Abstract
To study the effects of the nuclear receptors (NRs) HNF4α and COUP-TF1 on the life cycle of hepatitis B virus (HBV), the human hepatoma cell line Huh7 was transiently cotransfected with plasmids containing the HBV genome and encoding these two NRs. Overexpression of HNF4α and COUP-TF1 led to a 9-fold increase and a 7- to 10-fold decrease, respectively, in viral DNA synthesis. These two NRs also exhibited distinct modes of regulation of viral transcription. Overexpression of HNF4α led to a more-than-10-fold increase in synthesis of the pregenomic RNA but to only a 2- to 3-fold increase in synthesis of the pre-C and S RNAs. Moreover, the NR response element within the pre-C promoter, NRREpreC, played the major role in activation of pregenomic RNA synthesis by HNF4α. On the other hand, overexpression of COUP-TF1 led to an over-10-fold repression of synthesis of both pre-C and pregenomic RNAs mediated through either NRREpreC or NRREenhI. HNF4α and COUP-TF1 antagonized each other's effects on synthesis of pregenomic RNA and viral DNA when they were co-overexpressed. A naturally occurring HBV variant which allows for binding by HNF4α but not COUP-TF1 in its NRREpreC exhibited significantly higher levels of synthesis of pregenomic RNA and viral DNA than wild-type HBV in coexpression experiments. Last, deletion analysis revealed that non-NRRE sequences located within both the C and pre-S1 regions are also essential for maximum activation of the pregenomic promoter by HNF4α but not for repression by COUP-TF1. Thus, HNF4α and COUP-TF1 function through different mechanisms to regulate expression of the HBV genes.
Viral hepatitis B is a serious health hazard in many parts of the world. Its pathogen, human hepatitis B virus (HBV), is a member of the hepadnavirus family. Its circular genome consists of a partially double-stranded DNA, 3.2 kb in length, with four open reading frames (ORF) encoding the surface proteins (large S, middle S, and S), core proteins (pre-C and C), reverse transcriptase (P protein), and X protein. Synthesis of the HBV RNAs is under the control of the pre-C, pregenomic, S, pre-S1, and X promoters and is regulated by two enhancer elements, enhancer I and enhancer II. All HBV transcripts utilize a common polyadenylation signal located within the region encoding the C protein. The presence of a posttranscriptional regulatory element allows for the export of the HBV mRNAs without splicing. The pregenomic RNA transcribed from the pregenomic promoter plays pivotal roles in viral replication, serving both as the template for viral DNA synthesis through reverse transcription and as the mRNA encoding the C and P proteins, components of the nucleocapsid. On the other hand, the biological functions of pre-C RNA and its encoded pre-C protein are still unknown (10, 11).
Nuclear receptors (NRs) comprise a large family of transcription factors with similar domain structures and high sequence similarity in their DNA-binding regions. NRs bind to their respective NR response elements (NRREs) in a sequence-specific manner to regulate transcription from nearby promoters (38, 43). The binding of specific ligands to their NRs modulates both the affinities of NRs for their NRREs and their effects on transcription (13, 43). The orphan NRs are a subgroup of NRs whose ligands, if they exist, are not yet known (2, 22).
The hepatocyte nuclear factor 4α (HNF4α), a liver-enriched NR, is essential for hepatocyte differentiation during liver development. It plays important roles in regulating metabolic pathways in liver and in maintaining the hepatic phenotype in hepatoma cell lines (16, 25, 33). HNF4α forms homodimers and binds to DR1 elements, i.e., sequences containing tandem repeats resembling the consensus half-site sequence 5′-AGGTCA-3′ separated by 1 nucleotide (nt) (8, 20). While its physiological ligands remain unknown, fatty acyl-coenzyme A thioesters have been reported to be able to function as ligands (17).
The chicken ovalbumin upstream promoter transcription factors COUP-TF1 and COUP-TF2 were initially identified as transcriptional activators of the chicken ovalbumin promoter (1, 41, 42). They are ubiquitous transcription factors abundant in a variety of tissues including liver and play key roles in regulating organogenesis, neurogenesis, and cell differentiation during embryonic development in animals (28, 29). The COUP-TFs bind to a variety of NRREs including DR1s, resulting in transcriptional repression. They can also repress transcription by forming heterodimers with other NRs (6, 24). Ligands for the COUP-TFs, if any, remain unknown.
The HBV genome contains a glucocorticoid response element situated within the S ORF (39, 40) and three NRREs situated within the pre-C promoter (NRREpreC), enhancer I (NRREenhI), and enhancer II (NRREenhII) (see Fig. 7A). NRREpreC and NRREenhI can be bound by several NRs, including HNF4α, the COUP-TFs, retinoid X receptor (RXR), human testicular receptor 2 (TR2), and the peroxisome proliferator-activated receptors (PPARs); NRREenhII is bound only by HNF4α (12, 15, 18, 19, 30, 46, 47). These NRREs play critical roles in regulating expression and replication of HBV. We found that point mutations introduced into both NRREpreC and NRREenhI dramatically reduce synthesis of HBV RNA and DNA in the hepatoma cell line Huh7 (48). Interestingly, the CH variant of HBV, frequently found in chronic hepatitis B patients, contains point substitution mutations of A to T at nt 1764 and G to A at nt 1766 in one of the half-sites of NRREpreC. Introduction of these mutations into the wild-type HBV genome results in a modest increase in viral DNA synthesis in transiently transfected hepatoma cell lines (reviewed in reference 4). We (48) and others (37) reported that these two point mutations abolish the binding of NRREpreC by PPARα-RXRα and COUP-TF1 but have no effect on its binding by HNF4α. Although the CH variant is the most extensively studied among the naturally occurring HBV variants, the molecular mechanism of its striking preponderance in chronic hepatitis B patients remains unknown. Thus, we also examined here whether the low affinity of COUP-TF1 for binding NRREpreC contributes to the CH variant's growth advantage in the livers of patients chronically infected with HBV.
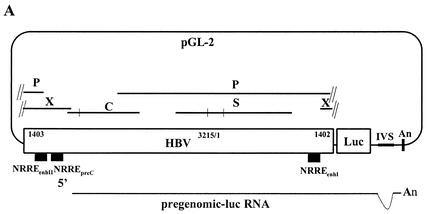
FIG. 7.
Effect of NRREenhII on synthesis of pregenomic RNA in Huh7 cells. (A) Schematic diagram of pWTluc, a plasmid containing a full-length HBV genome inserted into the luciferase (luc) expression vector pGL2 (Promega). Large open rectangle, HBV genome (the numbers indicate the inserted nucleotide residues in HBV); solid lines, locations of the P, S, C, and X ORF; solid rectangles, locations of the NRREs in enhancer I, enhancer II, and the pre-C promoter. The luc gene (small open rectangle) is not drawn to scale. Small horizontal and vertical bars to the right of the luc gene, intron (IVS) and polyadenylation signal (An). Shown at the bottom is the structure of the chimeric pregenomic RNA synthesized from this plasmid. (B) Autoradiogram showing the results of primer extension analysis of the pre-C and pregenomic (preg) RNAs accumulated in Huh7 cells cotransfected with the indicated plasmids. Numbers at the bottom are means ± standard errors of the data relative to the wild-type (WTluc) obtained from three experiments similar to the one for which results are shown.
