Abstract
Infection of T lymphocytes by the cytopathic retrovirus feline leukemia virus subgroup T (FeLV-T) requires FeLIX, a cellular coreceptor that is encoded by an endogenous provirus and closely resembles the receptor-binding domain (RBD) of feline leukemia virus subgroup B (FeLV-B). We determined the structure of FeLV-B RBD, which has FeLIX activity, to a 2.5-Å resolution by X-ray crystallography. The structure of the receptor-specific subdomain of this glycoprotein differs dramatically from that of Friend murine leukemia virus (Fr-MLV), which binds a different cell surface receptor. Remarkably, we find that Fr-MLV RBD also activates FeLV-T infection of cells expressing the Fr-MLV receptor and that FeLV-B RBD is a competitive inhibitor of infection under these conditions. These studies suggest that FeLV-T infection relies on the following property of mammalian leukemia virus RBDs: the ability to couple interaction with one of a variety of receptors to the activation of a conserved membrane fusion mechanism. A comparison of the FeLV-B and Fr-MLV RBD structures illustrates how receptor-specific regions are linked to conserved elements critical for postbinding events in virus entry.
Mammalian leukemia retroviruses, recently designated “γ-retroviruses” (gammaretroviruses) (22), have been widely dispersed by infection and by vertical transmission through the germ line. Glycoproteins that protrude from the gammaretrovirus membrane mediate entry into the cell during infection. These glycoproteins are trimers of heterodimers composed of surface (SU) and transmembrane (TM) subunits. The SU subunit contains an N-terminal domain that binds to receptor (6, 7, 15) and a C-terminal region that is disulfide bonded to the TM subunit, which mediates fusion between the virus and cell membranes. More than 10 subgroups of gammaretroviruses, distinguished by the specificity of their receptor-binding domains (RBDs), have been identified (44).
Initial hypotheses for the mechanism of cell entry by gammaretroviruses supposed, by analogy to the hemagglutinin envelope protein of influenza virus (43, 50), that the SU subunit functions as a clamp to suppress the membrane fusion activity of TM (20). In this scheme, binding of RBD to receptor would dissociate the three SU subunits, thereby releasing TM from a kinetically trapped, metastable conformation. Recent studies, however, require a modification of this view. Contrary to expectation, the addition of purified, monomeric RBD to the culture medium restores infection by mutant gammaretroviruses in which membrane fusion is uncoupled from receptor binding by deletion of the His residue in the conserved Ser-Pro-His-Gln motif near the N terminus of the viral RBD (4, 5, 27, 29). This observation excludes a simple model in which receptor binding activates the fusion machinery in TM by merely disrupting the quaternary structure of SU. To account for the transactivation activity of soluble RBD, it was initially proposed that virus binding to receptor primes the target cell for infection (29). Subsequent experiments suggested that receptor binding promotes an interaction between RBD and another region within the SU-TM complex, perhaps inducing conformational changes that result in fusion (4, 5, 27). In any case, RBD has a role in gammaretrovirus infection that follows receptor binding and that does not require the domain to be covalently attached to the virus.
The postbinding activity of soluble RBD extends beyond artificial experimental scenarios. It was recently reported that infection by feline leukemia virus subgroup T (FeLV-T), a T-cell-tropic retrovirus that causes immunodeficiency in cats (39, 41), depends on a “coreceptor” secreted from feline T lymphocytes (2). This factor, named FeLIX, is encoded by a defective endogenous provirus and comprises the RBD of feline leukemia virus subgroup B (FeLV-B) (18). Infection by FeLV-T also requires that the cells express Pit1 (2), the Na+-dependent phosphate symporter that serves as a receptor for FeLV-B (23, 36, 48). Pit1 is expressed more widely in host tissues than is FeLIX, so the cell type specificity of FeLV-T is restricted by FeLIX expression (2). In summary, the discovery of FeLIX indicates that soluble RBD can determine the outcome of retroviral challenge in vivo.
We previously determined by X-ray crystallography the structure of RBD from Friend murine leukemia virus (Fr-MLV), termed Fr-RBD (19), which binds the cationic amino acid transporter mCAT1 (1, 24, 49), and showed that it is a potent activator of membrane fusion by mutant virus envelope proteins in trans (4, 5). In this report, we present the structure of the RBD of FeLV-B, termed FeB-RBD, which binds Pit1, and show that FeB-RBD and Fr-RBD function like FeLIX in the activation of FeLV-T entry into cells. These results provide the opportunity to compare the structures and mechanisms of two RBDs that bind different receptors but trigger fusion and infection by the same gammaretroviruses. Through this comparison, we explore the mechanism of FeLV-T infection in particular and of gammaretrovirus infection in general.
MATERIALS AND METHODS
Protein expression and purification.
RBD proteins were expressed in insect cells by using a baculovirus vector and purified by using a protocol described previously for Fr-RBD (15). Briefly, FeB-RBD was subjected sequentially to nickel chelation chromatography, selective precipitation, cleavage of a 42-amino-acid C-terminal tag by using factor Xa, and purification by anion-exchange chromatography. The yield of purified FeB-RBD was 100 μg/liter of culture medium. N-terminal protein sequencing (Biopolymer Facility, Harvard Medical School) indicated that a 34-amino-acid signal peptide was removed during processing.
Cell lines and virus infection.
Human 293T kidney cells, AH927 feline embryonic fibroblasts (FeF; obtained from William Hardy), and Chinese hamster CHTG-derived cell lines expressing the receptors mouse CAT1 (mCAT1) and human Pit1 (hPit1) were propagated in Dulbecco’s modified Eagle’s medium-10% FCS. Human 293mCAT1 and CHTG-Pit1 cell lines have been described previously (5). The FeFmCAT1 cell line was prepared by selection in medium containing G418 (1 mg/ml) after transfection of 2 μg of pcDNA3-mCAT1, an expression plasmid encoding mCAT1 and neomycin phosphotransferase. Expression of mCAT1 was verified by demonstrating acquired susceptibility to infection by an ecotropic Fr-MLV vector (15).
The production of the retrovirus vectors expressing designated envelope glycoproteins has been described previously (5). Briefly, human 293 cells were transfected with the plasmids pMD.old.gagpol (20 μg) and pBABE-lacZ (20 μg) (33) and a pcDNA3 (Invitrogen)-based expression plasmid encoding the envelope glycoprotein (20 μg). FeLV-T-based retrovirus vectors were prepared by cotransfection of plasmid EECC(ψ−) encoding the packaging-defective FeLV-T provirus (20 μg) (10) and pBABE-lacZ (20 μg). Virus was harvested from supernatant 2 days after transfection and applied to indicator cells for 4 to 6 h. Infection of indicator cells was measured by assaying for acquired β-galactosidase activity 2 days later by incubation in the substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Virus titer was measured by endpoint dilution. FeLV-T virus in which RBD had been deleted was prepared by using a modified EECC(ψ−) plasmid as described previously (5).
Wild-type FeLV-T virus or modified FeLV-T in which RBD had been deleted was prepared by transfection of FeF cells with plasmid EECC (10) containing an intact FeLV-T provirus (20 μg). FeF fibroblasts (5 × 104) were exposed to FeLV-T containing supernatant (10 μl) and FeB-RBD (40 nM). After 4 h, culture medium containing virus was removed and replaced with fresh medium, and FeB-RBD was replenished. After 7 days, the cells were removed with Versene-phosphate-buffered saline and replated (1:3) in fresh medium containing FeB-RBD. FeF cells exposed to FeLV-T, but not to FeB-RBD, served as a control. After 24 h, photomicrographs were prepared by using phase-contrast microscopy at ×400 magnification.
Provirus DNA measurement.
DNA was isolated (DNeasy tissue kit; Qiagen) from infected cells, and the number of acquired proviruses was determined by quantitative real-time PCR by using an iCycler IQ detection system (Bio-Rad). The following primers and probe used for detection of the FeLV provirus are located in gag: forward primer (GGCACGAGCCCATAATCAAG), reverse primer (CCCATTCGGCCTCACATAAG), and probe (6-FAM-TGTCGAGGTCCGGAAAAAGAAATGGATT-BHQ-1; Biosearch Technologies). In a typical assay, 200 ng of genomic DNA was amplified in triplicate (Platinum Quantitative PCR SuperMix-UDG; Invitrogen) in reaction mixtures containing forward and reverse primers (400 nM each) and probe (200 nM). Amplification was performed by denaturing the template at 94°C for 20 s, followed by primer annealing at 60°C (20 s) and extension at 72°C (30 s) for 44 cycles. In each case, a standard curve was generated by spiking uninfected 293mCAT1 cell DNA with linear plasmid DNA containing the target gene at calculated concentrations of 0.01, 0.1, 1, 10, and 100 copies per haploid genome. Calculations were based on an approximation of 3 × 109 bp per haploid human genome. The limit of detection for FeLV provirus DNA is 0.01 provirus copy/haploid cell genome.
