Abstract
The multidomain RNA replication protein 1a of brome mosaic virus (BMV), a positive-strand RNA virus in the alphavirus-like superfamily, plays key roles in assembly and function of the viral RNA replication complex. 1a, which encodes RNA capping and helicase-like domains, localizes to endoplasmic reticulum membranes, recruits BMV 2a polymerase and viral RNA templates, and forms membrane-bound, capsid-like spherules in which RNA replication occurs. cis-acting signals necessary and sufficient for RNA recruitment by 1a have been mapped in BMV genomic RNA2 and RNA3. Both signals comprise an extended stem-loop whose apex matches the conserved sequence and structure of the TΨC stem-loop in tRNAs (box B). Mutations show that this box B motif is crucial to 1a responsiveness of wild-type RNA2 and RNA3. We report here that, unexpectedly, some chimeric mRNAs expressing the 2a polymerase open reading frame from RNA2 were recruited by 1a to the replication complex and served as templates for negative-strand RNA synthesis, despite lacking the normally essential, box B-containing 5′ signal. Further studies showed that this template recruitment required high-efficiency translation of the RNA templates. Moreover, multiple small frameshifting insertion or deletion mutations throughout the N-terminal region of the open reading frame inhibited this template recruitment, while an in-frame insertion did not. Providing 2a in trans did not restore template recruitment of RNAs with frameshift mutations. Only those deletions in the N-terminal region of 2a that abolished 2a interaction with 1a abolished template recruitment of the RNA. These and other results indicate that this alternate pathway for 1a-dependent RNA recruitment involves 1a interaction with the translating mRNA via the 1a-interactive N-terminal region of the nascent 2a polypeptide. Interaction with nascent 2a also may be involved in 1a recruitment of 2a polymerase to membranes.
Brome mosaic virus (BMV) is a well-studied member of the large alphavirus-like superfamily of animal and plant positive-strand RNA viruses. The genome of BMV is divided into three capped RNAs (1, 41). RNA1 and RNA2 encode nonstructural proteins 1a and 2a, respectively, which direct RNA replication and contain domains conserved with other superfamily members (3, 16, 22). 1a contains an N-terminal domain with m7G methyltransferase and covalent GTP binding activities that are required for capping of viral RNA during RNA replication in vivo (2, 3, 26) and a C-terminal domain with all motifs of DEAD box RNA helicases (19). Mutations in the helicase-like domain cause strong defects in RNA replication (3). The central portion of 2a is similar to RNA-dependent RNA polymerases (5, 21). RNA3 is a bicistronic RNA encoding the 3a cell-to-cell movement protein and the coat protein. Both of these proteins are required for systemic infection of BMV's natural plant hosts but are dispensable for RNA replication in a single cell (4, 27, 35). The 3a protein is directly translated from the 5′-proximal 3a gene of RNA3, whereas the 3′-proximal coat gene is translated from a subgenomic mRNA, RNA4, produced from the negative-strand RNA3 replication intermediate.
Like that of most, if not all, eukaryotic positive-strand RNA viruses, BMV RNA replication occurs on membrane-associated complexes (13, 17, 20, 32-34). Besides providing essential enzymatic functions for RNA replication, 1a plays key roles in the assembly and function of the BMV RNA replication complex, which is associated with endoplasmic reticulum (ER) membranes. 1a localizes to the cytoplasmic face of ER membranes in the absence of other viral factors (33) by signals residing in the N-proximal half of the protein (11). In contrast, the 2a polymerase depends on 1a for recruitment to the site of replication through direct interaction between the N terminus of 2a and the C-terminal helicase-like domain of 1a (9, 24, 25). 1a also recruits viral RNA templates into replication (9, 22, 40). The assembly, structure, and function of the BMV RNA replication complex exhibit close parallels with retrovirus capsids, with the BMV RNA replication proteins 1a and 2a and certain cis-acting replication signals recapitulating the functions of Gag, Pol, and RNA packaging signals in conventional retrovirus and foamy virus cores (37). To assemble the replication complex and replicate RNA, 1a induces formation of membrane-enveloped, capsid-like spherules that partially bud into the ER lumen, invaginating the outer ER membrane. When RNA template and 2a are coexpressed, they are sequestered into these spherules by interacting either directly or indirectly with 1a. Negative-strand RNA synthesis is then carried out inside the spherules by 1a and 2a. The resulting negative-strand RNA is retained in the spherule and used to synthesize progeny positive-strand RNA for export to the cytoplasm (37).
