much less than the amount of OmpR, which is the case in E. coli (37, 38), the steady-state level of [OmpR-P] is approximately independent of the concentration of EnvZ. Thus the output of the circuit will be insensitive to variations in the expression level of EnvZ within a range such that EnvZ is less abundant than OmpR. In addition, if the total amount of OmpR is much larger than the concentrations Ct and Cp, which are related to the Michaelis constants for the phosphotransfer and phosphatase reactions, respectively, then the level of [OmpR-P] is also approximately independent of the total amount of OmpR.
Construction of Two-Color Fluorescent Reporter Strains.
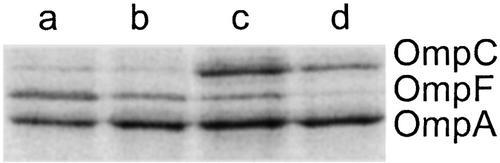
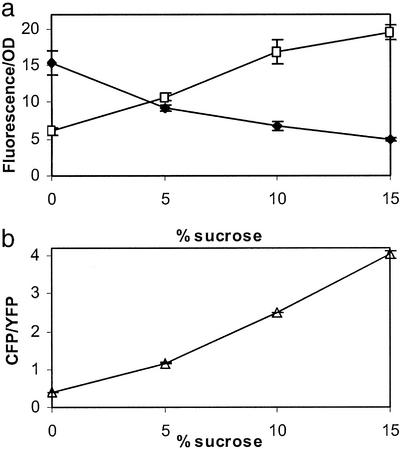
Ideally, to test the predictions of the model described above, we would like to measure directly [OmpR-P]. Unfortunately, at present we do not have a means to reliably quantify the extent of OmpR phosphorylation in cultures. We therefore used the relative transcription of ompF and ompC as an indirect measure of changes in the level of OmpR-P. To rapidly and accurately quantify the transcription of ompF and ompC with as little disruption of the EnvZ/OmpR circuit as possible, we constructed strains that contain chromosomal transcriptional (operon) fusions of the gene for CFP (cfp) to ompC and the gene for YFP (yfp) to ompF. A promoterless cfp with a ribosome-binding site was integrated into the chromosome at the ompC locus after the ompC stop codon and before the ompC transcription termination signal. In the same strain, a promoterless yfp with a ribosome-binding site was also inserted in the chromosome after the ompF stop codon and before the ompF transcription terminator. The resulting strain, MDG131, shows similar porin osmoregulation to the parental strain MC4100 (Fig. 2). There appears to be a small decrease in the level of porin expression for MDG131 relative to MC4100, which may be a result of a decrease in the mRNA stability of the transcriptional fusions. We have not observed any significant difference in growth rate between MDG131 and MC4100. For cultures grown in minimal media, background autofluorescence, as judged by cultures of the parental strain MC4100, was negligible in both fluorescence channels (data not shown). In addition, for cultures of MDG131 we have not been able to detect YFP fluorescence emission when exciting CFP at 434 nm (data not shown), and thus do not detect any resonance energy transfer between CFP and YFP. Fluorescence measurements of MDG131 cultures grown in varying concentrations of sucrose show an increase in CFP fluorescence and a decrease in YFP fluorescence with increasing osmolarity (Fig. 3).
Figure 2.
Coomassie-stained gels of cell envelope fractions showing that porin osmoregulation in MDG131 (lanes b and d) and MC4100 (lanes a and c) are comparable. Low osmolarity cultures (lanes a and b) were grown in medium A, and high osmolarity cultures (lanes c and d) were grown in medium A supplemented with 15% sucrose.
Figure 3.
Fluorescence measurements of ompF (YFP) and ompC (CFP) osmo-regulation in MDG131. Cultures were grown in minimal media with varying concentrations of sucrose. (a) YFP (⧫) and CFP (□) fluorescence (arbitrary units) normalized by culture optical density. (b) CFP/YFP fluorescence ratio (▵). Each error bar denotes the SD of three independent cultures.