Crystallization and structure determination.
Crystals of FeB-RBD (∼0.2 by ∼0.2 by ∼0.2 mm) were first grown by hanging-drop vapor diffusion over a well containing 0.2 M ammonium acetate, 0.1 M trisodium citrate dihydrate (pH 5.6), and 30% (wt/vol) polyethylene glycol 4000 (Hampton Research). Crystals for data collection were grown in a 9:1 (vol/vol) mixture of the above solution and glycerol. Crystals were transferred to a 1:1 (vol/vol) mixture of mineral oil and Parabar oil (Exxon) before flash freezing. Diffraction data were collected to a 2.5-Å resolution at 100 K on the ESRF ID14 1 beamline. Data were processed and scaled by using DENZO and SCALEPACK (38). Molecular replacement was performed by using AmoRe (34) with data to a 3.5-Å resolution. The search model consisted of the following regions of Fr-RBD: residues 9 to 49, 118 to 123, 133 to 165, 184 to 198, and 207 to 236. Residues in these regions that are not identical between the two viruses were changed to Ala in the search model. Original maps calculated from the molecular replacement solution were improved by using “dm” (14) with noncrystallographic symmetry averaging. The FeB-RBD structure was built by using O (http://origo.imsb.au.dk/∼mok/o). Structure refinement was done by using the “Crystallography and NMR System” (9), and no sigma cutoff was applied to the data. Noncrystallographic symmetry restraints were initially present during refinement but were removed when differences in the loop region after strand 4 became apparent. In one of the molecules in the asymmetric unit (molecule B), no residues are in disallowed regions of Ramachandran space. In the other molecule (molecule A), two serine residues (Ser134 and Ser150) at the distal ends of glycine- or serine/threonine-rich loops fall into disallowed regions of Ramachandran space. Although the rough trace of the backbone in the vicinity of these residues was unambiguous in simulated annealing omit maps, the precise geometry of the backbone could not be determined definitively. The β-scaffold regions were compared by using lsqkab (13) for FeB-RBD residues 7 to 16, 24 to 46, 78 to 83, 90 to 124, 161 to 175, and 178 to 205. The corresponding sections of Fr-RBD are residues 8 to 17, 24 to 46, 116 to 121, 131 to 165, 183 to 197, and 201 to 228. Crystallographic and refinement statistics are presented in Table 1.
TABLE 1.
Crystallographic and refinement statistics
| Parameter | Value |
|---|---|
| Temperature (°K) | 100 |
| Wavelength (Å) | 0.934 |
| Space group | P6522 |
| Cell dimensions | |
| a = b (Å) | 93.4 |
| c (Å) | 231.5 |
| Resolution range (Å) (outer bin) | 20-2.5 (2.59-2.5) |
| No. of unique reflections | 21,072 |
| Redundancy | 9 |
| Rsyma | 0.061 (0.192) |
| 〈I〉/〈σ〉 | 25.4 (9.4) |
| Completeness (%) | 99.7 (100) |
| No. of atoms | |
| Total | 3,539 |
| Protein | 3,202 |
| Carbohydrate | 148 |
| Water | 178 |
| Polyethylene glycol | 11 |
| Rwork/Rfreeb | 0.216/0.264 |
| RMSD | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.32 |
| Average B factorsc (Å2) | 47 |
Rsym = _j|Ij-〈I〉|/_|〈I〉| where Ij is the intensity measurement for reflection j, and 〈I〉 is the mean intensity for multiply recorded reflections.
Rwork,free = _‖Fobs|-|Fcalc‖/|Fobs| where the working and free R factors are calculated by using the working and free reflection sets, respectively. The free reflections are from a random 7% of the total.
The average B factors for the two molecules in the asymmetric unit were 44 and 50 Å2.
Coordinates.
The coordinates for FeB-RBD have been deposited in the Protein Data Bank (accession code 1LCS).
RESULTS
FeB-RBD is functionally equivalent to FeLIX.
The N-terminal 245 residues of the SU subunit of FeLV-B correspond to analogous regions of ecotropic Fr-MLV and amphotropic 4070 gammaretroviruses that contain discrete RBDs (6, 7, 15). This segment of FeLV-B SU, which is 28 residues shorter than FeLIX on the C-terminal end and differs at five amino acid positions (2), was expressed in insect cells by using an established protocol (15). The signal peptide was efficiently processed to yield FeB-RBD, a monomeric product of 211 residues that begins with the sequence Asn-Pro-Ser-Pro. Throughout the text, residue numbers refer to the mature FeB-RBD protein.
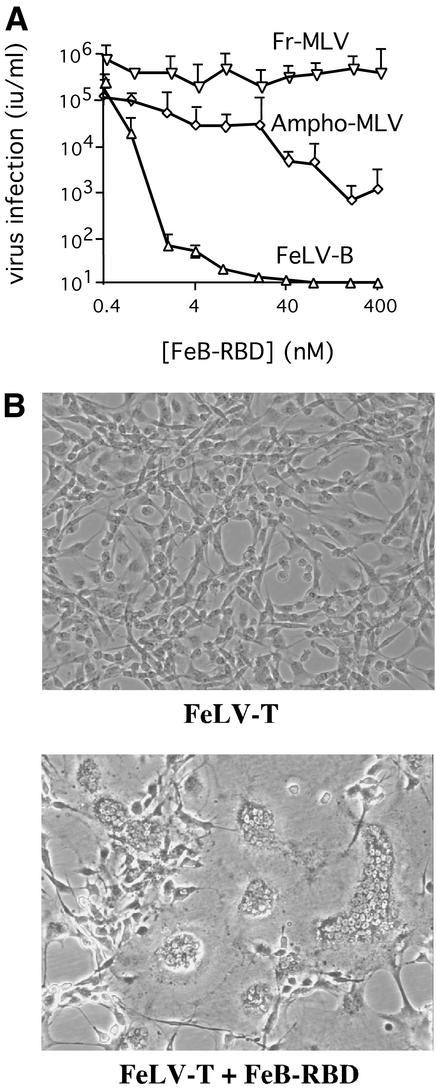
The receptor-binding activity of FeB-RBD was assessed by a competition assay using human 293mCAT1 cells. Addition of increasing concentrations of purified FeB-RBD to the culture medium progressively blocked infection by FeLV-B but not infection by Fr-MLV (Fig. 1A). This observation indicates that FeB-RBD binds to Pit1 and not to mCAT1 receptor. At high concentrations, FeB-RBD also partially blocked infection by amphotropic MLV, indicating that FeB-RBD has a low binding affinity for Pit2 receptor (40).
FIG. 1.
FeB-RBD binds to receptor and activates FeLV-T infection. (A) Recombinant FeB-RBD was expressed and purified. Receptor-binding activity was assessed by a competition assay using FeLV-B, Fr-MLV, and amphotropic MLV. Serial dilutions of each virus were applied to permissive human 293mCAT1 cells, and infection was measured as a function of FeB-RBD concentration (0 to 400 nM) added to the medium at the time of infection. The virus titer was determined byendpoint dilution in triplicate and is expressed as infectious units/milliliter ± one standard error. (B) FeB-RBD (40 nM) was added to the culture medium of one of two flasks containing FeF fibroblasts (5 × 105 cells) exposed to FeLV-T at a multiplicity of infection of 0.01. After 7 days, both the FeB-RBD-treated and the untreated control cells were removed by using Versene-phosphate-buffered saline and then replated (1:3) in fresh medium. Fresh FeB-RBD was added to the medium of the previously treated cells. After a further 24 h, the cells were examined and photographed through a phase-contrast microscope. Magnification, ×400.