BMV RNA replication and subgenomic RNA synthesis depend on cis-acting signals in the 5′, 3′, and intergenic sequences within RNAs 1, 2, and 3 (reviewed in reference 41). Negative-strand RNA synthesis is promoted by conserved, 3′-terminal, tRNA-like structures (8, 12), whereas correct positive-strand initiation requires 5′-terminal sequences (41) and a nontemplate guanylate added to the 3′ end of the negative strand in vitro (38). The intergenic region of RNA3 contains two overlapping cis signals: the subgenomic mRNA promoter (14) and an approximately 150-nucleotide (nt) recognition element, which contains a box B motif that is conserved with the TΨC loop of tRNAs (15). The same box B motif is found in the 5′ untranslated regions (UTRs) of RNAs 1 and 2. In the absence of 2a protein and hence RNA replication, 1a protein acts through these box B motifs and flanking sequences to recruit the BMV RNAs into the membrane-bound spherular replication complexes, strikingly increasing the in vivo stability of RNA2 and RNA3 (10, 40).
The 1a-responsive sequences in the RNA3 intergenic and RNA2 5′-terminal regions both form extended stem-loops (7, 10) that present the box B motif at the apex as a 7-nt hairpin loop, exactly matching the conserved TΨC stem-loop in tRNAs (7). Deletion, partial deletion, or mutation of these RNA2 or RNA3 box B elements or their flanking sequences dramatically impairs 1a responsiveness, negative-strand synthesis, and replication of these RNAs (10, 15, 30, 31, 39, 40). However, here we report that, unexpectedly, some RNA2 derivatives expressing the 2a polymerase open reading frame (ORF) were highly responsive to 1a and served as templates for negative-strand RNA synthesis, despite lacking the normally essential, box B-containing 5′ signal. We also find that this box B-independent 1a responsiveness depends on high-efficiency, in cis translation of the N-terminal half of 2a. Since this 2a region interacts directly with 1a and preserving this 1a-2a interaction was essential for the RNA to be 1a responsive, these and other results imply that this pathway for 1a-dependent RNA recruitment involves 1a interaction with the nascent 2a peptide associated with the translating mRNA.
MATERIALS AND METHODS
Yeast methods.
Yeast strain YPH500 (MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1) was used throughout. Plasmid DNAs were introduced into yeast cells by using the Frozen-EZ yeast transformation kit (Zymo Research) in accordance with the manufacturer's protocol. Yeast cultures were grown at 30°C in defined synthetic medium selective for the desired plasmids, with either 2% glucose or 2% galactose as a carbon source (6).
Plasmids and plasmid constructions.
Standard procedures were used for all DNA manipulations (36). DNA fragments generated by PCR were confirmed by DNA sequencing, and the overall structures of all plasmids were confirmed by restriction analysis. Laboratory designations for plasmids are given in parentheses if they are different from names used in this paper.
BMV 1a was expressed from pB1CT19 (23), a 2μm plasmid containing the HIS3 selectable marker gene and 1a ORF flanked by the yeast ADH1 promoter and ADH1 polyadenylation site.
RNA2 was expressed from pB2NR3, a centromeric plasmid containing the LEU2 selectable marker and a full-length RNA2 cDNA flanked by the yeast GAL1 promoter and a self-cleaving hepatitis delta virus ribozyme (10). RNA2ΔA (pB2NR3ΔSL1) and RNA2ΔAB (pB2NR3ΔSL12) were made by PCR with 5′ primers extending from the first nucleotide of RNA2 through and beyond the desired mutations and a common 3′ primer complementary to RNA2 nt 817 to 890. PCR products containing the desired mutations then were digested with NcoI and used to replace the SnaBI-NcoI fragment in pB2NR3.