The strain MDG131 and its derivatives possess a number of advantages over many of the frequently used β-galactosidase reporter strains. Both the ompF and ompC genes remain intact in MDG131, which avoids potential changes in cell physiology caused by loss of one of the porins. In addition, fluorescence measurements are rapid and less prone to handling errors compared with enzymatic assays. Finally, because separate reporters for ompF and ompC transcription are present within the same cells, ratios of CFP to YFP fluorescence provides a measure of the relative transcriptional activity of ompC to ompF that is independent of growth rate, total cell mass, or other factors that can change the total protein content within the cells.
OmpC/OmpF Transcription Is Insensitive to Variations in the Level of EnvZ.
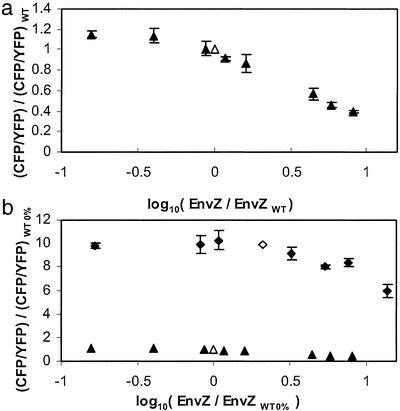
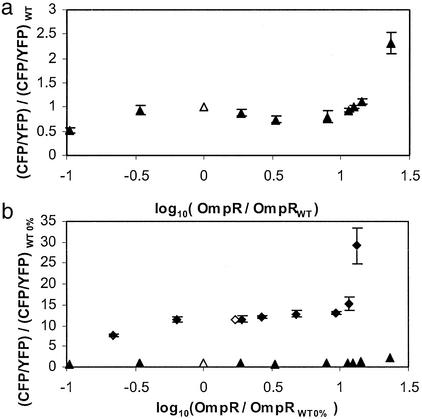
The model described above predicts that the output of the EnvZ/OmpR circuit is insensitive to variations in the total concentration of EnvZ ([EnvZ]T) within the range such that [EnvZ]T ≪ [OmpR]T. To test this, we perturbed the regulatory circuit by varying the expression level of EnvZ. We used the envZ− fluorescent reporter strain MDG135, which was transformed with the EnvZ expression plasmid pEnvZ (25). By varying the concentration of IPTG in the culture medium we were able to control the expression of EnvZ over a wide range from ≈10-fold below to 10-fold above WT levels. OmpR levels did not change significantly over this range of EnvZ (data not shown). The WT level of EnvZ was defined to be the expression level in an envZ+ strain containing a control plasmid, MDG131/pEB5, and was used to normalize bands on Western blots. The transcriptional activity of ompF and ompC was determined by measurements of YFP and CFP, respectively, and normalized by the corresponding (WT) fluorescence of cultures of MDG131/pEB5 grown under the same conditions. In the low osmolarity regime, in which cultures were grown in minimal media without sucrose, we found that the ratio of ompC transcription to ompF transcription remained fairly constant over roughly an order of magnitude change in EnvZ concentration (Fig. 4a). Near-WT levels of porin expression are maintained even when the amount of EnvZ in the cells is almost one-tenth of the WT value. As [EnvZ] is increased above the WT level, there is a relatively slow logarithmic decrease in CFP/YFP fluorescence. The fluorescence ratio decreases by 20% and 50% when the amount of EnvZ is roughly 2- and 5-fold above WT, respectively.
Figure 4.
(a) CFP/YFP fluorescence ratios versus EnvZ levels for cultures of MDG135/pEnvZ at low osmolarity (minimal medium) with varying amounts of IPTG. Fluorescence and EnvZ levels were normalized by WT values, which were determined from MDG131/pEB5. (b) Similar measurements for growth at high osmolarity (minimal medium with 15% sucrose). Fluorescence and EnvZ levels were normalized by the low osmolarity WT values (WT 0%) as in a. For reference, the low osmolarity data are also shown in b. ▴, MDG135/pEnvZ 0% sucrose; ▵, MDG131/pEB5 0% sucrose; ⧫, MDG135/pEnvZ 15% sucrose; ◊, MDG131/pEB5 15% sucrose. The WT level of EnvZ at high osmolarity is ≈2.1 times higher than the level at low osmolarity. Each error bar denotes the SD of three independent cultures.