The activity of FeB-RBD in supporting replication of FeLV-T was assessed in feline FeF cells, which do not produce FeLIX. Massive syncytium formation and cell death were observed among FeF cells exposed to both FeLV-T and FeB-RBD and then maintained with FeB-RBD in the medium for 7 days (Fig. 1B, bottom). No apparent changes in cell growth or morphology were observed among FeF cells exposed to FeLV-T alone (Fig. 1B, top). Using PCR, the FeLV-T provirus was only detected in genomic DNA from cells treated with both FeLV-T and RBD (data not shown). These studies indicate that, in vitro, FeB-RBD is functionally analogous to FeLIX in supporting FeLV-T replication.
Crystallization and structure determination of FeB-RBD.
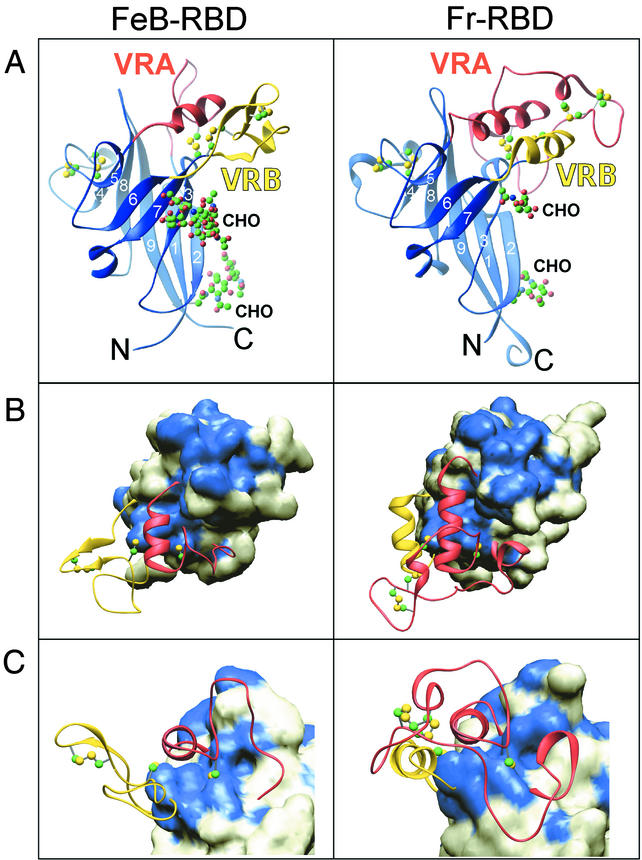
Crystals of FeB-RBD were obtained from a sparse matrix crystallization screen (Hampton Research). Diffraction data were collected to 2.5-Å resolution from crystals grown under modified conditions (see Materials and Methods). The crystals were of space group P6522, with unit cell dimensions a = b = 93.4 Å and c = 231.5 Å and contained two molecules per asymmetric unit. The FeB-RBD structure (Fig. 2A) was solved by molecular replacement by using a search model derived from the β-sandwich of the Fr-RBD structure (125 residues in total) (19). Sequence identity between Fr-RBD and FeB-RBD in this region is 52%; nonidentical residues were replaced by alanine to generate the search model. The remaining 103 residues of Fr-RBD, comprising an associated subdomain formed by two interstrand segments termed variable region A (VRA) and VRB, were omitted from the molecular replacement because both segments differ markedly in length and sequence from the corresponding regions of FeB-RBD.
FIG.2.
FeB-RBD (left) and Fr-RBD (right) structures. This figure was prepared by using Ribbons (11). (A) Ribbon diagram of the two RBD structures, with conserved scaffolds in blue, VRA in red, and VRB in yellow. The strands are numbered according to their order in the primary sequences. Cysteine residues and carbohydrates are shown in a ball-and-stick representation (see Fig. 3). (B) The molecular surfaces of the β-sandwich scaffolds are shown beneath ribbon representations of the variable regions. The view is appoximately from the top of the structure as seen in panel A. Residues on the surfaces of the scaffolds that are identical between FeB-RBD and Fr-RBD are colored blue, and nonidentical positions are white. Cysteine residues in the variable regions are shown in a ball-and-stick representation. Carbohydrates are not shown. (C) The view of the structures rotated 90° around the x axis from their orientations in panel B.
Using maps calculated from the molecular replacement solution, the remaining side chains of FeB-RBD were readily added to the lopsided β-sandwich (a six-stranded sheet packed against a three-stranded sheet) common to Fr-RBD and FeB-RBD (Fig. 2A). Based on the refined FeB-RBD model, the root mean square deviation (RMSD) in Cα positions of β-sandwich residues is 0.87 and 0.80 Å between Fr-RBD and each of the two FeB-RBD molecules in the asymmetric unit. The RMSD for Cα positions between the two FeB-RBD molecules is 0.47 Å over the same residues.
The electron density corresponding to the two N-linked carbohydrate chains in the domain also appeared in the original maps. The position of the first glycosylated Asn residue in each virus (Asn9 in FeB-RBD and Asn12 in Fr-RBD) is conserved. Although the position of the second is not conserved (Asn24 in FeB-RBD versus Asn168 in Fr-RBD), the two Asn residues are separated by <10 Å in the superposed domains, and the carbohydrates cover the concave face of each β-sandwich (Fig. 2A). Two N-acetylglucosamine (GlcNAc) residues were modeled at each of three glycosylation sites in the two molecules in the crystal asymmetric unit. At the fourth site (Asn24 in one of the molecules), a linear chain composed of two GlcNAc residues, two mannose units, and an additional GlcNAc was modeled, although the temperature factors of the last three carbohydrate residues in this chain are high (average B factor, ∼103 Å2).
The electron density for the 30 residues in VRA and the 35 residues in VRB of FeB-RBD was poor in the original maps calculated by using phases from the molecular replacement solution, even after noncrystallographic symmetry averaging. Therefore, the VRA and VRB sections of the FeB-RBD structural model were built iteratively from the ends of the strands during refinement of the β-sandwich scaffold. The final model (Fig. 2), verified by using simulated annealing omit maps, reveals that VRA of FeB-RBD contains a short proline-rich loop followed by a 2.5-turn helix composed entirely of amino acids with long side chains, including three Arg residues. VRB contains a β-hairpin with a turn of sequence Gly-Gly-Gly and a disulfide bond connecting the two strands. The β-hairpin is followed by a 19-residue loop rich in Ser, Thr, and Gly. A disulfide bond between Cys residues from each end closes the loop.
Overall, the FeB-RBD model spans residues 4 to 208. The electron density for residues 1 to 3 and 209 to 211 is not visible, and residues 4 through 6 are in different conformations in the two molecules of the asymmetric unit. In the “A” conformation, the side chain of the critical His5 residue required for activation of fusion interacts with Asp34. In the “B” conformation, steric clashes between Gln6 and a neighboring molecule prevent this interaction. In the context of RBD alone, the conserved Ser-Pro-His-Gln motif is poorly constrained.
Structural differences between receptor-specific subdomains.
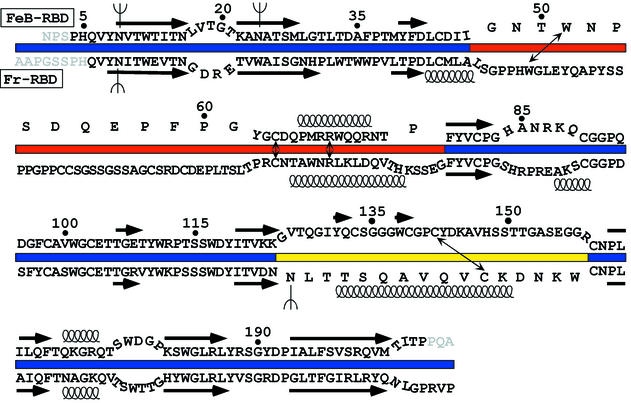
Although the VRA regions from both FeB-RBD and Fr-RBD are composed of a loop followed by a helix, the loops head in opposite directions approximately eight residues after emerging from the β-sandwich (Fig. 2C). In Fr-RBD, the loop is composed of 50 residues (Fig. 3) and encircles the entire subdomain. During this transit, the loop makes at least five hydrogen-bonding or salt bridge interactions with VRB. In contrast, the VRA loop in FeB-RBD is shorter (19 residues), is more compact, and does not contact VRB. The only structurally superposable segments of the polypeptide backbone within VRA are two residues preceding the helix N-cap (Cys64 and Asp65 in FeB-RBD) and the first two turns of the helix. Inspection of the amino acids C terminal to the last Cys residue in VRA of other gammaretroviruses reveals a suitable helix N-cap (Thr or Pro) followed closely by amino acids with high helix propensity (Arg, Lys, Ala, Gln, Leu), suggesting that the putative receptor-binding subdomains of all gammaretroviruses contain a helix in a similar position. Despite this congruity, only a single position in the VRA helices of FeB-RBD and Fr-RBD is occupied by the same amino acid (Arg70 in FeB-RBD and Arg104 in Fr-RBD; Fig. 3). Furthermore, Fr-RBD contains an additional helical turn.