G2aA was expressed from pB2YT5 (generously provided by M. Ishikawa), which is based on Ycplac111, a yeast centromeric plasmid that contains the LEU2 selectable marker gene and multiple cloning sites from pUC19 (18). G2aT was expressed from pB2YT5T, which was made by replacing the MluI-SphI fragment of pB2YT5 with the corresponding fragment from pB2NR3.
pB2YT5 was digested with BstBI, NcoI, and MluI, blunt-ended with T4 DNA polymerase, and religated to yield G2aA-fs1 (pB2YT5-M1), G2aA-fs4 (pB2YT5-M2), and G2aA-fs6 (pB2YT5-M3), respectively. Several intermediate plasmids were created to facilitate the construction of pB2YT5 derivatives G2aA-fs2 (pB2YT5-M4A), G2aA-fs3 (pB2YT5-M5), and G2aA-fs5 (pB2YT5-M6). pBSM1 is a pBluescript II SK(+) derivative whose polylinker was modified by KpnI and EcoRI digestion, blunt ending, and religation to remove these sites. The BamHI-PstI fragment from pB2YT5 was inserted into pBSM1 to yield pBSM1YT5, which was then digested with NheI, AvaI, and KpnI, blunt ended, and religated to yield pBSM1YT5-M4A, -M5, and -M6, respectively. Sequence analysis revealed one plasmid (pBSM1YT5-M4B) from the NheI treatment bearing a 3-nt insertion instead of the predicted 4-nt insertion. Finally, the BamHI-PstI fragment in pB2YT5 was replaced with the corresponding fragments from pBSM1YT5-M4A, -M4B, -M5, and -M6 to yield G2aA-fs2, -if2, -fs3, and -fs5, respectively.
To make G2aA-GFP (pB2YT2-G3), which contains a TRP1 selectable marker, the green fluorescent protein (GFP) gene was initially inserted into pB2YT5 between the 2a ORF and the polyadenylation signal. A unique NotI restriction site was added downstream from the 2a ORF but preceding the BamHI site by PCR-mediated mutagenesis to yield pB2YT5-NotI. Then, a BamHI-PstI (blunt-ended) fragment containing the GFP gene was inserted into pB2YT5-NotI by replacing the BamHI-NotI (blunt-ended) fragment to yield pB2YT5-G3. Finally, the BamHI-PstI fragment from pB2YT5-G3 was used to replace the corresponding fragment of pB2YT2 (generously provided by M. Ishikawa) to yield G2aA-GFP.
All in-frame deletions (G2aAΔ1 [pB2YT5-D13], G2aAΔ2 [pB2YT5-D14], G2aAΔ3 [pB2YT5-D15], G2aAΔ4 [pB2YT5-D16], G2aAΔ5 [pB2YT5-D17], G2aAΔ6 [pB2YT5-D18], G2aAΔ7 [pB2YT5-D19], and G2aAΔ8 [pB2YT5-D191]) were G2aA derivatives that were created by PCR-mediated mutagenesis (10).
RNA and protein analysis.
Total yeast RNA isolation, Northern blot analysis, two-cycle RNase protection assays, total protein extraction, and Western blot analysis were performed as described previously (9, 10). For cell fractionation, yeast cells were grown in synthetic galactose medium for 48 h to induce RNA expression, harvested at mid-log phase, and converted to spheroplasts as described previously (9). Spheroplasts were osmotically lysed by pipetting up and down in extraction buffer (50 mM Tris [pH 8.0] and 10 mM ribonucleoside vanadyl complex for RNA fractionation and 50 mM Tris [pH 8.0], 10 mM EDTA, 10 mM dithiothreitol, 5 mM benzamidine, 2 mM phenylmethylsulfonyl fluoride, and aprotinin, leupeptin, and pepstatin A at 10 μg/ml each for protein fractionation). The resulting lysate was centrifuged for 10 min at 10,000 × g. The supernatant was removed and retained, and the pellet was washed once with extraction buffer and resuspended to the original lysate volume in extraction buffer. Nucleic acids were isolated from these fractions by phenol-chloroform extraction and analyzed by Northern blotting.
RESULTS
1a induces membrane association of RNA2 derivatives lacking the 5′-terminal stem-loop.
When yeast cells are converted into spheroplasts, lysed, and centrifuged at low speed, a soluble supernatant and a membrane-containing pellet fraction can be obtained. In the absence of 1a expression, BMV RNAs, like most if not all yeast mRNAs and rRNAs, are recovered mainly in the supernatant fraction (10, 37). However, 1a coexpression induces specific membrane association of BMV RNAs that can be directly visualized by cell fractionation (Fig. 1). Multiple results link this 1a-induced membrane association to recruitment of BMV RNAs into replication (10, 37, 40). Thus, cell fractionation provides a direct measure of an early step of RNA replication that precedes negative-strand RNA synthesis, which additionally requires the polymerase-like 2a protein.