Tang et al. (36, 37) reported that overexpression of HNF4α and PPARα-RXRα enables replication of the HBV genome in the nonhepatic cell line NIH 3T3. They hypothesized that these two NRs allow the pregenomic promoter to be switched from an inactive to an active state in the liver. However, we (unpublished data) and others (15) observed that overexpression of HNF4α in the hepatoma cell line Huh7 only slightly increases synthesis of the pregenomic RNA from subgenomic HBV plasmids. To resolve this seeming discrepancy, we set out to study the effects of overexpressed HNF4α on transcription and replication of the wild-type HBV genome and a naturally occurring variant HBV genome. Since competition between HNF4α and COUP-TFs for binding to NRREs has been implicated in the regulation of expression of many genes in the liver (9, 21, 45), we also examined whether this competition plays a role in the regulation of HBV expression. We report here the effects of HNF4α and COUP-TF1 and mutations in the NRREs to which they bind on HBV RNA and DNA synthesis. We show that these two NRs play opposing roles in transcription by distinct modes of action, antagonizing each other's effects on HBV transcription and replication to regulate HBV expression.
MATERIALS AND METHODS
Plasmid constructs.
Plasmid pWT contains 1.2 tandem copies of the adr subtype of the HBV genome from nt 1403 to 1990 in plasmid pSP65 (Promega). Plasmids pNRREpreC− and pNRREenhI− are variants of pWT containing base substitution mutations in NRREpreC and NRREenhI, respectively, that do not alter the coding of any HBV proteins. Plasmid pNRREpreC−,enhI− contains mutations in both NRREs. Plasmid pCH contains the base pair substitution mutations A to T and G to A at nt 1764 and 1766 in the NRREpreC of pWT. The construction of these plasmids has been previously described (48). Both copies of NRREpreC were mutated in pNRREpreC−, pNRREpreC−,enhI−, and pCH.
Plasmids pΔ5 and pΔ6 were constructed by deletion of BstEII-to-XhoI and BglII fragments from pWT, respectively. Plasmid pC− is a variant of pWT containing a 1-bp insertion of T-A between nt 1953 and 1954 (X. Yu, unpublished data). This naturally occurring frameshift mutation at codon 17 in the C ORF results in premature termination of translation for the core proteins at codon 21. Plasmid pC−/S− contains additional base substitution mutations at nt 206, 221, 222, and 224, introduced by PCR-based site-directed mutagenesis (5), resulting in nonsense mutations at codons 14, 20, and 21 in the S ORF that abolish synthesis of all of the surface proteins but that do not alter the ORF encoding the P protein.
Plasmids pWTluc and pWT′luc (see Fig. 7A and 8A) were constructed by insertion of linearized full-length HBV genomes with different sequence permutations into the multiple cloning sites in pGL2 (Promega). Deletion mutants pΔ1luc through pΔ4luc were derived from pWTluc by deletion of DNA fragments from the region between nt 2436 and 1402 (see Fig. 8A). Plasmids pΔ5luc and pΔ6luc were derived from pWTluc and contain the same internal deletions as pΔ5 and pΔ6 (see Fig. 8A). Plasmid pΔ7luc was constructed by deletion of nt 1992 to 1097 from pWT′luc. Plasmid pΔ8luc was constructed by introduction of the point mutations present in pNRREpreC− into the NRREpreC in pΔ7luc. Plasmids pNRREpreC−luc and pNRREenhII−luc were constructed by introduction of the point mutations from pNRREpreC− and pNRREenhII−, respectively, into plasmid pWTluc.
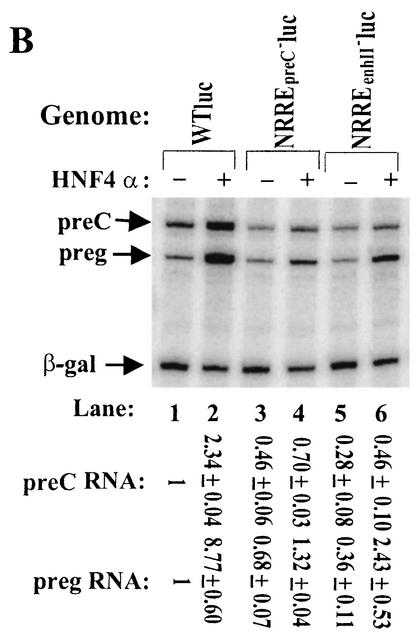
FIG. 8.
Mapping of regions in the HBV genome essential for transcriptional activation by HNF4α. (A) Schematic diagrams of the deletion variants of pWTluc and pWT′luc studied here. Plasmid pWT′luc is identical to pWTluc except for the permutation of the end points of the full-length HBV genome inserted into pGL2. Solid rectangles, locations of the three NRREs in pWTluc and pWT′luc; ∗ (in pΔ8luc), base substitutions in nt 1759, 1765, and 1768 in the NRREpreC− mutant previously described (48). preg, pregenomic. (B and C) Effects of deletions on activation of pregenomic RNA synthesis by HNF4α. WT, wild type. (D) Lack of effect of absence of C and S proteins on activation of synthesis of pregenomic RNA by HNF4α. Autoradiograms show the results of primer extension assays used to quantify the pre-C and pregenomic RNAs synthesized in Huh7 cells cotransfected with the indicated plasmids. The sequences of plasmids pC− and pC−/S− are described in Materials and Methods. Numbers in panels A and D indicate the amounts of the viral RNAs synthesized from the indicated HBV plasmid when HNF4α was overexpressed relative to the amounts synthesized from the same HBV plasmid when it was cotransfected in parallel with the parental expression plasmid. These data are means ± standard errors of the data obtained from three experiments similar to the one for which results are shown here and in Fig. 9.
The expression plasmids for rat HNF4α and human COUP-TF1, pCDMHNF4, and pRSVCOUP-TF1 and their parental plasmids, pCDM8 and pRSV0, respectively, have been previously described (46, 47).
Oligonucleotides and competition EMSAs.
The competition electrophoretic mobility shift assays (EMSAs) were performed as previously described (44, 49). The NRREpreC 25-bp synthetic oligonucleotide probe, the NRREenhI 29-bp synthetic oligonucleotide probe, and the NRREenhII 30-bp synthetic oligonucleotide probe were previously described (47). The nucleotide sequence of the mutant NRREenhII oligonucleotide probe was the same as the wild-type sequence except for the changes shown in Fig. 6A. The recombinant HNF4α protein was synthesized in a coupled transcription-translation rabbit reticulocyte lysate system (Promega).
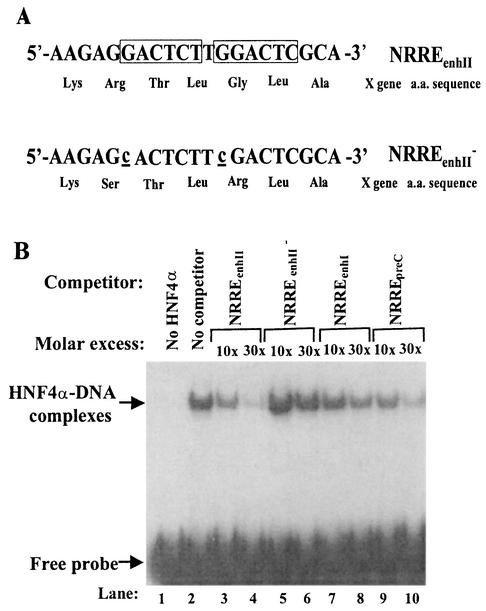
FIG. 6.
Effects of point mutations in NRREenhII on its affinity for HNF4α. (A) Sequences of NRREenhII and the point mutations introduced into it. The imperfect direct repeats of the NR half-site sequence are boxed. The base pair changes introduced by mutagenesis are underlined and in lowercase. Both are missense mutations in the overlapping X ORF. The amino acid changes are shown beneath the nucleotide sequences. (B) Competition EMSAs to determine the relative affinities of HNF4α for binding wild-type and mutant NRREs. Radiolabeled oligonucleotides containing the wild-type NRREpreC sequence were used as probes, and unlabeled double-stranded oligonucleotides containing the indicated wild-type and mutant NRREs were used as the competitors. Arrows, positions of the DNA-protein complexes and free DNA probe.
