Abstract
Muscle is an attractive target for gene delivery because of its mass and because vectors can be delivered in a noninvasive fashion. Adeno-associated virus (AAV) has been shown to be effective for muscle-targeted gene transfer. Recent progress in characterization of AAV serotype 1 (AAV1) and AAV6 demonstrated that these two AAV serotypes are far more efficient in transducing muscle than is the traditionally used AAV2. Since all cis elements are identical in these vectors, the potential determinants for their differences in transducing muscle appear to be located within the AAV capsid proteins. In the present study, a series of AAV capsid mutants were generated to identify the major regions affecting AAV transduction efficiency in muscle. Replacement of amino acids 350 to 736 of AAV2 VP1 with the corresponding amino acids from VP1 of AAV1 resulted in a hybrid vector that behaved very similarly to AAV1 in vitro and in vivo in muscle. Characterization of additional mutants carrying smaller regions of the AAV1 VP1 amino acid sequence in the AAV2 capsid protein suggested that amino acids 350 to 430 of VP1 function as a major tissue tropism determinant. Further analysis showed that the heparin binding domain and the major antigenic determinants in the AAV capsid region were not necessary for the efficiency of AAV1 transduction of muscle.
Adeno-associated virus (AAV) is a defective parvovirus that requires a helper virus to complete its lytic infection. Its single genome contains approximately 4,700 nucleotides flanked by two palindromic inverted terminal repeats (ITRs). The ITR is the only AAV sequence retained in the recombinant AAV vector, which is a sufficient cis element for AAV replication, rescue, packaging, and integration. The AAV genome contains two open reading frames encoding four Rep and three Cap proteins. All three structural proteins, as well as the Rep proteins, share identical sequences in their C termini. In packaged virus, VP3 is the major component and the ratio of VP1, VP2, and VP3 is close to 1:1:8 (2, 20).
Currently, there are six known AAV serotypes (19). AAV serotype 2 (AAV2), AAV3, and AAV5 are believed to be of human origin since antibodies against them are quite prevalent in the human population. In contrast, antibodies to AAV1 and AAV4 are very common in nonhuman primates (21). Newly isolated AAV6 appears to be a hybrid recombinant between AAV1 and AAV2 (25, 35); however, its origin remains unknown. Although initial gene transfer studies of AAV were largely based on AAV2, recent studies revealed that other serotypes showed a better performance than did AAV2 in different tissues (1, 4, 7, 9, 23, 35). In particular, AAV1 is a much more efficient vector for gene delivery to muscle than is AAV2. This phenomenon was first observed by initial studies using human α1-antitrypsin and erythropoietin genes as reporter genes and subsequently confirmed in the study by Chao et al. using the factor IX gene as the reporter gene (4, 35). The improvement of AAV1 over AAV2 is approximately 10- to 100-fold with the same genome dose and the identical vector construct. This dramatic increase cannot be ascribed to the viral DNA packaged into the capsid because all the vectors are pseudotyped AAV vectors with the AAV2 ITR. Since the rep gene used for AAV1 vector production was also of AAV2 origin, it would not have contributed to such differences in transduction even if the Rep proteins were associated with packaged virions in some ways (16). Thus, the only possible viral elements contributing to such enhancement are located in the AAV1 capsid.
Although we mention only the VP1 protein in describing the results in this study, those mutations would also appear in VP2 and/or VP3 depending on the location of the mutation in VP1. VP1 is only a minor structural protein in the AAV capsid. The major component is actually VP3. However, since all three proteins share the same reading frame, the alterations in various segments would have caused similar changes in VP2 and/or VP3. Changes in vector performance would have to be combined effects of VP1, VP2, and VP3.
The capsid gene of the virus determines the properties of viral particles such as tissue tropism and antigenic properties. The infectivities of AAV1 and AAV2 in muscle are drastically different, although the capsid genes of AAV1 and AAV2 share considerable homology and identity. As shown in Fig. 1B, 83.3% of amino acids in VP1 are identical and 88.9% of them are similar between AAV1 and AAV2 (27, 35). The nonhomologous amino acids consist of only 11.1% of the total amino acid sequences. Interestingly, these nonhomologous amino acids are not uniformly distributed throughout the AAV VP1 gene. They form clusters in several regions exhibiting greater diversities: amino acids 22 to 43, 136 to 163, 191 to 208, 448 to 477, 547 to 558, and 576 to 602. Our hypothesis is that those nonidentical amino acids determine not only antigenic properties of different serotypes but also tissue tropism and hence their performance in transduction. The remarkable differences between AAV1 and AAV2 in transducing muscle provided a possible assay to identify those amino acids. To test this hypothesis, our approach was to move individual regions from AAV1 and AAV2 to make hybrid AAV2/1 capsid vectors and then compare their performance to those of AAV1 vector and AAV2 vector in muscle. Since a series of plasmids had to be constructed, a universal system to name them was adopted (Fig. 1A). In this new naming system, the origins of the helper's rep sequence and cap sequences are clearly identified by numbers following the letter H. The original AAV1 and AAV2 helpers, p5E18 and p5E18(AV1), are therefore named pH21 and pH22, respectively (35). AAV2 helper pH22 contains both rep and cap genes from AAV2, whereas pH21 contains an AAV2 rep and an AAV1 cap gene.