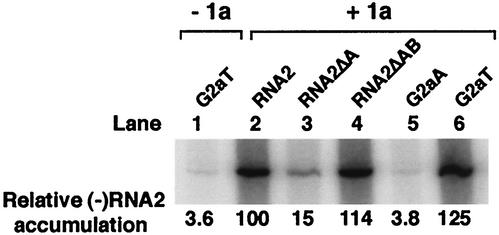
FIG. 1.
RNA2 5′ box B-dependent and 2a ORF-dependent 1a-induced membrane association. (A) Sequences and predicted secondary structure of the RNA2 5′ UTR. The conserved box B motif is boxed, and the three subdomains discussed in Results, which are defined based on alignment with the RNA1 5′ UTR, are indicated. (B) On the left are schematic diagrams of RNA2 and its derivatives. Membrane association abilities of these RNAs in the absence (−1a) or presence (+1a) of 1a were assessed by cell fractionation. Yeast cells expressing these RNAs with or without 1a were spheroplasted and lysed osmotically. The lysate then was centrifuged at 10,000 × g to yield a membrane-containing pellet (P) fraction and a supernatant (S) fraction. RNA was isolated from each fraction by phenol-chloroform extraction, and equal percentages of each fraction were analyzed by Northern blotting to detect positive-strand RNA2. Representative Northern blots are shown at the right.
The 5′ UTR of BMV RNA2 can be divided into three subdomains based on alignment with RNA1 5′ UTR (Fig. 1). Subdomain A, consisting of the first 46 nt, is 92% identical between RNA1 and RNA2; subdomain B is a 31-nt region specific to RNA2; and subdomain C is a pyrimidine-rich sequence that is 80% identical between RNA1 and RNA2 (28). Subdomain A has the potential to form a stem-loop structure, which is conserved between RNA1 and RNA2 of bromoviruses and cumumoviruses. Previous studies have shown that deleting all or part of subdomain A or creating base substitution mutations within subdomain A dramatically inhibits the 1a responsiveness of RNA2, including the ability of 1a to recruit RNA2 into a membrane-associated state (Fig. 1, RNA2ΔA) (10). However, unexpectedly, we found that some RNA2 derivatives lacking subdomain A also were highly responsive to 1a. For instance, an RNA2 derivative with both subdomains A and B deleted, RNA2ΔAB, was efficiently recruited to the membrane by 1a, as shown by cell fractionation (Fig. 1). Further studies (Fig. 1B) (also see below) revealed that this 1a responsiveness was conferred by sequences within the 2a ORF) of RNA2. As shown in Fig. 1B, RNA2 derivatives either with the 5′ UTR sequence replaced by the yeast GAL1 5′ UTR (G2aT) or with both the 5′ UTR and 3′ tRNA-like sequences replaced by the yeast GAL1 5′ UTR and ADH1 polyadenylation sequences (G2aA) were efficiently recruited to membrane by 1a. Previous studies have demonstrated that neither the yeast GAL1 5′ UTR nor the ADH1 polyadenylation signal confers 1a responsiveness by itself (10, 40). Thus, RNA2 derivatives retaining the 2a ORF as the only viral sequence can be 1a responsive independently of the 5′-terminal stem-loop structure. These findings and previous results thus imply that two independent pathways exist in RNA2 for 1a-dependent recruitment to membrane: the box B-containing 5′ terminal stem-loop-dependent pathway and an alternate 2a ORF-dependent pathway. The 2a ORF-dependent pathway appears to be cryptic or to function at very low levels in wt RNA2 and its derivatives retaining 5′ UTR segment B (Fig. 1B, RNA2ΔA). Deleting subdomains A and B simultaneously from RNA2 or replacing the entire RNA2 5′ UTR with the yeast GAL1 leader appeared to activate or enhance the functions of the 2a ORF-dependent pathway through mechanisms that are the subject of the studies presented below.
RNAs recruited through the 2a ORF-dependent pathway are functional templates for negative-strand RNA synthesis.