Cell line and transfections.
The human hepatoma cell line Huh7 was grown at 37°C with 5% CO2 in a 1:1 mixture of Dulbecco's modified Eagle's medium and F12 medium supplemented with 10% fetal bovine serum. Transient transfections were performed according to the calcium phosphate coprecipitation method (31). Cells in 60-mm-diameter dishes transfected with a total of 10 μg of plasmid DNA that included 3 μg of HBV plasmid, the amounts of expression plasmids for HNF4α and/or COUP-TF1 or their parental vectors indicated in the figure legends, 0.75 μg of the β-galactosidase (β-Gal) expression plasmid pEQ176 (32), and carrier pUC18 DNA. After incubation for 8 h with the calcium phosphate-DNA coprecipitates, the cells were washed and incubated in medium containing 5% fetal bovine serum until harvested.
Analysis of viral RNA.
Forty-eight hours after transfection, total cellular RNA was isolated with an RNeasy Mini kit (Qiagen). The relative amounts of the viral RNAs were determined by primer extension analysis as previously described (47). The primers for the pre-C, pregenomic, and S RNAs were the ones previously described (48). The primer for β-Gal RNA was the 17-mer 5′-GTTTTCCCAGTCACGAC-3′.
Analysis of viral DNA.
Forty-eight hours after transfection, the Huh7 cells were harvested. Viral nucleic acid was isolated from cytoplasmic nucleocapsids as previously described (34). The relative amounts of viral DNA were determined by Southern blot analysis (3). The radiolabeled DNA probe was prepared by using a random-primer labeling system (Amersham) with the full-length 3.2-kb HBV DNA genome as the template.
RESULTS
Effects of HNF4α and COUP-TF1 on HBV RNA and DNA synthesis.
HNF4α and COUP-TFs are known to activate and repress HBV transcription, respectively (12, 15, 30, 47). To better understand how interactions of these two NRs with NRREs in the context of the whole HBV genome affect viral transcription and replication, pWT, a plasmid containing 1.2 copies in tandem of the wild-type HBV genome and NRRE mutant variants of it were transfected in parallel into the hepatoma cell line Huh7 along with pCDMHNF4 or pRSVCOUP-TF1, plasmids expressing HNF4α and COUP-TF1, respectively, or their parental vectors. The relative quantities of the HBV RNAs accumulated in the cells by 48 h posttransfection were determined by primer extension analysis.
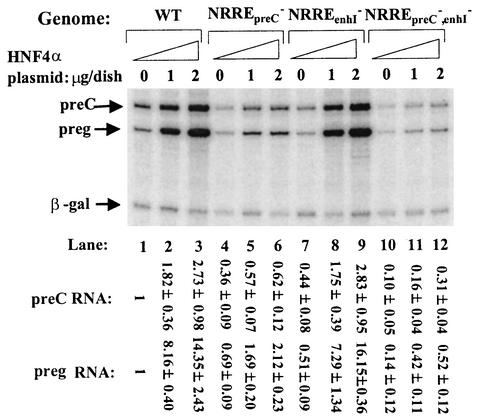
Overexpression of HNF4α led to a 14-fold increase in synthesis of pregenomic RNA from pWT (Fig. 1, lanes 1 to 3). This effect of HNF4α on viral RNA synthesis was largely dependent on the presence of a functional NRREpreC because synthesis of pregenomic RNA increased only 3-fold when this site was mutated (Fig. 1, lanes 4 to 6) yet synthesis of pregenomic RNA increased over 30-fold when NRREenhI was mutated (Fig. 1, lanes 7 to 9). When both NRREpreC and the NRREenhI were mutated, HNF4α activated synthesis of pregenomic RNA approximately threefold (Fig. 1, lanes 10 to 12). In contrast, overexpression of HNF4α led to a less-than-threefold activation of synthesis of pre-C RNA from either pWT (Fig. 1, lanes 1 to 3) or plasmids in which NRREpreC was mutated (Fig. 1, lanes 4 to 6 and 10 to 12). Overexpression of HNF4α also activated synthesis of the S RNAs only twofold from pWT, pNRREpreC−, and pNRREpreC−,enhI− (data not shown). With the basal level of synthesis of S RNA from pNRREenhI− being only one-fifth of that from pWT (48), overexpression of HNF4α led to a fivefold increase in synthesis of S RNAs from pNRREenhI− to the level equivalent to that synthesized from pWT without coexpressed HNF4α (data not shown). Overexpression of HNF4α had an even smaller effect on synthesis of large S and X RNAs (data not shown).
FIG. 1.
HNF4α activates synthesis of pre-C and pregenomic RNAs. Huh7 cells were cotransfected with pWT or the indicated NRRE mutant plasmid together with the indicated amount of HNF4α expression plasmid pCDMHNF4α or its parental plasmid. Shown is an autoradiogram of an 8 M urea-8% polyacrylamide gel indicating the results of primer extension analysis of the pre-C and pregenomic (preg) RNAs accumulated in the Huh7 cells by 48 h posttransfection. One-sixth of the RNA from a 60-mm-diameter dish of cells was used in each primer extension reaction. Arrows, positions of the viral and internal control β-Gal RNAs. Numbers at the bottom give the amounts of viral RNA in each lane relative to that synthesized from the wild-type (WT) genome in the absence of HNF4α overexpression. These numbers were determined with a PhosphorImager and normalized to the amount of β-Gal RNA present in the same sample and represent the means ± standard errors of the data obtained from three experiments similar to the one for which results are shown.
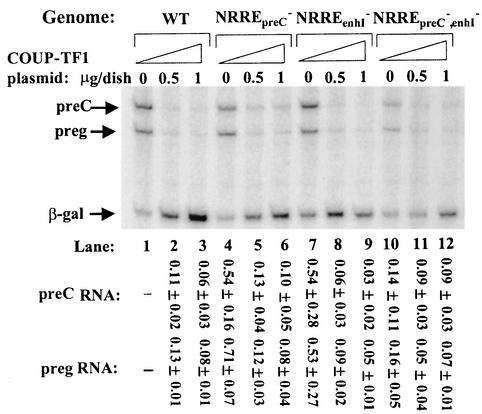
As expected, overexpression of COUP-TF1 led, instead, to repression of transcription from the HBV promoters. Interestingly, the overall reductions in synthesis of both pre-C and pregenomic RNAs were both approximately 10-fold and occurred when either NRREpreC or NRREenhI was still present (Fig. 2). Overexpression of COUP-TF1 led to fourfold and twofold reductions in synthesis of S and large S RNAs, respectively, from pWT (data not shown).
FIG. 2.
COUP-TF1 represses synthesis of pre-C and pregenomic RNAs. Huh7 cells were cotransfected with pWT or the indicated NRRE mutant plasmid together with the indicated amount of the COUP-TF1 expression plasmid pRSVCOUP-TF1 or its parental plasmid. Shown here is an autoradiogram of an 8 M urea-8% polyacrylamide gel indicating the results of primer extension analysis of the pre-C and pregenomic (preg) RNAs accumulated in the Huh7 cells by 48 h posttransfection. Since COUP-TF1 activated transcription of β-Gal from pEQ176, experiments were first separately performed with 0.75 μg of pEQ176 and 0, 0.5, and 1 μg of pRSVCOUP-TF1 to determine the effect of COUP-TF1 on synthesis of the β-Gal RNA in Huh7 cells. Quantitations were then performed as described in the legend to Fig. 1 except that the amount of the β-Gal RNA in each lane was first normalized to the amount in the absence of coexpressed COUP-TF1 in the transfection experiment mentioned above. Numbers at the bottom are means ± standard errors of the data relative to the wild type (WT) obtained from three experiments similar to the one for which results are shown.