FIG. 1.
(A) Illustration of the nomenclature for hybrid vectors. “H” is abbreviated from “helper.” The first numeral indicates the source of the rep gene. “2” means that the rep sequence is from AAV2. The second numeral indicates the source of the cap gene. “2” means that the cap sequence is from AAV2. The third numeral is the source of the hybrid cap sequence. The number after the hyphen indicates the exact sequence of the hybrid region. (B) Alignment of primary protein sequences of AAV1 and AAV2. Nonidentical amino acids are highlighted in the figure. The roman numbers indicate regions that were swapped between AAV1 and AAV2: I to VIII represent amino acids 1 to 112, 113 to 155, 156 to 212, 213 to 423, 424 to 480, 481 to 564, 565 to 669, and 670 to 736, respectively. Heparin binding regions are also marked.
For plasmid construction, we used PCR techniques to exchange fragments of the AAV1 capsid gene with the corresponding sequences of the AAV2 capsid gene. Plasmids pH21 and pH22 were used as templates for all other constructs. To generate pH221-1-351, a pH22 HindIII-BsiWI fragment, containing the N-terminal sequences of the AAV2 Cap protein, was excised and replaced with the corresponding fragment from pH21. The same was done vice versa to construct vector pH221-352-736. Hybrid vectors containing smaller regions of the AAV1 cap gene were generated by PCR amplification (Expand Long Template PCR system; Roche) of the desired sequences, representing the following AAV1 VP1 domains: amino acids 1 to 112, 113 to 155, 156 to 212, 213 to 423, 424 to 480, 481 to 564, 565 to 669, and 670 to 736 (Fig. 1B). Primers overlapping at the 5′ and 3′ ends of the amplified fragments were used to amplify the flanking sequences toward N and C termini, respectively, from the AAV2 VP1 gene, to obtain a fragment containing a complete cap gene. The resulting fragments were digested with HindIII and XbaI and replaced with the unmodified HindIII-XbaI fragment from pH22, which resulted in the pH221 series. To generate pH221-350-423, the HindIII-BsiWI fragment of pH221-213-423 was replaced with the HindIII-BsiWI fragment of pH22. The pH212 series helper plasmids were constructed by the same strategy. All plasmids were sequenced and confirmed to ensure that there were no mutations generated by the PCR.
The N terminus of VP1 does not contribute to AAV1 tissue tropism for muscle.
The first two vectors tested were pH221-1-351 and pH221-352-736. The plasmid pH221-1-351 carried a hybrid cap gene with amino acids 1 to 351 from AAV1 while pH221-352-736 carried a hybrid cap gene with amino acids 352 to 736 from AAV1. Recombinant AAV vectors with the human factor IX gene under the control of the cytomegalovirus (CMV) promoter were produced from AAV1 helper pH21, AAV2 helper pH22, pH221-1-351, and pH221-352-736. The vectors were produced by a modified triple plasmid transfection based on calcium phosphate precipitation, which has been described previously (3, 35). Vectors were purified by two rounds of CsCl gradient centrifugation, dialyzed against phosphate-buffered saline (PBS), and stored in PBS with 3% glycerol. To ensure that the same amounts of vectors were used for comparison, vector genome titers were determined by either slot blot hybridization with transgene probes or real-time quantitative PCR with the PRISM/7700 sequence detector (PE Applied Biosystems, Foster City, Calif.) (3). For real-time PCR titration of recombinant AAV preparations, the primer and fluorescent probe sets were selected from the factor IX gene. Primers and internal probe were designed to amplify 134 bp of the human factor IX sequence. The oligonucleotides with sequences TTC GAT CTA CAA AGT TCA CCA TCT ATA AC and AAA CTG GTC CCT TCC ACT TCA G were used as forward and reverse primers, respectively, and the sequence (5′→3′) 6-FAM-AAT CTC TAC CTC CTT CAT GGA AGC CAG CA-TAMRA tagged with 6-FAM fluorescent dye at the 5′ end and TAMRA quencher at the 3′ end was used as a probe. Reactions were performed according to the manufacturer's instructions. The final reaction mix consisted of 900 nM (each) primer; 200 nM probe; 200 M dATP, dCTP, and dGTP; 400 M dUTP; 3.5 mM MgCl2; 8% glycerol; and 1 U of uracil-N-glycosylase in 1× Taqman buffer containing the reference dye ROX and 0.25 U of AmpliTaq Gold polymerase (Perkin-Elmer, Norwalk, Conn.) in a total volume of 25 μl. The cycling conditions consisted of 40 cycles of 95°C for 15 s and 60°C for 1 min. AAV vector DNA samples were prepared by proteinase K digestion overnight, followed by heat inactivation at 100°C for 20 min.