Consistent with an essential role in RNA replication, prior studies have shown that RNA mutations blocking box B-dependent recruitment of BMV RNAs into the 1a-induced membrane-associated complex also block negative-strand RNA synthesis (10, 40). To test if RNAs recruited by 1a through the 2a ORF-dependent pathway also were assembled into functional RNA replication complexes, we examined whether such RNAs served as templates for negative-strand RNA synthesis. To assay for negative-strand RNA products, total RNA extracted from yeast cells was subjected to an RNase protection assay by hybridization with a 32P-labeled RNA probe complementary to 245 nt in the center of negative-strand RNA2 and treatment with single-strand-specific RNases A and T1. In keeping with previous findings (10), first, a strong negative-strand signal was detected in yeast coexpressing wild-type (wt) RNA2 and 1a (Fig. 2, lane 2). Second, a weak background signal was observed for all tested RNAs in cells lacking 1a expression, as illustrated in lane 1 of Fig. 2 for G2aA. As demonstrated previously, this background signal was produced by 1a- and 2a-independent mechanisms such as cryptic promoter-initiated transcription of the RNA2 cDNA in the direction opposite to GAL1-promoted transcription (10). Inhibiting 1a responsiveness reduced negative-strand RNA accumulation to a similar background level as illustrated for RNA2ΔA (Fig. 2, lane 3). Conversely, restoring 1a responsiveness by further deleting subdomain B (RNA2ΔAB) or by replacing the 5′ end with the GAL1 5′ UTR (G2aT) restored negative-strand RNA accumulation to the wt RNA2 level (Fig. 2, lanes 4 and 6). As expected, an RNA such as G2aA, lacking the viral 3′ tRNA-like structure containing the negative-strand promoter, was not a template for negative-strand RNA synthesis even though that RNA was efficiently recruited to membranes by 1a (Fig. 2, lane 5). Thus, these results demonstrated that RNAs recruited through the 2a ORF-dependent pathway could be functional templates for negative-strand RNA synthesis.
FIG. 2.
The 2a ORF-dependent pathway recruits RNA2 derivatives to functional RNA replication complexes. Negative-strand RNA2 accumulation was assayed by a two-cycle RNase protection assay (29) with equal amounts of total RNA extracted from yeast expressing each of the RNA2 derivatives shown in Fig. 1B in the presence (+1a) or absence (−1a) of 1a as indicated. After initial hybridization and RNase treatment to remove excess positive-strand RNA2, the remaining double-stranded RNA was denatured, hybridized with a 32P-labeled RNA probe corresponding to nt 1441 to 1685 of positive-strand RNA2, and treated with RNases A and T1. The reaction products were electrophoresed and autoradiographed. The relative strength of the negative-strand signal for each RNA2 derivative, averaged over three independent experiments, is shown at the bottom.
2a ORF-dependent 1a responsiveness is correlated with efficient 2a translation.
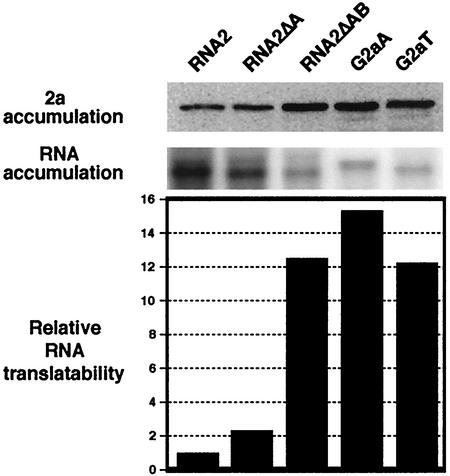
Previously (10), we found that wt RNA2 was translated poorly compared to a chimeric 2a-expressing RNA similar to G2aA, consisting of the 5′ yeast GAL1 UTR, the 2a ORF, and a 3′ poly(A) tail (Fig. 1B). To determine if high-level translation might be responsible for activating or enhancing the 2a ORF-dependent function of 1a responsiveness, the accumulation of 2a protein in yeast harboring each of the RNA2 derivatives described in Fig. 1B was compared by Western blotting of total protein extracts. At the same time, RNA accumulation also was compared by Northern blotting analysis of total RNA. The relative translational activity of each RNA then was calculated as the ratio between the levels of 2a protein and 2a mRNA. As shown in Fig. 3, RNA2ΔA, like wt RNA2, was translated poorly, while RNA2ΔAB, G2aA, and G2aT were translated at 12- to 15-fold-higher levels than wt RNA2 (Fig. 3). Thus, these results suggested that active translation of the RNA might be required for the 2a ORF-dependent pathway of 1a responsiveness. Additional experiments (see below) were used to further explore the role of translation in this process. As discussed further below, dependence on translation could be due to an active function played by 2a in this 1a responsiveness.