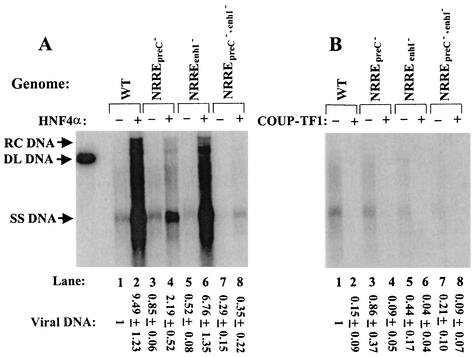
The effects of overexpression of HNF4α and COUP-TF1 on accumulation of viral DNA were assayed concurrently by Southern blot analysis. Cells transfected with pWT accumulated almost 10-fold more viral DNA when HNF4α was overexpressed (Fig. 3A, lane 2 versus lane 1). As expected, this increase in viral DNA synthesis was largely dependent on the presence of a functional NRREpreC (Fig. 3A, lanes 3 to 8). Likewise, cells transfected with either pWT or the NRRE mutants accumulated only 1/7 to 1/10 as much viral DNA when COUP-TF1 was overexpressed (Fig. 3B). Thus, synthesis of viral DNA correlated well with activation and repression of synthesis of pregenomic RNA by HNF4α and COUP-TF1, respectively.
FIG. 3.
Activation and repression of synthesis of viral DNA by HNF4α (A) and COUP-TF1 (B), respectively. Shown are autoradiograms of Southern blot analysis of viral DNA isolated from nucleocapsids present in the cytoplasm of Huh7 cells cotransfected with the indicated HBV genome and 2 μg of pCDMHNF4 (A) or 1 μg of pRSVCOUP-TF1 (B). One-third of the DNA from a 60-mm-diameter dish of cells was loaded in each lane. The total amount of viral DNA in each lane, from relaxed circular (RC) through single-stranded (SS) DNA, was quantified with a PhosphorImager. The smears between the specifically indicated DNA structures are the result of HBV DNA with incomplete, heterogeneous-length plus strands. Numbers at the bottom are means ± standard errors of the data relative to the wild type (WT) obtained from three experiments similar to the one for which results are shown. DL, duplex linear.
We conclude that HNF4α can play a major role in activating synthesis of pregenomic RNA and, therefore, viral DNA, largely via its interactions with NRREpreC. On the other hand, COUP-TF1 down-regulates synthesis of both pre-C and pregenomic RNAs and, consequently, viral DNA via interactions with either NRREpreC or NRREenhI. Thus, even though they function through the same NRREs, they regulate HBV expression through distinct modes of action as well as having opposing effects on transcription and replication.
Effects of HNF4α and COUP-TF1 on pregenomic RNA synthesis from a naturally occurring variant of HBV.
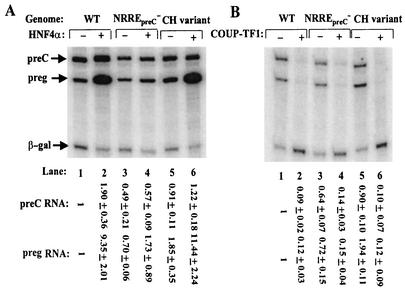
Since the two point substitution mutations in the NRREpreC of the naturally occurring HBV CH variant affect the binding to this site by COUP-TF1 and HNF4α differently (37, 48), we studied the effects of these mutations on regulation of HBV gene expression in cotransfection experiments after they were introduced into plasmid pWT. Overexpression of HNF4α led to nine- and sixfold increases in synthesis of pregenomic RNA from the wild-type and CH variant genomes, respectively (Fig. 4A, lane 2 versus lane 1 and lane 6 versus lane 5), with accumulations of pregenomic RNA ending up similar in amount (Fig. 4A, lane 2 versus lane 6). On the other hand, the mutations in pNRREpreC−, which abolish binding of NRREpreC by HNF4α as well as COUP-TF1 (48), resulted in only a twofold increase in the synthesis and accumulation of pregenomic RNA when HNF4α was overexpressed (Fig. 4A, lanes 3 and 4). Overexpression of COUP-TF1 repressed synthesis of pregenomic RNA from all three plasmids 5- to 15-fold (Fig. 4B), consistent with our above finding that either a functional NRREpreC or NRREenhI is sufficient for transcriptional repression by COUP-TF1. Thus, we conclude that the mutations present in the CH variant do not affect the accumulation of pregenomic RNA when HNF4α or COUP-TF1 is overexpressed.
FIG. 4.
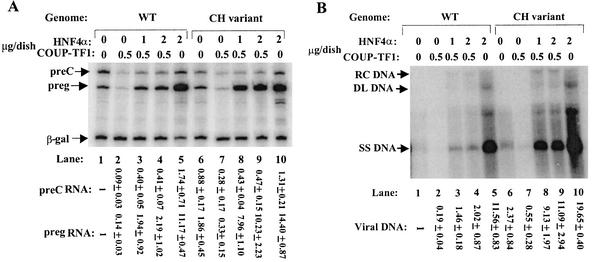
Effects of overexpression of HNF4α (A) and COUP-TF1 (B) on synthesis of pre-C and pregenomic RNAs from the wild-type and CH variant genomes. The autoradiograms show the results of primer extension assays used to quantify the pre-C and pregenomic (preg) RNAs synthesized in Huh7 cells cotransfected with 1 μg of pCDMHNF4 (A) or 0.5 μg of pRSVCOUP-TF1 (B). Quantitations were performed as described in the legends to Fig. 1 and 2. Numbers at the bottom are means ± standard errors of the data relative to the wild type (WT) obtained from three experiments similar to the one for which results are shown.
Effects of co-overexpression of HNF4α and COUP-TF1 on synthesis of pregenomic RNA and viral DNA.
To study the combined effects of the NRs on HBV transcription and replication, Huh7 cells were cotransfected with the wild-type or CH variant plasmid together with the indicated amounts of the HNF4α and COUP-TF1 expression plasmids. In cells transfected with pWT, repression of synthesis of pregenomic RNA by COUP-TF1 was overcome by overexpression of HNF4α (Fig. 5A, lane 2 versus lanes 3 and 4), resulting in a twofold overall increase in synthesis of pregenomic RNA (Fig. 5A, lane 4 versus lane 1). This increase is far less than the 11-fold increase observed when only HNF4α was overexpressed (Fig. 5A, lane 5). In cells transfected with pCH, HNF4α overcame the repression by COUP-TF1 more readily, resulting in a fivefold overall increase in synthesis of pregenomic RNA (Fig. 5A, lane 6 versus lanes 8 and 9).
FIG. 5.
Effects of co-overexpression of HNF4α and COUP-TF1 on synthesis of pre-C and pregenomic (preg) RNAs and viral DNA from wild-type (WT) HBV and the CH variant. Huh7 cells were cotransfected with 3 μg of pWT or pCH, the indicated amounts of HNF4α and COUP-TF1 expression plasmids, and their respective parental vectors at a total concentration of 10 μg of DNA per 60-mm-diameter dish. (A) Autoradiogram showing the results of primer extension assays used to quantify the pre-C and pregenomic RNAs. (B) Autoradiogram showing the results of Southern blot assays used to quantify cytoplasmic viral DNA. RC, DL, and SS are as defined in the legend for Fig. 3. Quantitations were performed as described in the legend to Fig. 1. Numbers at the bottom are means ± standard errors of the data relative to the wild type obtained from three experiments similar to the one for which results are shown.