We also made similar measurements for growth at high osmolarity, in which the culture medium contained 15% sucrose. Once again we found that ompC/ompF transcription is relatively insensitive to the expression level of EnvZ, particularly for levels below WT, and that the ratio decreases slowly (logarithmically) for EnvZ levels above WT (Fig. 4b). In Fig. 4b we have normalized the fluorescence and EnvZ levels by the corresponding low osmolarity WT values. We find that the WT level of EnvZ at high osmolarity is larger than the level at low osmolarity by a factor of ≈2.1, which is similar to the factor of ≈1.7 reported in ref. 38. Fig. 4b also includes the low osmolarity data to provide a sense of scale for the change in ompC/ompF transcription between low and high osmolarity relative to the variation in transcription at fixed osmolarity.
OmpC/OmpF Transcription Is Insensitive to Variations in the Level of OmpR.
To test whether the EnvZ/OmpR circuit is robust with respect to OmpR, we constructed the plasmid pEB15, which carries a copy of ompR that lacks its upstream regulatory region and is under control of the lac promoter (see Materials and Methods). When we transformed the ompR− strain MDG133 with pEB15 we were able to vary OmpR levels over almost 2.5 orders of magnitude (≈10-fold below to ≈30-fold above WT levels) by adjusting the concentration of IPTG in the growth medium. The levels of EnvZ did not change significantly over this range except at the very highest levels of OmpR, for which there was an ≈2-fold drop in EnvZ levels (data not shown). WT was taken to be an ompR+ strain that contained a control plasmid, MDG131/pEB16, and was used to normalize OmpR Western blots and fluorescence ratios. For growth at low osmolarity, ompC/ompF transcription shows little change as the expression of OmpR varies over roughly 1.5 orders of magnitude (Fig. 5a). CFP/YFP fluorescence decreases at the lowest level of OmpR. At high OmpR (>10 times the WT level) the fluorescence ratio increases sharply. Curiously, we find that the curve of CFP/YFP fluorescence as a function of OmpR is not monotonic but instead increases, then decreases and then increases again (Fig. 5a) as OmpR levels run from low to high; however, the magnitude of this modulation is relatively small.
Figure 5.
(a) CFP/YFP fluorescence ratios versus OmpR levels for cultures of MDG133/pEB15 at low osmolarity (minimal medium) with varying amounts of IPTG. Fluorescence and OmpR levels were normalized by WT values, which were determined from MDG131/pEB16. (b) Similar measurements for growth at high osmolarity (minimal medium with 15% sucrose). Fluorescence and OmpR levels were normalized by the low osmolarity WT values (WT 0%) as in a. For reference, the low osmolarity data are also shown in b. ▴, MDG133/pEB15 0% sucrose; ▵, MDG131/pEB16 0% sucrose; ⧫, MDG133/pEB15 15% sucrose; ◊, MDG131/pEB16 15% sucrose. The WT level of OmpR at high osmolarity is ≈1.7 times higher than the level at low osmolarity. Each error bar denotes the SD of three independent cultures.
The results for cultures of MDG133/pEB15 grown at high osmolarity are similar to the results for low osmolarity cultures (Fig. 5b). In Fig. 5b we have normalized the fluorescence and OmpR levels by the low osmolarity WT values and have included the low osmolarity data on the same plot. We find the WT expression level of OmpR at high osmolarity is larger than the expression level at low osmolarity by a factor of ≈1.7, which is consistent with the results in ref. 38. Again, the ompC/ompF transcription ratio is relatively independent of the expression level of OmpR over ≈2 orders of magnitude. There is a decrease in the ratio at concentrations of OmpR around one-tenth the WT concentration, as well as a sharp increase in the ratio as the concentration is increased beyond roughly five times the WT OmpR concentration.