FIG. 3.
Sequence alignment of FeB-RBD (top) (18) with Fr-RBD (bottom) (19). To correspond to the color scheme in Fig. 2, the sequence of the VRA region is displayed astride a red bar, VRB is indicated by a yellow bar, and the β-scaffold is indicated by a blue bar. The sequences of regions with structurally superposable polypeptide backbones are juxtaposed more closely, whereas the sequences from regions that are structurally distinct are separated. Sequences that form β-strands in the conserved scaffolds are indicated by arrows above the FeB-RBD sequence and below the Fr-RBD sequence. Springs represent α-helices. Thin, double-headed arrows point to the four positions in each RBD variable region that are occupied by the same amino acid in the two viruses and have structurally conserved roles. N-linked glycosylation sites are illustrated by forks (Ψ). Some FeB-RBD amino acid residues are numbered as reference points. Residues at the termini that cannot be modeled are shown in gray.
The VRB region of FeB-RBD is longer (35 residues) than that of Fr-RBD (16 residues). In Fr-RBD, VRB contains a helix that packs in antiparallel orientation to the VRA helix. FeB-RBD VRB has no helix; instead, in the analogous position it contains the β-hairpin oriented perpendicularly to the VRA helix (Fig. 2B).
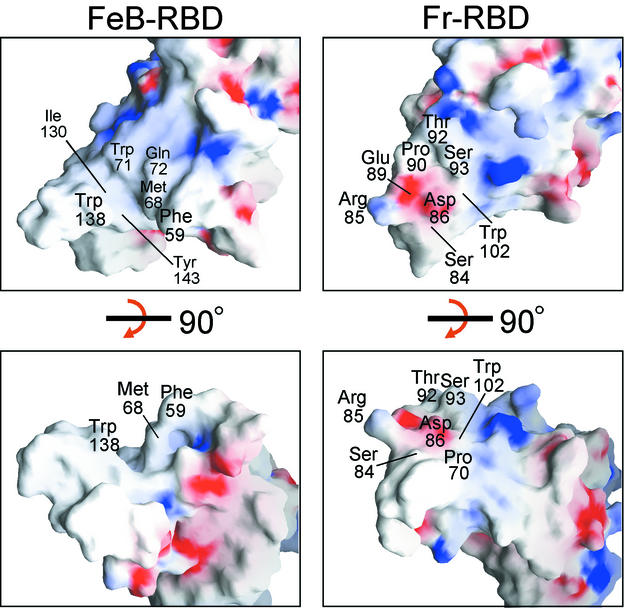
The putative receptor-binding surfaces of FeB-RBD and Fr-RBD are dramatically different (Fig. 4). In Fr-RBD, the side chains of residues Ser84 and Asp86 in the VRA loop and Trp102 in the VRA helix have been implicated in direct binding to mCAT1 (16). In this region of Fr-RBD VRA, the loop passes over the N-terminal end of the helix, forming a roughly convex, hydrophilic surface. In contrast, the VRA helix of FeB-RBD is not covered by the VRA loop and forms the bottom of a deep groove lined with exposed hydrophobic groups, including Phe59, Met68, Trp71, Ile130, Trp138, and Tyr143. Amino acids that modulate receptor binding in FeB-RBD have been localized to this groove (30, 37, 45-47). In the FeB-RBD crystals, Pro58 and Phe59 from a neighboring molecule penetrate into the groove, demonstrating that it can accommodate hydrophobic peptides (Fig. 5A).
FIG. 4.
Surface contours and charge density of FeB-RBD (left panels) compared to Fr-RBD (right panels). In the top panels, the RBDs are viewed in the same orientation as in Fig. 2B. The structures were then rotated 90° around the x axis to obtain the view in the bottom panels, which corresponds to Fig. 2C. Regions of basic potential (>10.6 kBT/e for FeB-RBD, >9.5 kBT/e for Fr-RBD) are in blue; acidic regions (<−9.8 kBT/e for FeB-RBD, −11.2 kBT/e for Fr-RBD) are in red. Some surface-exposed amino acids in the variable subdomains are labeled. The figure was generated by using GRASP (35).
FIG. 5.
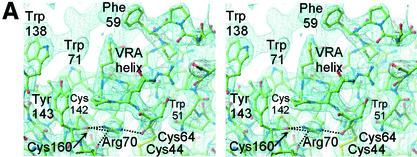
VRA helix and the interface between the receptor-binding surface and the conserved interaction with the β-scaffold. (A) A 2Fo-Fc electron density map calculated with CNS (9) is displayed at 1.2σ around the final model of FeB-RBD. Dotted lines indicate interactions between the guanidino group of Arg70 and backbone carbonyl groups in the vicinity. Unmodeled electron density toward the upper left corresponds to the region containing Pro68 and Phe59 in a symmetry-related molecule. (B) Schematic diagram of the interactions made by the four consensus residues, Trp51, Cys64, Arg70, and Cys142, at the interface between the variable subdomain and the conserved scaffold of FeB-RBD. The view is from the top of panel A. Interactions are shown in projection and are not to scale. Red balls represent backbone carbonyl oxygen atoms, yellow balls are cysteine sulfur atoms, blue balls are side chain nitrogen atoms, and green balls are side chain carbon atoms. Although the side chains of the Arg70 residue in each of the two molecules of the asymmetric unit of FeB-RBD superpose, they make slightly different contacts with adjacent residues. It is not clear whether these differences reflect real alternatives or limitations of the diffraction data. The composite of all interactions is shown in this figure. The corresponding residues in Fr-RBD are marked in Fig. 3.
Although the sequences, lengths, and structures of VRA and VRB differ between Fr-RBD and FeB-RBD (Fig. 3), they compensate for one another in certain aspects. In Fr-RBD, VRA is longer than VRB, but the converse is true in FeB-RBD. In each virus, the longer segment is composed of ∼40% Gly, Ser, and Pro residues. The loop in FeB-RBD VRB overlaps a portion of the circuitous VRA loop in Fr-RBD when the two RBDs are superposed. VRA and VRB together occupy roughly the same region of space in the two viruses, so that the RBDs have similar shape and volume.
Receptor-bound RBDs are functionally interchangeable in transactivation.
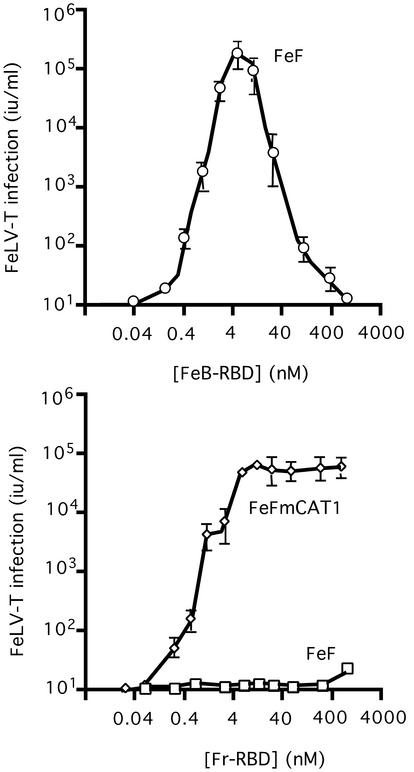
We complemented the structural comparison of FeB-RBD and Fr-RBD with a functional comparison and discovered that, like FeB-RBD, Fr-RBD also supports FeLV-T infection. An FeLV-T vector-based assay (10) was used to measure a single round of infection (Fig. 6). FeLV-T infection of feline FeF cells, which naturally express Pit1 receptor, was supported by FeB-RBD (top panel), and also by Fr-RBD (bottom panel) after expression of mCAT1 receptor. At the optimal concentration (4 nM), FeLV-T infection was only fivefold greater (3 × 105 IU/ml) in cells exposed to FeB-RBD than Fr-RBD (6 × 104 IU/ml). These studies indicate that FeLV-T infection does not strictly depend on a coreceptor specific for Pit1.