Accumulation of viral DNA in the cytoplasm of the transfected cells correlated well with the synthesis of pregenomic RNA. In cells transfected with pWT, overexpression of HNF4α in the presence of overexpressed COUP-TF1 led to a twofold increase in synthesis of viral DNA (Fig. 5B, lane 1 versus lanes 3 and 4), one-fifth of the increase observed when only HNF4α was overexpressed (Fig. 5B, lane 5). In cells transfected with pCH, overexpression of HNF4α in the presence of overexpressed COUP-TF1 led to a four- to fivefold increase in synthesis of viral DNA (Fig. 5B, lane 6 versus lanes 8 and 9), nearly one-half of the increase observed when only HNF4α was overexpressed (Fig. 5B, lane 10).
Thus, we conclude that increased expression of HNF4α overcomes the repressive effects of COUP-TF1 on synthesis of pregenomic RNA and viral DNA. Furthermore, HNF4α appears to compete more efficiently with COUP-TF1 for binding to the NRREs in the HBV genome in cells transfected with the CH variant than in cells transfected with wild-type HBV, possibly explaining the mutant's selective advantage for growth.
Effects of NRREenhII on activation of synthesis of pregenomic RNA by HNF4α.
To determine the contribution of HBV's third NRRE, NRREenhII, we introduced two point mutations into the two half-site sequences of this NRRE. Competition EMSAs showed that these mutations reduce NRREenhII's affinity for HNF4α approximately ninefold (Fig. 6B; data not shown). Unfortunately, these mutations also introduce two missense mutations into the overlapping X ORF (Fig. 6A). To overcome a potential problem in interpretation of data that could arise from synthesis of a mutant X protein, the NRREenhII point mutations were introduced into plasmid pWTluc (Fig. 7A) instead of into pWT. Since pWTluc lacks an intact X ORF, the missense mutations in the NRREenhII− mutant plasmid should be without consequence to the synthesis of functional X protein.
While the amount of pregenomic RNA synthesized from plasmid pNRREenhII−luc under endogenous levels of HNF4α was only one-third of the amount observed with pWTluc (Fig. 7B, lane 5 versus lane 1), overexpression of HNF4α still led to a sevenfold activation of synthesis of pregenomic RNA (Fig. 7B, lane 6 versus lane 5). By comparison, overexpression of HNF4α activated synthesis of pregenomic RNA from pNRREpreC−luc only twofold (Fig. 7B, lane 4 versus lane 3), to a level only one-sixth to one-seventh of the level observed with pWTluc (Fig. 7B, lane 2). Therefore, we conclude that NRREenhII plays a significant role in regulating synthesis of pregenomic RNA; however, its contribution is not as great as that of NRREpreC.
Regions essential for activation of pregenomic RNA synthesis by HNF4α.
Previous studies on the effect of overexpression of HNF4α on synthesis of pregenomic RNA in Huh7 cells showed activation of at most twofold (15; X. Yu, unpublished data); synthesis of pre-C RNA was actually repressed approximately fourfold or more when HNF4α was highly overexpressed (47). These results seem at odds with the ones reported here. However, one major difference between these two sets of experiments is that the ones presented here were performed with an intact, full-length HBV genome while the prior studies were performed with a subgenomic fragment containing only nt 1403 to 1991 of the HBV genome (47). Thus, HBV sequence elements in addition to the known NRREs may also play roles in the up-regulation of the pregenomic and pre-C promoters by HNF4α.
To test this hypothesis, we constructed a series of deletion variants of HBV (Fig. 8A). To insure proper polyadenylation and export of pre-C and pregenomic RNAs transcribed from these truncated HBV genomes, all of these HBV mutant genomes were introduced into pWTluc, a plasmid which contains a luciferase ORF upstream of an intron and a polyadenylation signal (Fig. 7A). Huh7 cells were transfected in parallel with pWTluc and its variants. Deletion of the 3′ half of the HBV genome did not adversely affect activation of synthesis of pregenomic RNA by HNF4α; if anything, it increased activation (Fig. 8B, lanes 1 to 6). However, removal as well of nt 2830 to 3068 resulted in activation by HNF4α of synthesis of pregenomic RNA being reduced to only 2.5-fold the synthesis in the absence of HNF4α activation (Fig. 8B, lanes 7 and 8). Additional removal of nt 2437 to 2829 did not have a further adverse effect on activation (Fig. 8B, lane 9 and 10). We also constructed an internal deletion mutant, pΔ5luc (Fig. 8A), to test whether removal of nt 2830 to 139 had the same effect as deletion of nt 2830 onward on transcriptional activation by HNF4α. Synthesis of pregenomic RNA from pΔ5luc was activated by HNF4α only 3.5-fold (Fig. 8B, lanes 11 and 12). The previously described HBV deletion mutant in which nt 1992 to 2435 were removed (46) was also introduced into pWTluc to generate pΔ6luc (Fig. 8A). Overexpression of HNF4α led to a less-than-twofold activation of the synthesis of pregenomic RNA from pΔ6luc (Fig. 8C, lanes 3 and 4).
To eliminate the possibility that the above findings were an artifact of the use of HBV-luc chimeras, this experiment was repeated with pΔ5 and pΔ6, plasmids created by deletion of nt 2830 to 139 and nt 1992 to 2435, respectively, from pWT, the original nonchimeric plasmid. The effects of overexpression of HNF4α on synthesis of pre-C and pregenomic RNA were essentially identical to the effects observed with the chimeric genomes (data not shown).
Thus, two regions in the HBV genome distal to the enhancer II and pre-C and pregenomic promoters are essential for high-level activation of synthesis of pregenomic RNA by HNF4α: one maps between nt 1992 and 2435 within the coding region for the C protein; the other maps between nt 2830 and 3068 within the coding region for pre-S1. However, neither of these two regions is required for the repression of synthesis of pre-C, pregenomic, and S RNAs by COUP-TF1 (data not shown).
One hypothesis to explain the above finding is that the viral proteins C and large S, abolished in these variants, play roles in this transcriptional up-regulation by HNF4α. To test this hypothesis, we constructed plasmids pC− and pC−/S−. Plasmid pC− contains a naturally occurring 1-bp insertion near the 5′ end of the C ORF, abolishing synthesis of core proteins. Plasmid pC−/S− contains, in addition, three in-frame translation termination codons within the S ORF, abolishing synthesis of all three surface proteins. Lack of synthesis of the C and S proteins had no significant effect on activation of synthesis of pregenomic RNA by HNF4α (Fig. 8D). Thus, these two regions likely function in cis in activation of transcription from the pregenomic promoter by HNF4α.
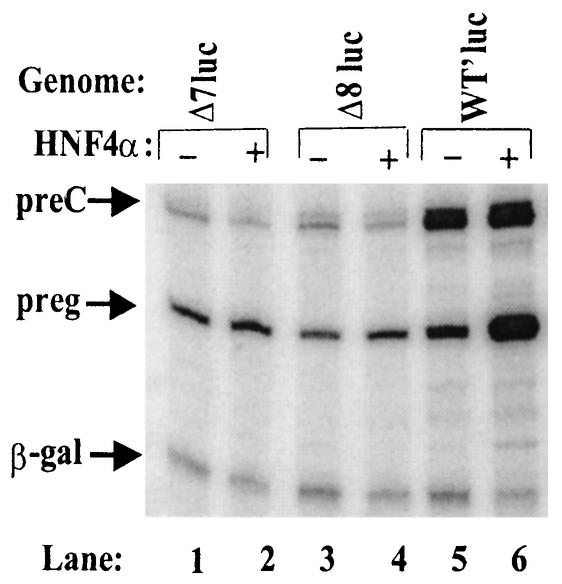
To demonstrate that these non-NRRE, cis-acting regulatory elements are also essential for high-level activation of the pregenomic promoter by HNF4α even when the NRREs are adjoined in their natural context, we constructed plasmid pWT′luc and its deletion variants pΔ7luc and pΔ8luc (Fig. 8A). Cotransfection experiments showed that, while HNF4α activated synthesis of pregenomic RNA from pWT′luc 4.5-fold (Fig. 9A, lanes 5 to 6), it only slightly activated synthesis of pregenomic RNA from either pΔ7luc or pΔ8luc (Fig. 9, lanes 1 to 4).