Discussion
Within the context of the EnvZ/OmpR two-component system it has been argued on the basis of a simplified model that the bifunctional nature of EnvZ results in a circuit output (the concentration of OmpR-P) that is insensitive to variations in the total concentrations of EnvZ ([EnvZ]T) and OmpR ([OmpR]T) (13, 15). The central assumptions of this model, however, are in conflict with more recent work in which the regions within EnvZ responsible for the autokinase, phosphotransfer, and phosphatase activities were shown to overlap and interact (25–29). We have therefore constructed a more complex model (Fig. 1) in which the enzymatic activities of EnvZ do not function independently. The steady-state behavior of this model can be solved analytically and we find that the concentration of OmpR-P depends on both [EnvZ]T and [OmpR]T. Thus, the model predicts that in general the output of a circuit with a bifunctional enzyme as in Fig. 1 will be sensitive to the concentrations of the regulatory proteins. It has been known for some time, however, that the expression level of EnvZ is significantly lower than that of OmpR in E. coli. Recently, the numbers of EnvZ and OmpR molecules per (E. coli) cell have been determined to be ≈100 and 3,500, respectively, corresponding to an EnvZ-to-OmpR ratio of 3% (38). Thus, the EnvZ/OmpR circuit operates in the limit [EnvZ]T ≪ [OmpR]T. We find that when we take this limit in our model the leading behavior for the steady-state concentration of OmpR-P is independent of [EnvZ]T (see Eq. 1 above). Thus, the model predicts that the circuit output will be insensitive to variations in the concentration of [EnvZ]T within a range that satisfies [EnvZ]T ≪ [OmpR]T. In addition, in the limit that [OmpR]T is not only much larger than [EnvZ]T but also large relative to the concentrations Cp and Ct, which characterize the phosphotransfer and phosphatase reactions (see above), the concentration of OmpR-P is insensitive to variations in [OmpR]T.
If the model in Fig. 1 is a correct description of the EnvZ/OmpR system, then the circuit output should be robust with respect to variations in the concentration of EnvZ, as long as the total amount of EnvZ in the cell is much less than the total amount of OmpR. The behavior with respect to variations in OmpR levels, on the other hand, also depends on the amount of OmpR relative to Cp and Ct (see above). Because we do not know the values of these parameters we cannot make a sharp prediction regarding the dependence on [OmpR]T. However, the model can account for robustness with respect to OmpR provided the total amount of OmpR is sufficiently high. To test robustness we have perturbed the regulatory circuit outside of its usual steady-state operating regime under laboratory conditions by varying the expression levels of EnvZ and OmpR. We used the relative transcription of ompC and ompF as an indirect measure of variations in OmpR-P levels (see the discussion below). To accurately and easily measure transcription of porin genes we constructed two-color fluorescent reporter strains that contain chromosomal transcriptional (operon) fusions of cfp and yfp to ompC and ompF, respectively. With these strains, the ratio of CFP to YFP fluorescence provides a rapid and sensitive measure of ompC/ompF transcription.
We find that the ompC/ompF transcription ratio is relatively insensitive to variations in the expression level of EnvZ. As EnvZ levels span the range from much lower to much higher than WT, CFP/YFP fluorescence is relatively flat (EnvZ independent) and then slowly decreases (logarithmically) with increasing EnvZ. These results are consistent with the model in Fig. 1, which predicts that [OmpR-P] is insensitive to [EnvZ]T in the limit [EnvZ]T ≪ [OmpR]T. When [EnvZ]T is sufficiently large so that this limit no longer applies, the model predicts that the steady-state value of [OmpR-P] decreases with increasing [EnvZ]T (data not shown). We should note, however, that for the data in Fig. 4, the YFP fluorescence remained relatively constant over the full range of [EnvZ]T investigated, whereas CFP fluorescence accounted for the decrease in the CFP/YFP ratio at high EnvZ (data not shown). Thus, it is possible that this decrease is not caused by a drop in [OmpR-P] levels but may instead be caused by other undetermined factors that regulate ompC expression.