FIG. 6.
FeLV-T infection is not strictly dependent on FeLIX. (Top) Feline FeF cells were exposed to serial dilutions of a retroviral vector derived from FeLV-T provirus, EECC(Ψ−) (10), and encoding Escherichia coli β-galactosidase. A specified concentration of purified FeB-RBD (0.04 to 1,000 nM) was added to the medium of each plate. After 4 h, medium containing virus and RBD was replaced with fresh medium. After an additional 48 h, cells expressing β-galactosidase were counted, and the virus titer (in international units/milliliter) was calculated by endpoint dilution. The data are the means ± 1 standard error of three independent experiments. (Bottom) FeLV-T infection was measured as a function of Fr-RBD concentration (0.01 to 1,000 nM) of FeF cells and FeF cells that stably express the Fr-RBD receptor, mCAT1, by using the protocol described for panel A.
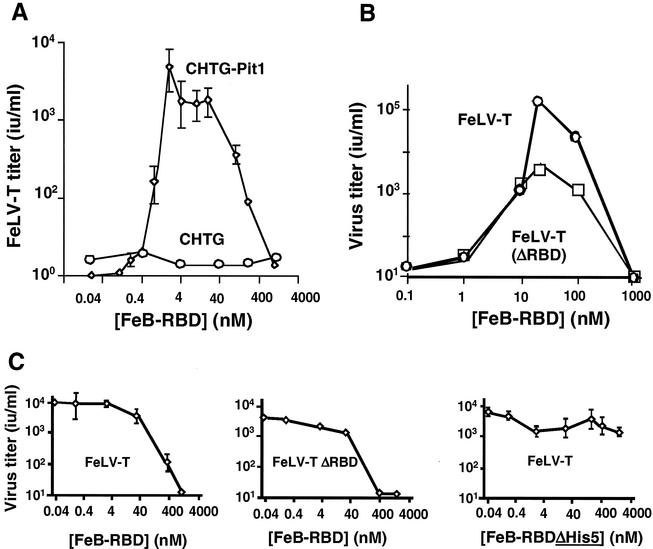
A biphasic relationship between RBD concentration and FeLV-T infection was specifically observed for FeB-RBD but not for Fr-RBD-mediated transactivation. In previous studies, this profile occurred when binding of virus RBD and trans-activating RBD to the same receptor was required for infection (4). This possibility was tested for FeLV-T by using the Chinese hamster-derived cell line CHTG, which is nonpermissive for FeLV-B (Fig. 7A) or Fr-MLV infection (12). FeLV-T infection of these cells was not supported by FeB-RBD until after introduction of a functional human Pit1 receptor (Fig. 7A). On these cells, the relationship between FeB-RBD concentration and FeLV-T infection was also biphasic. We tested the hypothesis that high concentrations of FeB-RBD competitively inhibit FeLV-T infection by blocking binding of FeLV-T RBD to Pit1 or to other cell receptors. RBD was deleted from the FeLV-T envelope glycoprotein, and the profile of FeB-RBD-dependent infection was reexamined on CHTG-Pit1 cells. FeLV-T (ΔRBD) infection was restored by FeB-RBD (Fig. 7B). Peak infection was reduced, but the biphasic relationship between infection and FeB-RBD concentration was observed. This finding indicates that high concentrations of FeB-RBD do not inhibit FeLV-T infection by competing with FeLV-T RBD for binding to Pit1 or to other membrane receptors.
FIG. 7.
Virus and host requirements for FeLV-T infection. (A) FeB-RBD-activated FeLV-T infection is dependent on the Pit1 receptor. CHTG or CHTG-Pit1 cells that stably express human Pit1 receptor were exposed to serial dilutions of a retroviral vector derived from FeLV-T provirus, EECC(Ψ−), and encoding E. coli β-galactosidase. A specified concentration of purified FeB-RBD (0.01 to 1,000 nM) was added to the medium of each plate. After 4 h, medium containing virus and RBD was replaced with fresh medium. After an additional 48 h, cells expressing β-galactosidase were counted, and the virus titer (in international units/milliliter) was calculated by endpoint dilution. The data are the means ± 1 standard error of three independent experiments. (B) The presence of FeLV-T RBD is not required for FeB-RBD-dependent transactivation. The infectious titers of FeLV-T and FeLV-T (ΔRBD) in which RBD has been deleted from the virus envelope glycoprotein were measured as a function of FeB-RBD concentration on CHTG-Pit1 cells as described above. (C) The effect of FeB-RBD concentration on Fr-RBD-dependent FeLV-T infection of CHTG-mCAT1 cells stably expressing the Fr-RBD receptor, mCAT1, was measured. In each example, cells were exposed to virus and Fr-RBD (40 nM) and the indicated concentration of FeB-RBD (left and center panels, 0.04 to 1,000 nM) or FeB-RBD (ΔHis5) in which the His5 residue was deleted (right panel, 0.04 to 4,000 nM). Under these conditions, the infectious titers of FeLV-T (left panel), FeLV-T (ΔRBD) (center panel), and FeLV-T (right panel) were measured as in panels A and B.
A search for additional targets for FeB-RBD was pursued. FeLV-T infection was established on CHTG cells by introduction of mCAT1 receptor and exposure to Fr-RBD. The relationship between infection and Fr-RBD concentration is similar to that observed on FeF-mCAT1 cells (Fig. 6, top panel), but the peak titer is slightly lower (104 IU/ml) (data not shown). Unlike FeLV-T infection mediated by FeB-RBD, a biphasic relationship between infection and Fr-RBD concentration was not observed. FeLV-T infection of CHTG-mCAT1 cells was established by using an optimal concentration of Fr-RBD (40 nM), and the effects of FeB-RBD titration were determined (Fig. 7C). FeB-RBD inhibited Fr-RBD-dependent FeLV-T (left panel) and FeLV-T (ΔRBD) infection (center panel), indicating that FeB-RBD blocks transactivation by Fr-RBD/mCAT1. Since CHTG-mCAT1 cells lack functional receptors for FeB-RBD, we conclude that its target is a postreceptor step in the Fr-RBD-dependent FeLV-T infection mechanism.
The postreceptor function of RBD is dependent on the histidine residue in the conserved Ser-Pro-His-Gln motif near the N terminus (3, 29), and therefore FeB-RBD lacking this residue (ΔHis5) was prepared and tested for inhibitory activity. The recombinant FeB-RBD (ΔH5) protein competitively inhibited FeLV-B infection but did not support FeLV-T infection, thus confirming that it retained Pit1 binding activity but is defective in postbinding activation of the fusion machinery (data not shown). Unlike FeB-RBD, FeB-RBD (ΔH5) did not markedly inhibit Fr-RBD-dependent FeLV-T infection (Fig. 7C, right panel). The measured titer of Fr-RBD-dependent FeLV-T infection remained within 20% of the maximum in the presence of a concentration of FeB-RBD (ΔH5) fourfold greater than required for FeB-RBD (1 μM) to inhibit FeLV-T infection by 1,000-fold. These findings indicate that FeB-RBD, but not FeB-RBD (ΔH5), binds with low affinity to a target that is recognized with high affinity by Fr-RBD/mCAT1 and FeB-RBD/Pit1 during transactivation. The conserved histidine residue (His5 in FeB-RBD and His8 in Fr-RBD) is located on a flexible arm that is located outside the structural domains of both RBDs. In this position, the four-amino-acid segment containing the histidine residue is available for interaction with binding sites on the host cell or viral membrane or, as suggested by mapping studies of transactivation (27), to the C-terminal segment of the FeLV-T envelope glycoprotein SU subunit.