To avoid the complication of the immune response, immunodeficient CD4-knockout mice were used for human factor IX expression in muscle. The performance of these vectors was studied by administering 1011 particles per animal to CD4-knockout mice intramuscularly. Blood samples obtained at various time points by retro-orbital bleeding were assayed for circulating human factor IX in mouse plasma. Levels of human factor IX were measured by enzyme-linked immunosorbent assay (ELISA) as described previously (15). Representative results from week 4 data are shown in Fig. 2A.
FIG. 2.
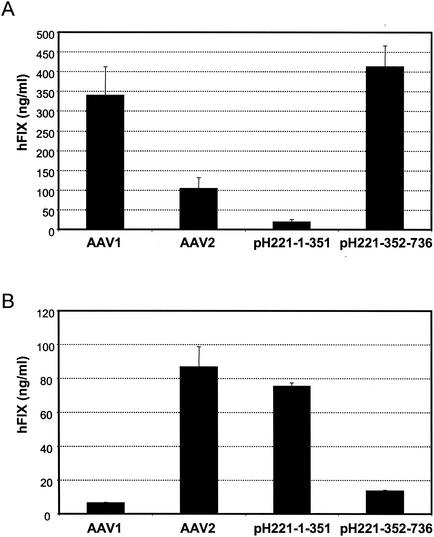
(A) Performance of vectors based on pH221-1-351 and pH221-352-736 in vivo in muscle. Vectors with the human factor IX gene made from AAV1 helpers pH21, pH221-1-351, and pH221-352-736 and AAV2 pH22 were administered to immunodeficient CD4-knockout mice at 1011 particles per animal intramuscularly. The blood obtained by retro-orbital bleeding at week 4 was assayed for human factor IX (hFIX) secretion in mouse plasma. Shown in the figure is the expression level at week 4. (B) The same vectors as in panel A were used in vitro to infect COS cells at a multiplicity of infection of 5,000. The supernatants were harvested at 24 h postinfection. The expression level of human factor IX (hFIX) was measured by ELISA.
As expected, AAV1-based vectors performed better than did AAV2 in muscle (Fig. 2A). Interestingly, the vector containing the first half of the sequence of AAV1 (pH221-1-351) functioned well in cultured COS cells (Fig. 2B). However, this did not lead to a high level of transgene expression in muscle (Fig. 2A). In contrast, vector pH221-352-736, containing the second half of the AAV1 capsid proteins, worked exceptionally well in muscle compared to the low levels of transgene expression in cultured COS cells. Together the in vitro and in vivo results suggest that the second half of the VP1 gene was responsible for the performance of AAV1 vectors in muscle and that the critical elements in AAV1 which determine its tropism are located in sequences in the second half of VP1 downstream of the BsiWI site.
Main region in AAV1 capsid protein affecting tissue tropism for muscle.
To identify specific regions in the AAV1 capsid protein that contribute to the tropism for muscle transduction, hybrid AAV1 and AAV2 helper plasmids with the AAV1 VP1 amino acid sequence 1 to 112, 113 to 155, 156 to 212, 213 to 423, 424 to 480, 481 to 564, 565 to 669, or 670 to 736 substituting for the corresponding AAV2 sequence were constructed. These amino acid sequences represent various clusters of nonidentical amino acid residues between AAV1 and AAV2 (Fig. 1B). Vectors based on these helpers were made by transfection. Unexpectedly, the vector yields from the helpers with AAV1 VP1 amino acids 481 to 564 and 565 to 669 were approximately 10-fold lower than those of AAV1 or AAV2 vectors. Despite the low yield, infectious particles resistant to DNase treatment were formed. This result was confirmed in several preparations with various vector plasmids (data not shown). We concluded that not all capsid regions can be freely swapped from one serotype to another despite their high degree of homology.