Recently, it was found that OmpR-P has extremely low binding affinity toward a cytoplasmic fragment of EnvZ (EnvZc) (39). If EnvZc has the same phosphatase activity as the full-length membrane-integrated EnvZ, this would raise important questions regarding the mechanism of OmpR-P dephosphorylation in vivo. However, it is also quite possible that the phosphatase activity of EnvZc in vitro is significantly less than the corresponding activity of EnvZ in vivo. It is interesting to compare our results with what would be expected from a model in which the dominant mode of OmpR-P dephosphorylation is caused by an EnvZ-independent mechanism (e.g., spontaneous hydrolysis or a phosphatase other than EnvZ). In this case one would expect the level of OmpR-P to increase with increasing [EnvZ]T. This possibility contrasts with our observations that ompC/ompF transcription is relatively insensitive to changes in [EnvZ]T and in fact slowly decreases with increasing [EnvZ]T (Fig. 4).
We also find that ompC/ompF transcription is insensitive to the expression level of OmpR (Fig. 5). At the lowest concentrations of OmpR for which we made measurements (roughly one-tenth of the WT level), CFP/YFP fluorescence decreases. Again, as was the case for high EnvZ, this is the regime in which the expression level of EnvZ is comparable to the level of OmpR and hence the limit [EnvZ]T ≪ [OmpR]T does not apply. Interestingly, as was the case for high EnvZ, the decrease in CFP/YFP fluorescence is caused by a decrease in CFP without any significant change in YFP (data not shown). When the level of OmpR is increased roughly 10 times above WT, the ratio of ompC/ompF transcription rapidly increases. In this case the increase is caused by both an increase in CFP and a decrease in YFP (data not shown) and is therefore consistent with an increase in the level of [OmpR-P]. Such an increase could be caused by cross-talk from alternative phospho-donors. When we include a small term in our model that allows for EnvZ-independent phosphorylation of OmpR, we find a similar increase in the concentration of OmpR-P (data not shown). It is also possible that very high levels of unphosphorylated OmpR could activate ompC and repress ompF.
We have used the relative transcription of ompC and ompF as an indirect measure of the levels of OmpR-P. Our primary assumption is that conditions in which ompC/ompF transcription does not change correspond to conditions in which [OmpR-P] is constant. It is possible, however, that the robust behavior that we have observed is not caused by the circuit depicted in Fig. 1 but instead derives from the mechanism of OmpR-P control of porin transcription, which is not well understood (7–11). Thus it is possible that moderate changes in [OmpR-P] have little effect on ompC/ompF transcription and that it is only very large changes that result in activation of ompC and repression of ompF (12–14). Although we cannot rule out this possibility, it seems unlikely given that we find robustness at both high and low osmolarity (corresponding to high and low levels of OmpR-P) and given that the change in ompC/ompF transcription as a function of osmolarity is quite gradual (Fig. 3). In either case, our observations that porin transcription is insensitive to the level of the regulatory proteins EnvZ and OmpR provides important constraints on models of the EnvZ/OmpR circuit. With further progress in elucidating the mechanism of OmpR regulation of porin expression it will be possible to translate OmpR-P levels into rates of transcription of ompF and ompC. Incorporating OmpR–DNA binding and porin expression explicitly will enable more stringent tests of quantitative models of EnvZ/OmpR signaling.