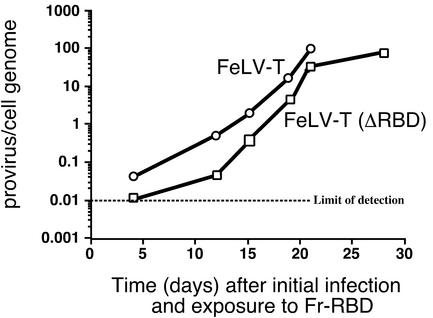
To test the requirements for pathogenesis, FeLV-T was prepared in which RBD was deleted from the envelope glycoprotein and the properties of FeLV-T (ΔRBD) were examined on human 293mCAT1 cells in the presence of Fr-RBD. Replication and spread of FeLV-T (ΔRBD) through the culture were monitored by using quantitative PCR to measure the accumulation of FeLV-T proviruses in cell DNA and also by inspection for cell injury as in Fig. 1A. Replication of FeLV-T (ΔRBD) was supported by Fr-RBD, but the observed rate of provirus accumulation per cell was slightly delayed compared to FeLV-T (Fig. 8). The endpoint of >95% cell-cell fusion (as shown in Fig. 1A) was achieved by FeLV-T-exposed cells in 21 days (101 proviruses/hg) and by FeLV-T (ΔRBD)-exposed cells in 28 days (156 proviruses/hg). Therefore, establishment of transactivation is sufficient for FeLV-T replication and cell-cell fusion, and absolute requirements for FeB-RBD/FeLIX activity and for virus RBD binding to receptor are excluded.
FIG. 8.
Transactivation is sufficient for FeLV-T replication and pathogenesis. Human 293-mCAT1 cells were exposed to either FeLV-T or FeLV-T in which RBD had been deleted from the virus envelope glycoprotein (FeLV-T [ΔRBD]) and cultured in the continuous presence of Fr-RBD in the medium (20 nM). Cells were passaged every 3 or 4 days, and Fr-RBD was replaced. Cells were inspected for virus-induced cell-cell fusion, and the experiment was stopped when >95% of cells were present in large syncytia that did not survive passage (day 21 for FeLV-T and day 28 for FeLV-T [ΔRBD]). At intervals after initial infection, cell DNA was prepared and used as a template for quantitative PCR measurement of acquired FeLV-T proviruses by using specific primers derived from the FeLV-T gag gene. These measurements were calibrated against a standard curve obtained by spiking 293mCAT1 cell DNA with a linear plasmid containing the FeLV-T provirus plasmid, EECC, at calculated concentrations of 0.01, 0.1, 1, 10, and 100 copies/haploid genome. The number of FeLV-T proviruses per 293mCAT1 cell haploid genome is shown (average of triplicate measurements from a single experiment) as a function of number of days after initial exposure to FeLV-T. The lower limit of FeLV-T provirus detection is 0.01 copy per haploid genome.
Adapting variable subdomains to a conserved scaffold and transactivation mechanism.
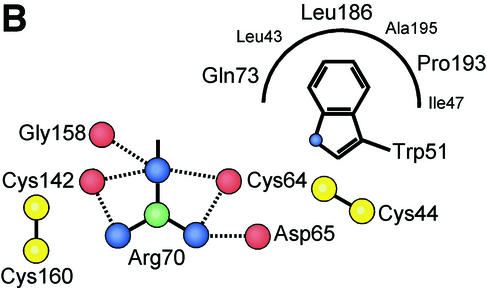
The ability of FeB-RBD and Fr-RBD to bind to distinct receptors but converge upon and activate a conserved membrane fusion mechanism prompted an examination of the interface between the structurally variable and the conserved regions in these domains. In part, this interface is minimized because VRA and VRB project from the ends of β-strands, so they interact largely with each other and less so with the β-scaffold. A few contacts, however, are made between the variable subdomains and the scaffolds. We identified four residues in the variable subdomain with structurally conserved roles (Fig. 5). In FeB-RBD, these residues are Trp51, Cys64, and Arg70 in VRA and Cys142 in VRB. In Fr-RBD, the analogous residues are Trp56, Cys98, Arg104, and Cys178. These residues are aligned along an axis through the variable subdomain that is approximately perpendicular to the axis defined by the strands of the β-sandwich scaffold. The superposable Cys residues make disulfide bonds to conserved Cys residues located at the edge of the scaffold. The key Arg residue occupies the space between the structurally conserved VRA and VRB Cys residues, forming contacts, in one case water mediated, with their backbone carbonyl groups (Fig. 5). The shared Trp residue is on the opposite side of the VRA Cys from the Arg. Its side chain packs into a conserved hydrophobic pocket in the scaffold formed from residues at the C terminus of β-strand 8, at the N terminus of β-strand 9, and following β-strand 3. In the two viruses, the Trp side chains are in different rotamers and are contributed by nonoverlapping regions of the polypeptide backbone. For this reason, the designation of this Trp as a consensus residue based on alignment of γ-retroviral amino acid sequences (7) was fortuitous. In fact, the shared Arg residue has been repeatedly misaligned with neighboring Arg residues (7; J. M. Cunningham, unpublished observations). The coincidence of the Trp, Arg, and two Cys residues in otherwise-variable regions of all gammaretrovirus RBDs, as well as their locations and contacts in the FeB-RBD and Fr-RBD structures, indicate that they constitute a conserved motif that attaches the receptor-specific subdomains to the scaffolds.
DISCUSSION
Structural basis for receptor diversity in mammalian leukemia viruses.
Gammaretroviruses are distinguished by differences in receptor usage and pathogenicity (40, 44). The first atomic-resolution comparison of receptor binding domains from two gammaretroviruses that bind to distinct receptors is described here. We report the structure of the RBD from FeLV-B and compare it to the previously determined structure of the analogous domain from Fr-MLV (19). For each, receptor specificity is determined by a subdomain formed by two loops (VRA and VRB) that protrude from a conserved scaffold. The sequence and organization of these loops in each RBD create dramatically different binding surfaces. In Fr-RBD, a prominent feature is two antiparallel α-helices, one in VRA and one in VRB, that interact extensively. In contrast, VRB in FeB-RBD lacks a helix and does not contact VRA. The predominant structural feature in FeB-RBD is a deep groove lined with the side chains of residues in VRA and in VRB that are implicated in binding to Pit1 or related Pit2 receptors (8, 32, 37, 45-47). The formation of receptor-specific subdomains by the two loops that extend from the scaffold indicates how receptor diversity is accommodated within the otherwise conserved structural framework of mammalian leukemia virus envelope glycoproteins.
Overlapping target for activation of virus fusion by distinct RBDs.
Left unanswered by this structural comparison is how receptor binding triggers the activation of the fusion mechanism, which is conserved among these viruses. An insight into this problem has been provided by studies of FeLV-T infection. Specifically, we confirm that FeLV-T is defective, and we report that infection is restored by the addition of either FeB-RBD or Fr-RBD to the cell culture medium. Transactivation of FeLV-T infection depends on the binding of each RBD to its cognate receptor and to a common or overlapping target. The latter conclusion is based on the observation that high concentrations of FeB-RBD block Fr-RBD/mCAT1-dependent transactivation. The target for inhibition is likely to be located on the non-RBD portion of the FeLV-T envelope glycoprotein, but the possibility that the target is an unidentified cofactor that functions in concert with RBD is not excluded. In summary, mammalian leukemia virus RBDs function as transducers that create a common signal for activation of fusion through binding to one of a number of distinct receptors.
Properties of FeLV-T that favor transactivation.
The studies of FeB-RBD were motivated by the striking observation of Andersen et al. that FeLV-T infection and pathogenesis are strictly limited to feline T cells that secrete the coreceptor FeLIX (2). FeLIX is a truncated SU subunit containing an intact RBD encoded by an endogenous FeLV-B provirus. The dependence of FeLV-T on FeLIX was reminiscent of the behavior of mutant viruses for which infection depends on provision of RBD in trans (4, 27, 29). Indeed, inspection of the FeLV-T envelope glycoprotein sequence reveals that it lacks a critical histidine residue in the otherwise conserved Ser-Pro-His-Gln motif near the N terminus of SU that is absolutely required for activation of fusion by other mammalian leukemia viruses (3). Mutant viruses lacking the conserved histidine residue bind to surface receptors (4) but fail to infect in the absence of a wild-type RBD. We demonstrate that FeB-RBD, like FeLIX, restores FeLV-T infection. The amino acid sequence of the RBD portion of FeLIX differs from FeB-RBD at five positions that can be placed on the structure. Three are conservative changes at positions in the scaffold (I10V, L39M, and K112R). Both of the remaining changes are located in the VRA portion of the receptor-specific subdomain. The first, G55D, is in the loop, and the second, A77P, is at the C-terminal cap of the helix, adjacent to the N-terminal residue of the fifth β-strand. In our studies, FeB-RBD transactivation activity, like that of FeLIX, is strictly dependent on Pit1 receptor expression. However, it remains possible that a subtle difference in RBD receptor specificity caused by these changes was not recognized under the conditions of our experiments.