FIG. 3.
Translational activities of RNA2 derivatives. The accumulation of 2a protein in yeast harboring each of the RNA2 derivatives shown in Fig. 1B was compared by Western blotting of total protein extracts. At the same time, RNA accumulation of these RNA2 derivatives was compared by Northern blotting of total RNA from the same yeast cultures. The relative translational activity of each RNA was calculated as the ratio between the levels of 2a protein and 2a mRNA. The averages from three or more independent experiments are shown.
Translation of the N-terminal half of 2a is required for 2a ORF-dependent 1a responsiveness.
To explore whether translation of full-length 2a was required for the 2a ORF-dependent 1a responsiveness, we created a series of G2aA derivatives with frameshift mutations in the 2a ORF by restriction enzyme digestion, blunt ending, and religation (Fig. 4A). The number of nucleotides inserted or deleted and their position in RNA2, as well as the resulting number of wt 2a codons and out-of-frame codons translated, are indicated in Fig. 4A. For example, G2aA-fs1 bore a 2-nt insertion at RNA2 position 110, resulting in translation of 3 wt 2a codons followed by 18 out-of-frame codons. G2aA-if2 (Fig. 4B), bearing a 3-nt in-frame insertion at RNA2 position 198 without causing frameshift or early termination, was obtained serendipitously during the construction of G2aA-fs2. Western blotting analysis was performed to confirm that all mutants exhibited the expected translation phenotypes in yeast. Cells expressing G2aA-fs4 to -fs6 and G2aA-if2 contained 2a peptides of the predicted sizes, while cells expressing the smaller ORFs of G2aA-fs1 to -fs3 contained no detectable 2a-related peptides (Fig. 4C).
FIG. 4.
Effects of frameshift mutations on 2a ORF-dependent 1a responsiveness. (A) On the left are schematic diagrams of G2aA and its frameshift derivatives. The number of nucleotides inserted (i) or deleted (Δ) and their position (*) in RNA2 as well as the resulting number of in-frame 2a codons (preceding +) and out-of-frame codons (following +) translated are indicated. Membrane association abilities of these RNAs in the absence (−1a) or presence (+1a) of 1a were assessed by cell fractionation to measure RNA distribution in the pellet (P) and supernatant (S) fractions as described in the legend to Fig. 1. Representative Northern blots are shown. As a measure of 1a responsiveness, the difference between the percentages of pelletable RNA in the presence and the absence of 1a was calculated for each RNA. The data shown are averages from three or more independent experiments for each derivative. (B) Comparison of the 1a responsiveness of matched frameshift and in-frame insertion mutations, which differ by only a single nucleotide. (C) Western blotting of total protein extracts from yeast harboring each of the G2aA derivatives shown in panels A and B.
Cell fractionation assays showed that these frameshift mutations induced a progressive range of changes in 1a responsiveness, from complete loss to no significant change, which correlated with the length of 2a peptide or RNA translated (Fig. 4A). G2aA-fs1, -fs2, and -fs3, with fewer than 122 2a residues translated, lost 1a responsiveness completely. For these RNAs, no changes in RNA distribution between the pellet and supernatant fractions were detected in response to 1a expression. G2a-fs4 and -fs5, with 261 to 382 2a residues translated, displayed slight to moderate 1a responsiveness, while G2aA-fs6, with 527 2a residues translated, was as responsive to 1a as was G2aA. Because these frameshift mutations were at widely separated sites, these effects appeared likely to be via the encoded protein, not a cis-acting RNA element. In further support of this interpretation, we found that G2aA-if2, which differs from G2aA-fs2 by only a single nucleotide (i.e., the insertion of 3 versus 4 nt after position 198 [Fig. 4B]), was fully competent in 1a responsiveness, in contrast to G2aA-fs2's complete loss of 1a responsiveness. Similarly, while frameshift mutations G2aA-fs1 and G2aA-fs4 in the N-terminal region of 2a abolished 2a ORF-dependent 1a responsiveness (Fig. 4A), the same frameshift mutations previously were shown to have no effect on the 5′, terminal stem-loop-dependent 1a responsiveness (frameshifts B2fs1 and B2fs2 in reference 10). Thus, for G2aA and its derivatives, translating the N-terminal half of 2a was required for the RNA to be fully responsive to 1a in the absence of box B.