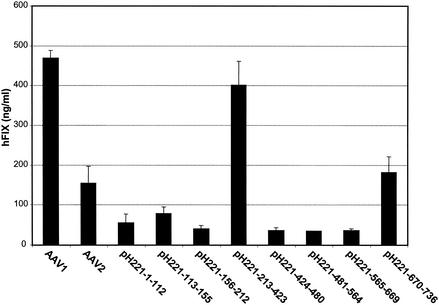
The above hybrid vectors expressing the human factor IX gene as a reporter gene were administered to CD4-knockout mice intramuscularly at a dose of 1011 particles per mouse. A representative peak expression profile of human factor IX in mouse plasma at week 6 is presented in Fig. 3. Again AAV1 outperformed AAV2. Only pH221-213-423 nearly reached the level of AAV1 vectors. However, pH221-670-736 was slightly higher than AAV2. Other hybrid vectors show levels of transgene expression comparable to or lower than that of AAV2, despite similar performances as AAV2 in vitro (data not shown). Although actual expression levels of factor IX in these experiments fluctuated over time, the relative differences in expression level remained unchanged for the experimental period (3 months). The result suggests that AAV1 VP1 amino acids 213 to 423 contain a determinant for muscle tropism.
FIG. 3.
Performances of AAV2 vectors with AAV1 epitopes in muscle. AAV-CMV-human factor IX vectors based on AAV2 with AAV1 epitopes were purified by CsCl gradient centrifugation and administered intramuscularly to CD4-knockout mice at a dose of 1011 viral genomes per mouse. Blood from mice was drawn at 6 weeks post-vector administration. The expression levels of human factor IX (hFIX) in mouse plasma were measured by ELISA. Each group consists of four mice. AAV1 and AAV2 vectors with the same transgene were used as controls.
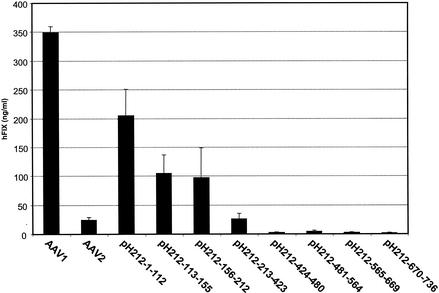
To confirm the above observation, another series of AAV1 and AAV2 hybrid helper plasmids were constructed. These vectors are pH212-1-112, pH212-113-155, pH212-156-212, pH212-213-423, pH212-424-480, pH212-481-564, pH212-565-669, and pH212-670-736. In these helper constructs, AAV1 VP1 sequences were replaced by the corresponding AAV2 sequence. In vitro studies using green fluorescent protein and human factor IX genes as reporter genes showed that all these hybrid vectors inherited the same poor efficiency of transduction of cultured cells as AAV1 (data not shown). After vectors based on these helpers were injected into the muscle of CD4-knockout mice (see Fig. 4), we observed a reversed pattern of expression compared to those in Fig. 3. The vectors based on pH212-1-112, pH212-113-155, and pH212-156-212 all had higher transduction levels than did AAV2, and the levels of expression from pH212-213-423 and pH212-670-736 were noticeably lower than those of their counterparts pH221-213-423 and pH221-670-763, respectively. This supports the finding that the C-terminal region of AAV1 VP1 is crucial for muscle transduction.
FIG. 4.
Performances of AAV1 vectors with AAV2 epitopes in muscle. AAV-CMV-human factor IX vectors based on AAV1 with AAV2 epitopes were purified by CsCl gradient centrifugation and administered intramuscularly to CD4-knockout mice at a dose of 1011 viral genomes per mouse. Blood from mice was drawn at 6 weeks post-vector administration. The expression levels of human factor IX (hFIX) in mouse plasma were measured by ELISA. Each group consists of four mice. AAV1 and AAV2 vectors with the same transgene were used as controls.
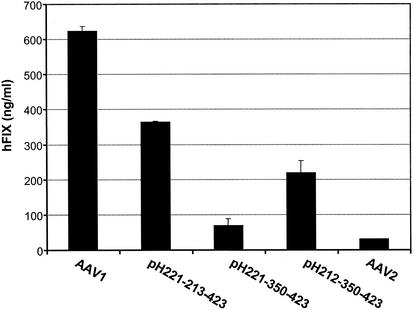
The initial experiment suggested that amino acids 350 to 736 would largely determine the performance of AAV1 in muscle (Fig. 2). On the other hand, amino acids 213 to 423 would improve AAV2's transduction to that of AAV1 (Fig. 3 and 4). It appeared that amino acids 350 to 423 would have the same effect in influencing AAV's tropism for muscle. Vectors based on pH221-350-423 and pH212-350-423 were made to examine this hypothesis. The in vivo results are summarized in Fig. 5. As demonstrated previously, pH221-213-423 vectors showed an approximately 8-fold increase over AAV2 compared to the AAV1 vectors' 15-fold increase over AAV2. The switch of amino acid sequence 350 to 423 from AAV2 to AAV1 (pH212-250-423) decreased AAV1 vector performance by approximately 60%. On the other hand, the switch of the same region from AAV1 to AAV2 improved the efficiency of AAV2 vectors by almost twofold. In summary, amino acids 350 to 423 corresponding to AAV1 VP1 contribute significantly to the high performance of AAV1 vectors in transduction of muscle.