It has been reported that SU subunits from other mammalian leukemia viruses, including some that bind to Pit1 or Pit2 receptors, do not support FeLV-T infection (25). However, the observation that Fr-RBD activates FeLV-T infection excludes an essential requirement for FeLIX/FeB-RBD. Our studies of FeB-RBD indicate that significant activation of FeLV-T infection occurs only within an intermediate range of RBD concentrations, raising the possibility that previous studies of other Pit1 binding proteins may have been performed outside this window. Moreover, successful transactivation is likely to depend on a number of additional factors, including receptor density on the surface membrane, affinity of virus and RBD for receptor and for each other, and possible confounding effects of virus RBD (4, 5, 27, 29). In our previous studies of mutant viruses, the transactivation activity of Fr-RBD exceeded that of other RBDs (4, 5). However, FeB-RBD-dependent FeLV-T infection is 5- to 50-fold greater than infection mediated by the optimal concentration of Fr-RBD. This observation indicates that the transactivation mechanism of FeLV-T is primed for efficient activation by FeB-RBD, possibly reflecting selection of FeLV-T in the presence of FeLIX. These studies indicate that FeLIX-dependent FeLV-T infection and pathogenesis is a naturally occurring example of the function of the transactivation mechanism revealed by the studies of mutant viruses in vitro (4, 5, 27, 29).
FeLV-T receptor requirement for transactivation.
The initial studies of defective mutant viruses indicated that virus binding to receptor is a prerequisite for transactivation (4, 29, 30). It has been proposed that Pit1 is the receptor for FeLV-T, as well as FeLIX (26). If true, the hamster isoform of Pit1 receptor must bind to the FeLV-T RBD during Fr-RBD/mCAT-dependent transactivation. Direct studies of hamster Pit1 will be required for a rigorous test of this conclusion. However, we have observed that infection of nonpermissive hamster cells by the mink cell focus-forming (MCF) class of mammalian leukemia viruses is conferred by expression of its receptor or, alternatively, by expression of mCAT1 receptor and exposure to Fr-RBD (D. L. Wensel, unpublished data). These experiments indicate that, unlike the mutant viruses studied previously, MCF virus binding to receptor is not required for transactivation of infection. Remarkably, viruses in the closely related xenotropic class of mammalian leukemia viruses are not targets for transactivation in the absence of receptor binding. Susceptibility to transactivation correlates with a deletion of six amino acid residues in the loop region of VRA of MCF, but not xenotropic, viruses that is associated with reduced binding affinity for receptor (Wensel, unpublished). A strikingly similar deletion is also present in the equivalent region of the VRA loop of FeLV-T (residues 95 to 100) compared to the closely related FeLV-A (61E) virus that is not susceptible to FeLIX (41). Based on these findings, we speculate that FeLIX-dependent FeLV-T infection may not require binding of FeLV-T to a cellular receptor. Consistent with this hypothesis, binding of FeLV-T SU to membranes of susceptible cells expressing functional Pit1 receptors has not been detected (26).
Possible relationship between transactivation and pathogenesis.
In addition to the changes in RBD, the C-terminal segment of FeLV-T SU contains a six amino acid insertion (DYLTAP388-393) that correlates with enhanced sensitivity to infection and pathogenesis (21, 41). It has been recently reported that exchange of small regions of the C-terminal segment of SU between closely related viruses sensitizes these chimeric proteins to transactivation, likely by destabilizing the prefusion conformation of the virus fusion machinery (28). Indeed, a low level of receptor-independent infection by these viruses was observed. We speculate that the loss of the critical histidine residue may have occurred in FeLV-T to prevent premature activation of a hair trigger fusion mechanism created by the destabilizing effect of the DYLTAP388-393 insertion into the C-terminal segment. In addition, the apparent destabilizing effect of the C-terminal insertion may accompany an increased affinity for binding to RBD and thereby explain the inhibitory effect of FeB-RBD on transactivation of FeLV-T infection by Fr-RBD/mCAT1.
At present, it is unclear if the proposed low-affinity binding of FeB-RBD to FeLV-T is reversible or if binding activates the virus fusion mechanism. In either case, the changes in FeLV-T compared to its FeLV-A parent reflect adaptation to a transactivation mechanism at the expense of a receptor-dependent mechanism. Consistent with this conclusion, the loss of receptor binding caused by deletion of RBD has little consequence for FeLV-T replication and cytopathicity on human 293 cells established by Fr-RBD/mCAT1-dependent transactivation. Therefore, binding of FeLV-T to receptor is not required for its replication or pathogenicity. One consequence of an entry mechanism that does not require direct binding of FeLV-T to receptor is that the subsequent production of its envelope glycoprotein by the infected cell will not prevent additional infection by downregulating receptor. This consequence of transactivation likely explains the acquisition of 50 to 100 FeLV-T proviruses/cell and the associated high level of envelope glycoprotein expression that causes cell-cell fusion (17, 41). It must be stated, however, that transactivation of other viruses lacking the critical N-terminal histidine residue is strictly dependent on receptor binding (4, 29, 30). Therefore, the role of virus binding to receptor in transactivation of FeLV-T infection remains an unresolved issue.
Evolution of receptor-dependent activation domains.
The integral role of the conserved N-terminal His residue in initiating membrane fusion and its location outside the RBD fold in multiple viruses suggest that, in the intact envelope glycoprotein, this residue is not truly part of the RBD but rather is part of the membrane fusion machinery that also includes TM and the C-terminal region of SU. The proximity of the first and last β-strands of the scaffold is consistent with the proposal that the SU subunit of gammaretroviruses may have arisen by insertion of RBD into an ancestral fusogenic protein composed of TM, the C-terminal portion of SU, and the N-terminal amino acids of SU, including the His residue. This scheme is analogous to the proposed evolution of the influenza virus hemagglutinin by insertion of the sialic acid-binding domain into a loop of the stem domain (42). Once a progenitor RBD was introduced into gammaretroviruses, the host range of these viruses may then have expanded by diversification of VRA and VRB. Our structural studies on Fr-RBD and FeB-RBD, as well as a comparison of RBD sequences from other gammaretroviruses, suggest that diversification of receptor specificity has been subject to certain structural constraints, which may include the presence of the VRA helix and the preservation of the four residues linking the variable subdomain to the conserved scaffold. It remains to be determined whether these apparent constraints reflect the conservation of a key functional element required for coupling of receptor binding to fusion activation or simply an architectural requirement for the folding and assembly of functional RBDs. In either case, it is notable that all gammaretrovirus receptors identified to date are multiple pass transmembrane proteins that, for the cases that have been studied, transport small molecules (40). Despite these shared properties, these receptors do not contain homologous sequences and are encoded by distinct genes even in prokaryotes. Therefore, it is unlikely that gammaretrovirus subgroups evolved by coexpansion of a progenitor receptor and virus. One proposal is that the similarities among gammaretrovirus receptors are indicative of their location in a restricted membrane compartment (31) enriched in a cofactor required for the activation step. A surface-exposed cluster of conserved aromatic residues adjacent to VRA in both FeB-RBD (Trp101, Tyr110, Trp111, and Tyr187) and Fr-RBD is an intriguing candidate either for interaction with membrane-bound components or for critical quaternary structural contacts involved in fusion activation.
Acknowledgments
We thank Maria Anderson and Julie Overbaugh for the EECC plasmids, Qing Yao for technical assistance, and Jason Smith and James Berger for suggestions on the manuscript.
This work was supported by the Howard Hughes Medical Institute (J.M.C.) and the Leukemia Research Foundation (D.F.). David Wensel is a Howard Hughes Medical Institute Predoctoral Fellow.