FIG. 9.
Maximal transcriptional up-regulation of the pregenomic promoter by HNF4α requires non-NRRE sequence elements as well. The autoradiogram shows the results of primer extension assays used to quantify the pre-C and pregenomic (preg) RNAs synthesized in Huh7 cells cotransfected with the indicated plasmids.
Thus, we conclude that inclusion of enhancers I and II and the pre-C and pregenomic promoters adjoined in their natural context with all three HNF4α-binding NRREs near each other is not sufficient for activation of synthesis of pregenomic RNA by HNF4α. In contrast, repression of synthesis of the pre-C and pregenomic RNAs by COUP-TF1 requires only the presence of the NRREs (data not shown).
DISCUSSION
Addressed here are the roles of HNF4α and COUP-TF1 in transcription and replication of HBV. We show that overexpression of HNF4α leads to a dramatic increase in synthesis of pregenomic RNA and a moderate increase in synthesis of pre-C RNA (Fig. 1), while overexpression of COUP-TF1 leads to a dramatic decrease in synthesis of both pre-C and pregenomic RNAs (Fig. 2). As expected from their effects on synthesis of pregenomic RNA, HNF4α and COUP-TF1 are a potent activator and repressor, respectively, of HBV DNA synthesis (Fig. 3). Overexpression of HNF4α and COUP-TF1 has effects on synthesis of pregenomic RNA from the naturally occurring CH variant of HBV similar to those on synthesis from the wild-type genome (Fig. 4). However, when these two NRs are co-overexpressed in transfected Huh7 cells, HNF4α reverses the repressive effect of COUP-TF1 on viral transcription more efficiently in the CH variant than in the wild-type genome (Fig. 5). Inactivation of NRREenhII also significantly reduced basal and HNF4α-activated synthesis of pregenomic RNA (Fig. 6 and 7). Deletion mapping revealed that the presence of cis-acting elements, in addition to the NRREs, is essential for activation of synthesis of pregenomic RNA by HNF4α but not for repression by COUP-TF1 (Fig. 8 and 9; data not shown).
HNF4α activation of synthesis of pregenomic RNA and viral DNA.
Overexpression of HNF4α led to a 14-fold increase in synthesis of pregenomic RNA from pWT, while it had minimum effects on synthesis of pre-C, S, pre-S1, and X RNAs (Fig. 1; data not shown). Given the fact that the HBV subviral particles, which contain the surface proteins and lipid only, are in 104- to 106-fold excess over Dane particles in the serum of HBV-infected patients (7), this preferential activation of synthesis of pregenomic RNA by HNF4α is an efficient and economic way for HBV to increase production of virions. Furthermore, limiting the activation of synthesis of pre-C RNA may also be beneficial because it avoids excessive production of the pre-C protein, which is known to repress HBV DNA synthesis (14, 23).
The mechanism of differential regulation of the closely nested pre-C and pregenomic promoters by HNF4α is unknown. One model is that binding of HNF4α to NRREpreC activates transcription of this promoter region but physically interferes partially with the formation of preinitiation complexes on the pre-C promoter because it overlaps directly this promoter's TATA box-like sequence (46). This model could also explain the similar observation that the binding of PPARs-RXRs to the NRREpreC also activates synthesis of pregenomic RNA while having little or no effect on synthesis of pre-C RNA (48).
Activation of synthesis of pregenomic RNA by HNF4α is primarily mediated through the NRREpreC. When both NRREpreC and NRREenhI were mutated, overexpression of HNF4α still activated synthesis of pregenomic RNA threefold, indicating that NRREenhII can partially compensate for the loss of function of these two other NRREs (Fig. 1, lanes 10 to 12). As previously reported (48), overexpression of PPAR-RXR in the presence of their respective ligands increases pregenomic RNA synthesis from pWT approximately 4- to 6-fold, not the 14-fold observed here with HNF4α (Fig. 1). Thus, HNF4α is likely a more potent activator of the pregenomic promoter than PPARs-RXRs in Huh7 cells. Our finding that NRREpreC is crucial for the activation of the pregenomic promoter by HNF4α is consistent with the report of Tang and McLachlan that NRREpreC mutants replicate considerably less well than wild-type HBV does in nonliver NIH 3T3 cells cotransfected with plasmids containing 1.3 tandem copies of the HBV genome and an HNF4α expression plasmid (36).
Interestingly, while activation of synthesis of pregenomic RNA by HNF4α is largely mediated by the NRREpreC (Fig. 1), activation of synthesis of S RNA by HNF4α is largely mediated by the NRREenhI (data not shown). Thus, activation of synthesis of HBV RNAs by HNF4α exhibits a position-dependent pattern, with the binding of HNF4α having a greater effect on transcription from the nearby promoter. However, when a nearby NRRE is inactivated, other NRREs in the HBV genome can partially compensate for its loss.
COUP-TF1 repression of synthesis of viral RNA and DNA.
Overexpression of COUP-TF1 in Huh7 cells repressed synthesis of pre-C and pregenomic RNAs similarly (Fig. 2) yet repressed synthesis of S and large S RNAs only moderately (data not shown). Therefore, like HNF4α, COUP-TF1 also modulates HBV replication by regulating synthesis of pregenomic RNA (Fig. 3). However, unlike what was found for HNF4α, regulation of transcription by COUP-TF1 does not seem to be position specific, i.e., COUP-TF1 repressed synthesis of pre-C, pregenomic, and S RNAs through interaction with either NRREpreC or NRREenhI (Fig. 2; data not shown). COUP-TF1 had very little effect on synthesis of pre-C, pregenomic, and S RNAs when both NRREpreC and NRREenhI, the two NRREs COUP-TF1 is known to bind, were mutated (Fig. 2, lanes 10 to 12; data not shown). This dependence on the presence of a functional NRRE in the genome and lack of dependence on sequences elsewhere in the genome (data not shown) indicates that COUP-TF1 represses transcription primarily, if not exclusively, via interactions with the NRREs.
Effects of the CH variant on competition between HNF4α and COUP-TF1 for regulation of HBV transcription and replication.
The mutations in the NRREpreC of the CH variant inactivate binding by COUP-TF1 and PPAR-RXR but leave binding by HNF4α unaffected (37, 48). Thus, as expected, activation of synthesis of pregenomic RNA by HNF4α was not affected in the CH variant because HNF4α can still bind NRREpreC (Fig. 4A, lanes 5 and 6). Repression of synthesis of this RNA by COUP-TF1 was also not affected because COUP-TF1 can still bind NRREenhI (Fig. 4B, lanes 5 and 6). However, when these two NRs were co-overexpressed in transfected Huh7 cells, HNF4α competed more efficiently with COUP-TF1 for the up-regulation of the pregenomic promoter in the CH variant than in the wild-type genome (Fig. 5A), leading to greater viral DNA synthesis (Fig. 5B).
Both HNF4α and COUP-TFs are potent transcriptional factors abundant in liver. Thus, their competition for binding to the NRREs in the HBV genome may be important for the regulation of HBV replication. The CH variant is frequently found in the livers of chronic hepatitis B patients (4). The widespread presence and persistence of the CH variant in these patients may be due to the fact that HNF4α up-regulates its replication in the presence of COUP-TFs more efficiently than it does the wild-type HBV.