FIG. 5.
AAV-CMV-human factor IX vectors based on AAV1, AAV2, pH221-213-423, pH221-350-423, or pH212-350-423 were purified by CsCl gradient centrifugation and administered intramuscularly to CD4-knockout mice at a dose of 1011 viral genomes per mouse. Blood from mice was drawn at 6 weeks post-vector administration. The expression levels of human factor IX (hFIX) in mouse plasma were measured by ELISA. Each group consists of four mice. AAV1 and AAV2 vectors with the same transgene were used as controls.
Characterization of receptor and antigenic properties of hybrid vectors.
The host cell receptors presumably determine the performance of individual AAV serotypes since vectors based on the various AAV serotypes generally differed only in the capsid protein. Several studies have suggested that each AAV serotype probably has its own unique receptors (8, 18, 25, 33). As reported elsewhere for AAV2, heparan sulfate proteoglycan (HSPG) and integrin αvβ5 FGFR1 could function as AAV receptors and coreceptors, respectively (22, 29, 30). The expression levels of these molecules in host cells have been found to affect the efficiency of AAV infection. Among the above molecules, HSPG has the strongest affinity to AAV2 virions. Heparin can not only inhibit AAV2 infection completely but also be used to purify AAV2 vectors by column chromatography (28, 36). The binding domains for heparin were shown previously to be located in the second half of the AAV capsid gene (34). AAV6 basically has the same capsid protein as that of AAV1, and it cannot bind to heparin (12). To determine whether heparin binding activity has an effect on the transduction efficiency of AAV1, AAV2, and hybrid vectors, the heparin binding activities of these vectors were examined. As shown in Table 1, vectors based on pH221-213-423 can be inhibited by heparin. This result suggested that vectors from pH221-213-423 inherited the same heparin binding properties from AAV2, which is consistent with previous studies showing that the heparin binding domain is in the C terminus of the AAV2 Cap protein (34). Vectors made from pH221-481-564 and pH221-565-669 could not be tested because of their low titer.
TABLE 1.
Virion properties of hybrid vectors based on AAV1 and AAV2a
| Vector | % Factor IX secretion
|
||
|---|---|---|---|
| Heparin | Anti-AAV2 | Anti-AAV1 | |
| AAV1 | 51.70 | 11 | 12.90 |
| AAV2 | 7.90 | 4.90 | 85.70 |
| pH221-1-112 | 5.70 | 4.30 | 70.80 |
| pH221-113-155 | 5.80 | 18.90 | 83.20 |
| pH221-156-212 | 21.30 | 14.70 | 83.60 |
| pH221-213-423 | 13.50 | 38 | 81.10 |
| pH221-670-763 | 13.30 | 16.90 | 85.80 |
Heparin and NABs against AAV were assayed in the following way. COS cells (105 cells/well) were seeded in 24-well plates and infected with AAV-CMV-human factor IX vectors (multiplicity of infection, 20,000) at 37°C for 1 h in serum-free Dulbecco's modified Eagle's medium containing 25 mM HEPES buffer. At 1 h postinfection, cells were washed with PBS and incubated in Dulbecco's modified Eagle's medium with 10% fetal bovine serum for 48 h and medium was assayed directly in an ELISA for human factor IX expression. To study the effect of heparin on the infectivity of AAV vectors, AAV vectors were preincubated with heparin (Sigma) at a final concentration of 200 μg/ml in serum-free Dulbecco's modified Eagle's medium containing 25 mM HEPES buffer at 37°C for 1 h before they were applied to the cells. To obtain NABs against AAV1 and AAV2, mouse plasma samples were collected 4 weeks after AAV1 or AAV2 vectors were administered to C57BL/6 mice intravenously. The effects of NABs against AAV1 and AAV2 on the hybrid vectors were assayed by preincubating the hybrid vectors with mouse sera containing NABs which were diluted 1:5 in Dulbecco's modified Eagle's medium containing 25 mM HEPES buffer. After the vectors and mouse sera were incubated for 30 min at room temperature, the vectors were applied to target cells. Supernatants were harvested at 48 h postinfection, and human factor IX in the samples was assayed in duplicates. The secretion of human factor IX into the medium was determined by ELISA. AAV1 and AAV2 values are shown as controls. The data are shown as the percentiles of factor IX secretion compared to the values in the absence of any inhibition (incubation with PBS) because of variations in expression for the individual vectors. The two vectors pH221-565-669 and pH221-481-564 were not included because their titers were very low.