REFERENCES
- 1.Albritton, L. M., L. Tseng, D. Scadden, and J. M. Cunningham. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659-666. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, M. M., A. S. Lauring, C. C. Burns, and J. Overbaugh. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828-1830. [DOI] [PubMed] [Google Scholar]
- 3.Bae, Y., S. M. Kingsman, and A. J. Kingsman. 1997. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J. Virol. 71:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnett, A. L., and J. M. Cunningham. 2001. Receptor binding transforms the surface subunit of the mammalian C-type retrovirus envelope protein from an inhibitor to an activator of fusion. J. Virol. 75:9096-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnett, A. L., R. A. Davey, and J. M. Cunningham. 2001. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc. Natl. Acad. Sci. USA 98:4113-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battini, J. L., O. Danos, and J. M. Heard. 1995. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J. Virol. 69:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battini, J. L., J. M. Heard, and O. Danos. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J. Virol. 66:1468-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boomer, S., M. Eiden, C. C. Burns, and J. Overbaugh. 1997. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J. Virol. 71:8116-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54(Pt. 5):905-921. [DOI] [PubMed] [Google Scholar]
- 10.Burns, C. C., M. Moser, J. Banks, J. P. Alderete, and J. Overbaugh. 1996. Identification and deletion of sequences required for feline leukemia virus RNA packaging and construction of a high-titer feline leukemia virus packaging cell line. Virology 222:14-20. [DOI] [PubMed] [Google Scholar]
- 11.Carson, M. 1997. Ribbons. Methods Enzymol. 277:493-505. [PubMed] [Google Scholar]
- 12.Chaudry, G. J., K. B. Farrell, Y. T. Ting, C. Schmitz, S. Y. Lie, C. J. Petropoulos, and M. V. Eiden. 1999. Gibbon ape leukemia virus receptor functions of type III phosphate transporters from CHOK1 cells are disrupted by two distinct mechanisms. J. Virol. 73:2916-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collaborative Computational Project. 1994. The CCP4 suite programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 14.Cowtan, K. 1994. A ccp4 density modification package. Joint CCP4 ESF-EACBM Newsl. Protein Crystallogr. 31:34-38. [Google Scholar]
- 15.Davey, R. A., C. A. Hamson, J. J. Healey, and J. M. Cunningham. 1997. In vitro binding of purified murine ecotropic retrovirus envelope surface protein to its receptor, mCAT-1. J. Virol. 71:8096-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davey, R. A., Y. Zuo, and J. M. Cunningham. 1999. Identification of a receptor-binding pocket on the envelope protein of Friend murine leukemia virus. J. Virol. 73:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donahue, P. R., S. L. Quackenbush, M. V. Gallo, C. M. de Noronha, J. Overbaugh, E. A. Hoover, and J. I. Mullins. 1991. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J. Virol. 65:4461-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elder, J. H., and J. I. Mullins. 1983. Nucleotide sequence of the envelope gene of Gardner-Arnstein feline leukemia virus B reveals unique sequence homologies with a murine mink cell focus-forming virus. J. Virol. 46:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fass, D., R. A. Davey, C. A. Hamson, P. S. Kim, J. M. Cunningham, and J. M. Berger. 1997. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science 277:1662-1666. [DOI] [PubMed] [Google Scholar]
- 20.Fass, D., and P. S. Kim. 1995. Dissection of a retrovirus envelope protein reveals structural similarity to influenza hemagglutinin. Curr. Biol. 5:1377-1383. [DOI] [PubMed] [Google Scholar]
- 21.Gwynn, S. R., F. C. Hankenson, A. S. Lauring, J. L. Rohn, and J. Overbaugh. 2000. Feline leukemia virus envelope sequences that affect T-cell tropism and syncytium formation are not part of known receptor-binding domains. J. Virol. 74:5754-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hull, R. 2001. Classifying reverse transcribing elements: a proposal and a challenge to the International Committee on Taxonomy of Viruses. Arch. Virol. 146:2255-2261. [DOI] [PubMed] [Google Scholar]
- 23.Kavanaugh, M. P., D. G. Miller, W. Zhang, W. Law, S. L. Kozak, D. Kabat, and A. D. Miller. 1994. Cell surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc. Natl. Acad. Sci. USA 91:7071-7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, J. W., E. I. Closs, L. M. Albritton, and J. M. Cunningham. 1991. Transport of cationic amino acids by the mouse ecotropic retrovirus receptor. Nature 352:725-728. [DOI] [PubMed] [Google Scholar]
- 25.Lauring, A. S., M. M. Anderson, and J. Overbaugh. 2001. Specificity in receptor usage by T-cell-tropic feline leukemia viruses: implications for the in vivo tropism of immunodeficiency-inducing variants. J. Virol. 75:8888-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauring, A. S., H. H. Cheng, M. V. Eiden, and J. Overbaugh. 2002. Genetic and biochemical analyses of receptor and cofactor determinants for T-cell-tropic feline leukemia virus infection. J. Virol. 76:8069-8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavillette, D., B. Boson, S. J. Russell, and F. L. Cosset. 2001. Activation of membrane fusion by murine leukemia viruses is controlled in cis or in trans by interactions between the receptor-binding domain and a conserved disulfide loop of the carboxy terminus of the surface glycoprotein. J. Virol. 75:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavillette, D., A. Ruggieri, B. Boson, M. Maurice, and F. L. Cosset. 2002.. Relationship between SU subdomains that regulate the receptor-mediated transition from the native (fusion-inhibited) to the fusion-active conformation of the murine leukemia virus glycoprotein. J. Virol. 76:9673-9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavillette, D., A. Ruggieri, S. J. Russell, and F. L. Cosset. 2000. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J. Virol. 74:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavillette, D., S. J. Russell, and F. L. Cosset. 2001. Retargeting gene delivery using surface-engineered retroviral vector particles. Curr. Opin. Biotechnol. 12:461-466. [DOI] [PubMed] [Google Scholar]
- 31.Lu, X., and J. Silver. 2000. Ecotropic murine leukemia virus receptor is physically associated with caveolin and membrane rafts. Virology 276:251-258. [DOI] [PubMed] [Google Scholar]
- 32.Lundorf, M. D., F. S. Pedersen, B. O'Hara, and L. Pedersen. 1998. Single amino acid insertion in loop 4 confers amphotropic murine leukemia virus receptor function upon murine Pit1. J. Virol. 72:4524-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titer retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navaza, J. 1994. Amore: an automated package for molecular replacement. Acta Crystallogr. A 50:157-163. [Google Scholar]
- 35.Nichols, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 36.Olah, Z., C. Lehel, W. B. Anderson, M. V. Eiden, and C. A. Wilson. 1994. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J. Biol. Chem. 269:25426-25431. [PubMed] [Google Scholar]
- 37.O'Reilly, L., and M. J. Roth. 2000. Second-site changes affect viability of amphotropic/ecotropic chimeric enveloped murine leukemia viruses. J. Virol. 74:899-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 39.Overbaugh, J., P. R. Donahue, S. L. Quackenbush, E. A. Hoover, and J. I. Mullins. 1988. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science 239:906-910. [DOI] [PubMed] [Google Scholar]
- 40.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65:371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohn, J. L., M. S. Moser, S. R. Gwynn, D. N. Baldwin, and J. Overbaugh. 1998. In vivo evolution of a novel, syncytium-inducing and cytopathic feline leukemia virus variant. J. Virol. 72:2686-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenthal, P. B., X. Zhang, F. Formanowski, W. Fitz, C. H. Wong, H. Meier-Ewert, J. J. Skehel, and D. C. Wiley. 1998. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature 396:92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 44.Sommerfelt, M. A., and R. A. Weiss. 1990. Receptor interference groups of 20 retroviruses plating on human cells. Virology 176:58-69. [DOI] [PubMed] [Google Scholar]
- 45.Sugai, J., M. Eiden, M. M. Anderson, N. Van Hoeven, C. D. Meiering, and J. Overbaugh. 2001. Identification of envelope determinants of feline leukemia virus subgroup B that permit infection and gene transfer to cells expressing human Pit1 or Pit2. J. Virol. 75:6841-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tailor, C. S., and D. Kabat. 1997. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J. Virol. 71:9383-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tailor, C. S., A. Nouri, and D. Kabat. 2000. A comprehensive approach to mapping the interacting surfaces of murine amphotropic and feline subgroup B leukemia viruses with their cell surface receptors. J. Virol. 74:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeuchi, Y., R. G. Vile, G. Simpson, B. O'Hara, M. K. Collins, and R. A. Weiss. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J. Virol. 66:1219-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, H., M. P. Kavanaugh, R. A. North, and D. Kabat. 1991. Cell surface receptor for ecotropic murine retroviruses is a basic amino acid transporter. Nature 352:729-731. [DOI] [PubMed] [Google Scholar]
- 50.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]