NRREenhII also plays an important role in regulation of synthesis of pregenomic RNA.
NRREenhII, initially identified by Guo et al. (15), has not been characterized extensively. HNF4α is the only NR known to bind it. We found that mutations in NRREenhII in the context of the whole HBV genome reduce synthesis of pregenomic RNA by two-thirds to three-fourths both in the presence and absence of overexpressed HNF4α (Fig. 7B). Thus, NRREenhII is important for maximal transcription from the pregenomic promoter, with activation of synthesis of pregenomic RNA by HNF4α involving interactions with both NRREpreC and the NRREenhII.
Non-NRRE, cis-acting regulatory elements are also required for the up-regulation of the pregenomic promoter by HNF4α.
We (unpublished data) and others (15) had noted previously that HNF4α activates synthesis of pregenomic RNA only 1.5- to 2-fold from a subgenomic HBV plasmid containing the entire enhancer II and pre-C and pregenomic promoters (nt 1403 to 1990) in Huh7 cells overexpressing HNF4α. It was reported that a negative regulatory element from nt 1612 to 1633 in HBV suppressed activation of the pre-C and pregenomic promoters by enhancer II or HNF4α (15, 26). However, we found that when nt 1403 to 1633 were removed from the subgenomic HBV plasmid described above, overexpression of HNF4α led to only an approximately threefold increase in synthesis of pregenomic RNA (data not shown). One hypothesis to explain the seeming disparity between the large activation by HNF4α observed with the full-length HBV genome and the small activation with the HBV subgenome is that sequence elements in addition to the NRREs play roles in this activation. Moreover, activation of synthesis of pregenomic RNA by HNF4α from the subgenomic plasmids pΔ7luc and pΔ8luc containing enhancer I, enhancer II, and the pre-C and pregenomic promoters was barely detectable (Fig. 9), indicating that these elements are not in these two enhancers. Analysis of deletion variants of HBV supports this hypothesis and indicates that sequences within the regions from nt 1992 to 2435 and nt 2830 to 3068 are crucial for the up-regulation of the pregenomic promoter by HNF4α (Fig. 8B and C). Analysis of plasmids which either prevent synthesis of the P and X proteins by interruption of their ORF as in pWTluc (Fig. 7A) or which contain frameshift and nonsense mutations in the C and S ORF of HBV ruled out the possible involvement of virus-encoded trans-acting factors. Thus, we conclude that it is cis-acting regulatory elements present within these two regions that play roles in activation by HNF4α. These elements are likely binding sites for other transcription factors that interact with HNF4α from a distance to facilitate transcription from a promoter near the NRRE.
Interestingly, these elements are not required for the moderate activation of synthesis of S, pre-C, and pregenomic RNAs by HNF4α in Huh7 cells (15) (data not shown). Thus, we hypothesize that there exist two mechanisms of activation of HBV RNA synthesis by HNF4α in Huh7 cells: basal activation and enhanced activation. The former, which results in 2- to 3-fold activation, requires only the presence of NRREs; the latter, which results in 5- to 10-fold or more activation, requires the presence of other sequence elements within the HBV genome. The enhanced transcriptional activation by HNF4α was observed here only for the pregenomic promoter. Similar activation of synthesis of pregenomic RNA and viral DNA from an adw2 HBV genome has also been observed (data not shown).
HNF4α regulates the expression of a large number of viral and cellular genes. Although it has been reported that HNF4α and other transcription factors bound to sites near NRREs can cooperatively activate transcription (27, 35), this is the first case implying that long-range interactions with regulatory proteins can also be crucial for transcriptional regulation by HNF4α.
In summary, we conclude that HNF4α and COUP-TF1 are potent regulators of HBV and exert opposite effects on transcription and replication. In addition, they function through distinct mechanisms, with the positions of the NRREs and the presence of other cis-acting regulatory elements being crucial for activation by HNF4α, while COUP-TF1 acts at a distance without the requirement for additional regulatory elements.
Acknowledgments
We thank Dan Loeb, Jeff Habig, Paul Lambert, and members of our laboratory for helpful discussions and comments on the manuscript.
This work was supported by Public Health Service research grants CA22443 and CA07175 from the National Cancer Institute.
REFERENCES
- 1.Bagchi, M. K., S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 1987. Purification and characterization of chicken ovalbumin gene upstream promoter transcription factor from homologous oviduct cells. Mol. Cell. Biol. 7:4151-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumberg, B., and R. M. Evans. 1998. Orphan nuclear receptors—new ligands and new possibilities. Genes Dev. 12:3149-3155. [DOI] [PubMed] [Google Scholar]
- 3.Brown, T. 1993. Southern blotting, p. 2.9.1. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Buckwold, V., and J. Ou. 1999. Hepatitis B virus C-gene expression and function: the lessons learned from viral mutants. Curr. Top. Virol. 1:71-81. [Google Scholar]
- 5.Butz, K., and F. Hoppe-Seyler. 1993. Transcriptional control of human papillomavirus (HPV) oncogene expression: composition of the HPV type 18 upstream regulatory region. J. Virol. 67:6476-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooney, A. J., X. Leng, S. Y. Tsai, B. W. O'Malley, and M.-J. Tsai. 1993. Multiple mechanisms of chicken ovalbumin upstream promoter transcription factor-dependent repression of transactivation by the vitamin D, thyroid hormone, and retinoic acid receptors. J. Biol. Chem. 268:4152-4160. [PubMed] [Google Scholar]
- 7.Dane, D. S., C. H. Cameron, and M. Briggs. 1970. Virus-like particles in serum of patients with Australia-antigen-associated hepatitis. Lancet i:695-698. [DOI] [PubMed]
- 8.Fraser, J. D., V. Martinez, R. Straney, and M. R. Briggs. 1998. DNA binding and transcription activation specificity of hepatocyte nuclear factor 4. Nucleic Acids Res. 26:2702-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galson, D. L., T. Tsuchiya, D. S. Tendler, L. E. Huang, Y. Ren, T. Ogura, and H. F. Bunn. 1995. The orphan receptor hepatic nuclear factor 4 functions as a transcriptional activator for tissue-specific and hypoxia-specific erythropoietin gene expression and is antagonized by EAR3/COUP-TF1. Mol. Cell. Biol. 15:2135-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganem, D., and R. J. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2969. In D. M. Knipe and P. M. Howley (ed.), Virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 11.Ganem, D., and H. E. Varmus. 1987. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 56:651-693. [DOI] [PubMed] [Google Scholar]
- 12.Garcia, A. D., P. Ostapchuk, and P. Hearing. 1993. Functional interaction of nuclear factors EF-C, HNF-4, and RXRα with hepatitis B virus enhancer I. J. Virol. 67:3940-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 14.Guidotti, L. G., B. Matzke, C. Pasquinelli, J. M. Shoenberger, C. E. Rogler, and F. V. Chisari. 1996. The hepatitis B virus (HBV) precore protein inhibits HBV replication in transgenic mice. J. Virol. 70:7056-7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, W., M. Chen, T. S. B. Yen, and J.-H. Ou. 1993. Hepatocyte-specific expression of the hepatitis B virus core promoter depends on both positive and negative regulation. Mol. Cell. Biol. 13:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayhurst, G. P., Y.-H. Lee., G. Lambert, J. M. Ward, and F. J. Gonzalez. 2001. Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 21:1393-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hertz, R., J. Magenheim, I. Berman, and J. Bar-Tana. 1998. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4α. Nature 392:512-516. [DOI] [PubMed] [Google Scholar]