AAV1 and AAV2 have distinct antigenic properties. To investigate what antigenic properties new hybrid vectors of AAV1 and AAV2 inherited from their parental capsids, we did inhibition assays using neutralizing antibodies (NABs) to AAV1 and AAV2 generated from C57BL/6 mice. All new hybrid vectors were incubated with AAV-NABs before infection of COS cells. The levels of transgene expression are shown in Table 1. As demonstrated previously, there were some crossover reactions of NAB to AAV2 on AAV1 since we observed the same level of reduction in transgene expression in the presence of AAV2 antibodies. It is interesting that the AAV2-NAB reduced the expression of the pH221-213-423 hybrid by only 60%. Under the same conditions, the AAV1 vectors retained only approximately 10% of the original expression and the AAV2 vectors could reach 5% of the original expression level in the presence of AAV2-NABs. The level of expression from other hybrid vectors fell between those of AAV1 and AAV2. In contrast, AAV1-NAB neutralized only the infectivity of the AAV1 vector and had little effect on AAV2 vectors and all hybrid vectors tested. This is evidenced by the fact that the expression levels are still maintained at approximately 80% of the original level in the presence of NAB to AAV1. Therefore, the major antigenic epitopes should be in amino acids 424 to 669 of the VP1 gene.
As shown in Table 1, vectors made from pH221-213-423 could still be inhibited by heparin as well as could AAV2. Since the identified heparin binding domain is in the C terminus of the AAV capsid, the heparin binding properties of this helper plasmid were not altered. Nevertheless, the performance of this vector was greatly improved with amino acids from AAV1 regions. A most likely conclusion is that the heparin binding properties or HSPG receptors do not contribute significantly to AAV1 transduction of muscle. Note that this result does not affect the role of HSPG receptors in the transduction of muscle by AAV2. Since pH221-213-423 vectors retained the antigenic properties of AAV2 and yet gained the ability of AAV1 to transduce muscle at high efficiency, it is likely that the epitopes for the B-cell response and the regions that interact with receptors were not linked to each other.
Although it is thought that receptor-mediated binding and entry into the target cells are the main reason for the differences in transduction of various AAV serotypes, other mechanisms could also play a role. In the case of AAV5, it has been shown elsewhere that there is no direct correlation between transduction efficiency and viral binding to muscle cells, which suggests that endocytic or intracellular pathways play a role in regulating AAV infection (10). Such barriers also affected the level of transduction by AAV2 in various cell lines as reported by Srivastava's group (13, 14). Other members of the parvovirus family also exhibited similar properties. For example, the VP1 N-terminal sequence of canine parvovirus could affect nuclear transport of virions and efficient cell infection (32). In the present study, although it is still unclear what is the exact mechanism by which AAV1 transduces muscle better than does AAV2, we were able to identify domains which are responsible for the differences in performance by a domain-swapping strategy.
One interesting observation is that not all these hybrid vectors could be generated at equal efficiencies. The yields of vectors from pH221-481-564 and pH221-565-669 vectors were low. In contrast, the exchange of the corresponding region of AAV2 with AAV1 still produced AAV vectors at normal yield. It appears that all amino acids need to be coordinated with each other in AAV packaging. This result is also in accordance with previous studies showing that not all epitope insertions could be tolerated in the positions that could be altered (11, 24, 34). Since several vectors could not be tested in vivo, it is hard to rule out their contribution to the transduction of muscle even though we were able to identify several important amino acids.
In this study, all clones were generated through molecular cloning, which has no natural selection for growth advantage. A novel marker rescue system to circumvent the decreased packaging efficiency has been proposed by Bowles et al. (2a). Those naturally occurring mutants based on homologous recombination would select for mutants with strong viability and complement those mutants that cannot be made by a cloning strategy (2a).