Expressing 2a in trans does not restore 1a responsiveness of frameshift RNAs.
Since frameshift mutations in the N-terminal portion of 2a abolished 2a ORF-dependent 1a responsiveness, we performed experiments to explore whether supplying 2a in trans could complement the defects of the affected frameshift RNAs. G2aA-fs1 and G2aA-fs2 were selected as targets for these complementation experiments because of their complete loss of 1a responsiveness and the limited size (3 or 32 amino acids [aa]) of the 2a peptides that they could express (Fig. 4A and C), since larger N-terminal 2a fragments that are able to interact with 1a (9) might exhibit dominant negative effects in the complementation experiments. These frameshift RNAs, like other RNA2 derivatives used in previous experiments, were expressed from plasmids containing the LEU2 selectable marker. To facilitate the complementation experiments, a 2a-expressing plasmid containing the TRP1 instead of the LEU2 selectable marker was created to ensure retention of both plasmids during yeast growth. In addition, the ORF of GFP was inserted between the 2a ORF and the polyadenylation signal to make the RNA larger than the frameshift RNAs, so that proper separation of the 2a-providing RNA and the frameshift RNAs could be achieved by agarose gel electrophoresis (Fig. 5).
FIG. 5.
Expressing 2a in trans fails to rescue 1a responsiveness of frameshift RNAs. (A) Schematic diagram of G2aA-GFP, which differs from Ga2A by the addition of the GFP ORF between the 2a ORF and the polyadenylation signal. (B) Membrane association levels of G2aA-fs1 or G2aA-fs2 mRNAs coexpressed with G2aA-GFP mRNA in the absence (−1a) or presence (+1a) of 1a. Cell fractionation was used to assay RNA distribution in the pellet (P) and supernatant (S) fractions as described in the legend to Fig. 1, and representative Northern blots are shown.
Western blotting analysis showed that 2a was expressed from G2aA-GFP to levels similar to those of G2aA (data not shown). G2aA-GFP RNA also was highly responsive to 1a expression, as shown by an approximately 10-fold increase in RNA accumulation in the pellet fraction in the presence of 1a compared to yeast lacking 1a (Fig. 5B). However, even in the presence of wt 2a, neither G2aA-fs1 nor G2aA-fs2 RNA showed a significant 1a-induced increase in membrane association (Fig. 5B). Thus, 2a did not complement the defects of frameshift RNAs when provided in trans. These results suggested that either the 2a protein itself was not involved in the observed 1a-RNA interactions or 2a acted only in cis on the RNA from which it was translated.
Deletion of the N-terminal region of 2a abolishes 1a responsiveness.
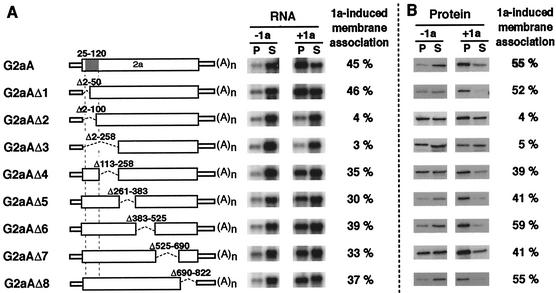
To further map the sequences responsible for 2a ORF-dependent 1a responsiveness, we created a series of in-frame deletions in G2aA RNA that collectively spanned the entire 2a ORF, and we tested the abilities of the resulting RNAs to associate with membranes in the presence of 1a (Fig. 6A). To separate 2a protein effects from possible effects of disturbing downstream translation of the RNA, these deletions were created without causing frameshifting or termination of the 2a ORF. G2aAΔ1 to G2aAΔ3 were N-terminal, progressive deletions lacking 2a codons 2 to 50, 100, and 258, respectively. While G2aAΔ1 RNA was as responsive to 1a as the parental G2aA RNA, G2aAΔ2 and -3 RNAs almost completely lost their 1a responsiveness. In addition, RNAs with internal and 3′-proximal deletions that collectively deleted 2a sequences from codon 113 to the end of 2a all retained levels of 1a responsiveness approaching that of G2aA (Fig. 6A). Thus, sequences encoding the N-proximal portion of 2a between aa 50 and 113 were specifically important for 2a ORF-dependent 1a responsiveness.