- 18.Huan, B., M. J. Kosovsky, and A. Siddiqui. 1995. Retinoid X receptor α transactivates the hepatitis B virus enhancer 1 element by forming a heterodimeric complex with the peroxisome proliferator-activated receptor. J. Virol. 69:547-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huan, B., and A. Siddiqui. 1992. Retinoid X receptor RXR α binds to and trans-activates the hepatitis B virus enhancer. Proc. Natl. Acad. Sci. USA 89:9059-9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, G., L. Nepomuceno, K. Hopkins, and F. M. Sladek. 1995. Exclusive homodimerization of the orphan receptor hepatocyte nuclear factor 4 defines a new subclass of nuclear receptors. Mol. Cell. Biol. 15:5131-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura, A., A. Nishiyori, T. Murakami, T. Tsukamoto, S. Hata, T. Osumi, R. Okamura, M. Mori, and M. Takiguchi. 1993. Chicken ovalbumin upstream promoter-transcription factor (COUP-TF) represses transcription from the promoter of the gene for ornithine transcarbamylase in a manner antagonistic to hepatocyte nuclear factor-4 (HNF-4). J. Biol. Chem. 268:11125-11133. [PubMed] [Google Scholar]
- 22.Kliewer, S. A., J. M. Lehmann, and T. M. Willson. 1999. Orphan nuclear receptors: shifting endocrinology into reverse. Science 284:757-760. [DOI] [PubMed] [Google Scholar]
- 23.Lamberts, C., M. Nassal, I. Velhagen, H. Zentgraf, and C. H. Schröder. 1993. Precore-mediated inhibition of hepatitis B virus progeny DNA synthesis. J. Virol. 67:3756-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leng, X., A. J. Cooney, S. Y. Tsai, and M.-J. Tsai. 1996. Molecular mechanisms of COUP-TF-mediated transcriptional repression: evidence for transrepression and active repression. Mol. Cell Biol. 16:2332-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, J., G. Ning, and S. A. Duncan. 2000. Mammalian hepatocyte differentiation requires the transcription factor HNF-4α. Genes Dev. 14:464-474. [PMC free article] [PubMed] [Google Scholar]
- 26.Lo, W.-Y., and L.-P. Ting. 1994. Repression of enhancer II activity by a negative regulatory element in the hepatitis B virus genome. J. Virol. 68:1758-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paré, J.-F., S. Roy, L. Galarneau, and L. Bélanger. 2001. The mouse fetoprotein transcription factor (FTF) gene promoter is regulated by three GATA elements with tandem E box and Nkx motifs, and FTF in turn activates the Hnf3β, Hnf4α, and Hnf1α gene promoters. J. Biol. Chem. 276:13136-13144. [DOI] [PubMed] [Google Scholar]
- 28.Pereira, F. A., M.-J. Tsai, and S. Y. Tsai. 2000. COUP-TF orphan nuclear receptors in development and differentiation. Cell. Mol. Life Sci. 57:1388-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu, Y., S. Y. Tsai, and M.-J. Tsai. 1994. COUP-TF: an orphan member of the steroid/thyroid hormone receptor superfamily. Trends Endocrinol. Metab. 5:234-239. [DOI] [PubMed] [Google Scholar]
- 30.Raney, A. K., J. L. Johnson, C. N. A. Palmer, and A. McLachlan. 1997. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J. Virol. 71:1058-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed, p. 16.14-16.20. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Schleiss, M. R., C. R. Degnin, and A. P. Geballe. 1991. Translational control of human cytomegalovirus gp48 expression. J. Virol. 65:6782-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spath, G. F., and M. C. Weiss. 1997. Hepatocyte nuclear factor 4 expression overcomes repression of the hepatic phenotype in dedifferentiated hepatoma cells. Mol. Cell. Biol. 17:1913-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staprans, S., D. D. Loeb, and D. Ganem. 1991. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J. Virol. 65:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stroup, D., and J. Y. Chiang. 2000. HNF4 and COUP-TFII interact to modulate transcription of the cholesterol 7 alpha-hydroxylase gene (CYP7A1). J. Lipid Res. 41:1-11. [PubMed] [Google Scholar]
- 36.Tang, H., and A. McLachlan. 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl. Acad. Sci. USA 98:1841-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang, H., A. K. Raney, and A. McLachlan. 2001. Replication of the wild type and a natural hepatitis B virus nucleocapsid promoter variant is differentially regulated by nuclear hormone receptors in cell culture. J. Virol. 75:8937-8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai, M.-J., and B. W. O'Malley. 1994. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 63:451-486. [DOI] [PubMed] [Google Scholar]
- 39.Tur-Kaspa, R., R. D. Burk, Y. Shaul, and D. A. Shafritz. 1986. Hepatitis B virus DNA contains a glucocorticoid-responsive element. Proc. Natl. Acad. Sci. USA 83:1627-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tur-Kaspa, R., Y. Shaul, D. D. Moore, R. D. Burk, S. Okret, L. Poellinger, and D. A. Shafritz. 1988. The glucocorticoid receptor recognizes a specific nucleotide sequence in hepatitis B virus DNA causing increased activity of the HBV enhancer. Virology 167:630-633. [PubMed] [Google Scholar]
- 41.Wang, L.-H., S. Y. Tsai, I. Sagami, M.-J. Tsai, and B. W. O'Malley. 1987. Purification and characterization of chicken ovalbumin upstream promoter transcription factor from HeLa cells. J. Biol. Chem. 262:16080-16086. [PubMed] [Google Scholar]
- 42.Wang, L.-H., N. H. Ing, S. Y. Tsai, B. W. O'Malley, and M.-J. Tsai. 1991. The COUP-TFs compose a family of functionally related transcription factors. Gene Expr. 1:207-216. [PMC free article] [PubMed] [Google Scholar]
- 43.Whitfield, G. K., P. W. Jurutka, C. A. Haussler, and M. R. Haussler. 1999. Steroid hormone receptors: evolution, ligands, and molecular basis of biologic function. J. Cell Biochem. 32-33(Suppl.):110-122. [DOI] [PubMed] [Google Scholar]
- 44.Wiley, S. R., R. J. Kraus, F. Zuo, E. E. Murray, K. Loritz, and J. E. Mertz. 1993. SV40 early-to-late switch involves titration of cellular transcriptional repressors. Genes Dev. 7:2206-2219. [DOI] [PubMed] [Google Scholar]
- 45.Yanai, K., K. Hirota, K. Taniguchi-Yanai, Y. Shigematsu, Y. Shimamoto, T. Saito, S. Chowdhury, M. Takiguchi, M. Arakawa, Y. Nibu, F. Sugiyama, K. Yagami, and A. Fukamizu. 1999. Regulated expression of human angiotensinogen gene by hepatocyte nuclear factor 4 and chicken ovalbumin upstream promoter-transcription factor. J. Biol. Chem. 274:34605-34612. [DOI] [PubMed] [Google Scholar]
- 46.Yu, X., and J. E. Mertz. 1996. Promoters for synthesis of the pre-C and pregenomic mRNAs of human hepatitis B virus are genetically distinct and differentially regulated. J. Virol. 70:8719-8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, X., and J. E. Mertz. 1997. Differential regulation of the pre-C and pregenomic promoters of human hepatitis B virus by members of the nuclear receptor superfamily. J. Virol. 71:9366-9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, X., and J. E. Mertz. 2001. Critical roles of nuclear receptor response elements in replication of hepatitis B virus. J. Virol. 75:11354-11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuo, F., and J. E. Mertz. 1995. Simian virus 40 late gene expression is regulated by members of the steroid/thyroid hormone receptor superfamily. Proc. Natl. Acad. Sci. USA 92:8586-8590. [DOI] [PMC free article] [PubMed] [Google Scholar]