Vectors with the C-terminal part of VP1 (amino acids 351 to 736) inherited the muscle tropism of AAV1. In combination with the observation that vectors made from pH221-213-423 exhibit AAV1's tropism for muscle, we draw the conclusion that amino acids 350 to 423 are important in determining AAV1 tropism. This hypothesis is confirmed by experimental results shown in Fig. 5. As seen in the alignment of AAV1 and AAV2, the differences between AAV1 and AAV2 lie in two amino acids: one at position 363I→V and the other at 427E→D. This result is not expected, as these two amino acids are not located in any of those regions clustered with nonidentical amino acids. In addition, these amino acids are considered to be structurally similar. Structural analysis of parvovirus capsid found that the degree of conservation of surface-exposed residues is lower than average (6, 26). However, the location of these amino acids would be consistent with the putative secondary structure of AAV capsid proteins predicted by Chapman and Rossmann (5, 6, 17, 31). According to their structural analysis of various parvovirus capsid proteins, AAV should have five putative loop regions. It is interesting that these two amino acids fall into the regions that were designated loop III (amino acids 346 to 392) and loop IV (amino acids 420 to 646). Our results may suggest that loop III and loop IV are major regions that determine AAV receptors and hence tissue tropism. On the other hand, simply substituting these two amino acid sequences in pH221-350-423 and pH212-350-423 failed to restore the full vector efficiency to that of AAV1 or pH221-213-423 (Fig. 5). Because of the complexity of the AAV capsid structure, additional elements may be contributing to the difference between AAV1 and AAV2. These results reveal the limitations of our present strategy, which takes into consideration only linear epitopes and primary amino acid sequences and thus may oversimplify the three-dimensional virion structure.
Acknowledgments
We thank Yuqing Wang, Antje Kaufhold, and Yi Zhang for assistance in vector administration. We thank Katherine High, Richard Jude Samulski, Roland Herzog, and Denise Sabatino for critical comments on the manuscript.
This study was supported by NIH grant R01HL069051 to W.X.
REFERENCES
- 1.Alisky, J. M., S. M. Hughes, S. L. Sauter, D. Jolly, T. W. Dubensky, Jr., P. D. Staber, J. A. Chiorini, and B. L. Davidson. 2000. Transduction of murine cerebellar neurons with recombinant FIV and AAV5 vectors. Neuroreport 11:2669-2673. [DOI] [PubMed] [Google Scholar]
- 2.Berns, K. I. 1995. Parvoviridae: the viruses and their replication, p. 1007-1041. In P. M. Howley (ed.), Fundamental virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 2a.Bowles, D. E., J. E. Rabinowitz, and R. J. Samulski. 2003. Marker rescue of adeno-associated virus (AAV) capsid mutants: a novel approach for chimeric AAV production. J. Virol. 77:423-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao, L., Y. Liu, M. J. During, and W. Xiao. 2000. High-titer, wild-type free recombinant adeno-associated virus vector production using intron-containing helper plasmids. J. Virol. 74:11456-11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao, H., Y. Liu, J. Rabinowitz, C. Li, R. J. Samulski, and C. E. Walsh. 2000. Several log increase in therapeutic transgene delivery by distinct adeno-associated viral serotype vectors. Mol. Ther. 2:619-623. [DOI] [PubMed] [Google Scholar]
- 5.Chapman, M. S., and M. G. Rossmann. 1995. Single-stranded DNA-protein interactions in canine parvovirus. Structure 3:151-162. [DOI] [PubMed] [Google Scholar]
- 6.Chapman, M. S., and M. G. Rossmann. 1993. Structure, sequence, and function correlations among parvoviruses. Virology 194:491-508. [DOI] [PubMed] [Google Scholar]
- 7.Chiorini, J. A., F. Kim, L. Yang, and R. M. Kotin. 1999. Cloning and characterization of adeno-associated virus type 5. J. Virol. 73:1309-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiorini, J. A., L. Yang, Y. Liu, B. Safer, and R. M. Kotin. 1997. Cloning of adeno-associated virus type 4 (AAV4) and generation of recombinant AAV4 particles. J. Virol. 71:6823-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson, B. L., C. S. Stein, J. A. Heth, I. Martins, R. M. Kotin, T. A. Derksen, J. Zabner, A. Ghodsi, and J. A. Chiorini. 2000. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc. Natl. Acad. Sci. USA 97:3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan, D., Z. Yan, Y. Yue, W. Ding, and J. F. Engelhardt. 2001. Enhancement of muscle gene delivery with pseudotyped adeno-associated virus type 5 correlates with myoblast differentiation. J. Virol. 75:7662-7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girod, A., M. Ried, C. Wobus, H. Lahm, K. Leike, J. Kleinschmidt, G. Deleage, and M. Hallek. 1999. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat. Med. 5:1438.. [DOI] [PubMed] [Google Scholar]