The model described here may also be an appropriate description of other two-component systems with bifunctional histidine kinases. Although in most cases there does not appear to be much information regarding the relative concentrations of the regulatory proteins, the model presented here predicts that systems for which the histidine kinase is in low abundance relative to the response regulator should exhibit robust behavior with respect to variations in the histidine kinase concentration. For the response regulator, robust behavior would also require that the response regulator concentration is high relative to the parameters Cp and Ct. Other concentration ranges for either of the regulatory proteins will lead to a more complex dependence of the phosphorylated response regulator on their expression level. In many two-component systems the transcription of the regulatory proteins is controlled autogenously or by other transcriptional regulators. For example, there is evidence that the operon consisting of ompR and envZ is subject to control by integration host factor, cAMP-catabolite activator protein, and the histone-like protein HNS (reviewed in ref. 40). At first sight, the fact that the regulatory proteins are under transcriptional control might appear to be inconsistent with robustness because there would be no point in changing their expression levels if there were no effect on the circuit output. However, there are a variety of explanations for why cells would modulate the expression of these regulatory proteins that are consistent with our results. In some cases, the circuit may only be robust with respect to the histidine kinase so that varying levels of the response regulator will modulate the circuit output as in Eq. 1. Alternatively, some cellular functions may be controlled by the relative amount of phosphorylated to unphosphorylated response regulator, which would again imply that varying response regulator levels would modulate the output. Finally, under certain growth conditions cross-regulation (e.g., from alternative phosphodonors such as acetyl phosphate or possibly from additional phosphatases) could make the circuit sensitive to the levels of the regulatory proteins.
The model in Fig. 1 is consistent with the experimental results reported here but is clearly a simplified description of the EnvZ/OmpR system. There are a number of aspects of the circuit that have not been included such as EnvZ dimerization (41) and binding of OmpR to EnvZ (38, 39), as well as possible conformational changes of OmpR (42) or additional enzymatic steps. Nevertheless, the model captures the basic features of the generally accepted view of EnvZ/OmpR signaling and is a reasonable starting point for building more sophisticated models and explaining qualitative features of the system such as robustness. At the same time, beyond its applicability to EnvZ/OmpR, the model provides an interesting mechanism for achieving robust behavior with a bifunctional enzyme that may be broadly applicable to other regulatory circuits within cells.
Supplementary Material
Acknowledgments
We thank T. J. Silhavy for anti-EnvZ and anti-OmpR antibodies and M. Elowitz for plasmids containing cfp and yfp. We also thank A. N. Binns, F. Daldal, T. J. Silhavy, S. M. Simon, members of the Simon lab, and M. van der Woude for helpful discussions. M.G. is grateful to S. M. Simon for support during the early stages of this work. This work was supported by National Science Foundation Grant MCB-0212925 (to M.G.) and a National Science Foundation graduate fellowship (to E.B.).
Abbreviations
- OmpR-P
phospho-OmpR
- IPTG
isopropyl-β-d-thiogalactoside
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Stock J B, Ninfa A J, Stock A M. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoch J A, Silhavy T J. Two-Component Signal Transduction. Washington, DC: Am. Soc. Microbiol. Press; 1995. [Google Scholar]
- 3.Egger L A, Park H, Inouye M. Genes Cells. 1997;2:167–184. doi: 10.1046/j.1365-2443.1997.d01-311.x. [DOI] [PubMed] [Google Scholar]
- 4.Stock A M, Robinson V L, Goudreau P N. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 5.Forst S A, Roberts D L. Res Microbiol. 1994;145:363–373. doi: 10.1016/0923-2508(94)90083-3. [DOI] [PubMed] [Google Scholar]
- 6.Mizuno T. J Biochem (Tokyo) 1998;123:555–563. doi: 10.1093/oxfordjournals.jbchem.a021972. [DOI] [PubMed] [Google Scholar]