- 12.Halbert, C. L., J. M. Allen, and A. D. Miller. 2001. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J. Virol. 75:6615-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen, J., K. Qing, H. J. Kwon, C. Mah, and A. Srivastava. 2000. Impaired intracellular trafficking of adeno-associated virus type 2 vectors limits efficient transduction of murine fibroblasts. J. Virol. 74:992-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen, J., K. Qing, and A. Srivastava. 2001. Infection of purified nuclei by adeno-associated virus 2. Mol. Ther. 4:289-296. [DOI] [PubMed] [Google Scholar]
- 15.Herzog, R. W., J. N. Hagstrom, S. H. Kung, S. J. Tai, J. M. Wilson, K. J. Fisher, and K. A. High. 1997. Stable gene transfer and expression of human blood coagulation factor IX after intramuscular injection of recombinant adeno-associated virus. Proc. Natl. Acad. Sci. USA 94:5804-5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kube, D. M., S. Ponnazhagan, and A. Srivastava. 1997. Encapsidation of adeno-associated virus type 2 Rep proteins in wild-type and recombinant progeny virions: Rep-mediated growth inhibition of primary human cells. J. Virol. 71:7361-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langeveld, J. P. 1993. B-cell epitopes of canine parvovirus: distribution on the primary structure and exposure on the viral surface. J. Virol. 67:765-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muramatsu, S., H. Mizukami, N. S. Young, and K. E. Brown. 1996. Nucleotide sequencing and generation of an infectious clone of adeno-associated virus 3. Virology 221:208-217. [DOI] [PubMed] [Google Scholar]
- 19.Murphy, F. A., C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers (ed.). 1995. Classification and nomenclature of viruses: sixth report of the International Committee on Taxonomy of Viruses, p. 169-175. Springer-Verlag, Vienna, Austria.
- 20.Muzyczka, N. 1992. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr. Top. Microbiol. Immunol. 158:97-129. [DOI] [PubMed] [Google Scholar]
- 21.Parks, W. P., D. W. Boucher, J. L. Melnick, L. H. Taber, and M. D. Yow. 1970. Seroepidemiological and ecological studies of the adenovirus-associated satellite viruses. J. Virol. 2:716-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qing, K., C. Mah, J. Hansen, S. Zhou, V. Dwarki, and A. Srivastava. 1999. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 5:71-77. [DOI] [PubMed] [Google Scholar]
- 23.Rabinowitz, J. E., F. Rolling, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabinowitz, J. E., W. Xiao, and R. J. Samulski. 1999. Insertional mutagenesis of AAV2 capsid and the production of recombinant virus. Virology 265:274-285. [DOI] [PubMed] [Google Scholar]
- 25.Rutledge, E. A., C. L. Halbert, and D. W. Russell. 1998. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 72:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson, A. A., B. Hebert, G. M. Sullivan, C. R. Parrish, Z. Zadori, P. Tijssen, and M. G. Rossmann. 2002. The structure of porcine parvovirus: comparison with related viruses. J. Mol. Biol. 315:1189-1198. [DOI] [PubMed] [Google Scholar]
- 27.Srivastava, A., E. W. Lusby, and K. I. Berns. 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Summerford, C. 1999. Viral receptors and vector purification: new approaches for generating clinical-grade reagents. Nat. Med. 5:587-588. [DOI] [PubMed] [Google Scholar]
- 29.Summerford, C., J. S. Bartlett, and R. J. Samulski. 1999. αvβ5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat. Med. 5:78-82. [DOI] [PubMed] [Google Scholar]
- 30.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsao, J., M. S. Chapman, M. Agbandje, W. Keller, K. Smith, H. Wu, M. Luo, T. J. Smith, M. G. Rossmann, R. W. Compans, et al. 1991. The three-dimensional structure of canine parvovirus and its functional implications. Science 251:1456-1464. [DOI] [PubMed] [Google Scholar]
- 32.Vihinen-Ranta, M., D. Wang, W. S. Weichert, and C. R. Parrish. 2002. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J. Virol. 76:1884-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walters, R. W., S. M. Yi, S. Keshavjee, K. E. Brown, M. J. Welsh, J. A. Chiorini, and J. Zabner. 2001. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J. Biol. Chem. 276:20610-20616. [DOI] [PubMed] [Google Scholar]
- 34.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao, W., N. Chirmule, S. C. Berta, B. McCullough, G. Gao, and J. M. Wilson. 1999. Gene therapy vectors based on adeno-associated virus type 1. J. Virol. 73:3994-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zolotukhin, S., B. Byrne, E. Mason, I. Zolotukhin, M. Potter, K. Chesnut, C. Summerford, R. Samulski, and N. Muzyczka. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6:973-985. [DOI] [PubMed] [Google Scholar]