- 7.Rampersaud A, Harlocker S L, Inouye M. J Biol Chem. 1994;269:12559–12566. [PubMed] [Google Scholar]
- 8.Pratt L A, Silhavy T J. Mol Microbiol. 1995;17:565–573. doi: 10.1111/j.1365-2958.1995.mmi_17030565.x. [DOI] [PubMed] [Google Scholar]
- 9.Huang K J, Igo M M. J Mol Biol. 1996;262:615–628. doi: 10.1006/jmbi.1996.0540. [DOI] [PubMed] [Google Scholar]
- 10.Head C G, Tardy A, Kenney L J. J Mol Biol. 1998;281:857–870. doi: 10.1006/jmbi.1998.1985. [DOI] [PubMed] [Google Scholar]
- 11.Bergstrom L C, Qin L, Harlocker S L, Egger L A, Inouye M. Genes Cells. 1998;3:777–788. doi: 10.1046/j.1365-2443.1998.00228.x. [DOI] [PubMed] [Google Scholar]
- 12.Forst S, Delgado J, Rampersaud A, Inouye M. J Bacteriol. 1990;172:3473–3477. doi: 10.1128/jb.172.6.3473-3477.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russo F D, Silhavy T J. J Mol Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- 14.Lan C Y, Igo M M. J Bacteriol. 1998;180:171–174. doi: 10.1128/jb.180.1.171-174.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russo F D, Silhavy T J. Trends Microbiol. 1993;1:306–310. doi: 10.1016/0966-842x(93)90007-e. [DOI] [PubMed] [Google Scholar]
- 16.Wadington C H. Nature. 1942;150:563–565. [Google Scholar]
- 17.Savageau M A. Nature. 1971;229:542–544. doi: 10.1038/229542a0. [DOI] [PubMed] [Google Scholar]
- 18.Barkai N, Leibler S. Nature. 1997;387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 19.Freeman M. Nature. 2000;408:313–319. doi: 10.1038/35042500. [DOI] [PubMed] [Google Scholar]
- 20.Morohashi M, Winn A E, Borisuk M T, Bolouri H, Doyle J, Kitano H. J Theor Biol. 2002;216:19–30. doi: 10.1006/jtbi.2002.2537. [DOI] [PubMed] [Google Scholar]
- 21.Sengupta A M, Djordjevic M, Shraiman B I. Proc Natl Acad Sci USA. 2002;99:2072–2077. doi: 10.1073/pnas.022388499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Dassow G, Meir E, Munro E M, Odell G M. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- 23.Alon U, Surette M G, Barkai N, Leibler S. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 24.Yi T M, Huang Y, Simon M I, Doyle J. Proc Natl Acad Sci USA. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsing W, Silhavy T J. J Bacteriol. 1997;179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsing W, Russo F D, Bernd K K, Silhavy T J. J Bacteriol. 1998;180:4538–4546. doi: 10.1128/jb.180.17.4538-4546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutta R, Qin L, Inouye M. Mol Microbiol. 1999;34:633–640. doi: 10.1046/j.1365-2958.1999.01646.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y, Qin L, Yoshida T, Inouye M. Proc Natl Acad Sci USA. 2000;97:7808–7813. doi: 10.1073/pnas.97.14.7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dutta R, Yoshida T, Inouye M. J Biol Chem. 2000;275:38645–38653. doi: 10.1074/jbc.M005872200. [DOI] [PubMed] [Google Scholar]
- 30.Kawaji H, Mizuno T, Mizushima S. J Bacteriol. 1979;140:843–847. doi: 10.1128/jb.140.3.843-847.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller J H. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 32.Donnenberg M S, Kaper J B. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casadaban M J. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 34.Morona R, Reeves P. J Bacteriol. 1982;150:1016–1023. doi: 10.1128/jb.150.3.1016-1023.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang R F, Kushner S R. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 36.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1998. [Google Scholar]
- 37.Comeau D E, Ikenaka K, Tsung K L, Inouye M. J Bacteriol. 1985;164:578–584. doi: 10.1128/jb.164.2.578-584.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai S J, Inouye M. J Biol Chem. 2002;277:24155–24161. doi: 10.1074/jbc.M110715200. [DOI] [PubMed] [Google Scholar]
- 39.Mattison K, Kenney L J. J Biol Chem. 2002;277:11143–11148. doi: 10.1074/jbc.M111128200. [DOI] [PubMed] [Google Scholar]
- 40.Pratt L A, Hsing W, Gibson K E, Silhavy T J. Mol Microbiol. 1996;20:911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y, Inouye M. Proc Natl Acad Sci USA. 1991;88:11057–11061. doi: 10.1073/pnas.88.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattison K, Oropeza R, Byers N, Kenney L J. J Mol Biol. 2002;315:497–511. doi: 10.1006/jmbi.2001.5222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.