FIG. 6.
Deletion analysis of 2a sequences required for 2a ORF-dependent 1a responsiveness. (A) On the left are schematic diagrams of G2aA and its in-frame deletion derivatives. The previously defined limits of resolution for the region encoding the 1a-interacting domain of the 2a protein are indicated with a shaded box. Membrane association abilities of these RNAs in the absence (−1a) or presence (+1a) of 1a were assessed by cell fractionation to measure RNA distribution in the pellet (P) and supernatant (S) fractions as described in the legend to Fig. 1. Representative Northern blots are shown. As a measure of 1a responsiveness, the difference between the percentage of pelletable RNA in the presence and the absence of 1a was calculated for each RNA. The data shown are averages from three or more independent experiments for each derivative. (B) Western blots showing parallel analysis of the membrane association levels of the corresponding 2a protein derivatives in the absence (−1a) or presence (+1a) of 1a. 2a and 2a derivatives were detected with a mixture of three monoclonal antibodies that recognize epitopes mapped to the C-terminal, central polymerase, and N-terminal regions of 2a (R. Hershberger and P. Ahlquist, unpublished results). As a measure of 1a-induced membrane association, the difference between the percentages of pelletable 2a protein in the presence and the absence of 1a was calculated for each mutant. The data shown are averages from three or more independent experiments for each derivative.
2a protein sequences within aa 25 to 120 mediate interaction with 1a for recruiting 2a polymerase into the membrane-associated RNA replication complex in vivo (10, 24, 25). To test for a possible correlation between the 1a-induced membrane association of the 2a protein derivatives and their mRNAs, we performed cell fractionation to assess the sedimentation of these 2a mutants in the presence or absence of 1a. The results are shown in Fig. 6B.
In the absence of 1a, 85 to 90% of wt 2a protein was recovered in the supernatant (Fig. 6B). However, for some 2a deletion mutants, 20 to 50% of the protein pelleted even in the absence of 1a. Prior cell fractionation and immunolocalization results with similar 2a deletion derivatives show that, unlike association with the membrane-localized 1a protein, this 1a-independent sedimentation is likely due to partial aggregation of the mutant proteins rather than membrane association (9). Accordingly, to provide a measure of the 1a responsiveness of each 2a protein derivative, we calculated the increase in the percentage of pelletable 2a protein upon coexpression of 1a (Fig. 6B).
Coexpression of 1a induced 55% of wt 2a protein to fractionate with the membrane-containing pellet. For 2a derivatives G2aAΔ1 and G2aAΔ4 to -8, 1a induced similar amounts of protein to become membrane associated (39 to 59%) (Fig. 6). However, 1a-induced membrane association for mutants G2aAΔ2 and -3 was inhibited to only 4 to 5%. Thus, comparing Fig. 6A and B, there was a strong correlation between the effect of 2a deletions on the 1a responsiveness of these RNA2 derivatives and the effect of the same deletions on the ability of the encoded 2a protein to interact with 1a.
DISCUSSION
In this report, we describe the discovery of a box B motif-independent pathway for 1a-mediated recruitment of BMV RNA2 to the replication complex, reflected by 1a-induced membrane association (Fig. 1) and negative-strand RNA synthesis (Fig. 2) by RNA2 derivatives lacking the box B-containing 5′ stem-loop. Multiple results imply that this new pathway of template recruitment requires the previously documented interaction between 1a and the N terminus of 2a (9, 24) to occur when the nascent 2a peptide is still associated with translated RNA. First, 1a responsiveness required high-efficiency translation of the RNA, thus increasing the number of nascent 2a proteins available for interaction with 1a (Fig. 3). Second, multiple small mutations in the N-terminal region of 2a causing minimal changes in the RNA but inducing translational frameshifts inhibited 1a responsiveness of the RNAs, while a similar in-frame insertion did not (Fig. 4). Third, providing 2a in trans did not restore 1a responsiveness of RNAs with frameshift mutations (Fig. 5). Fourth, only those deletions in the N-terminal region of 2a that abolished 2a interaction with 1a abolished 1a responsiveness of the RNA (Fig. 6). Figure 7 combines these and other results into a mechanistic model for the box B-independent pathway of RNA2 recruitment. When RNA2 template is efficiently translated and the N terminus of 2a emerges from the translating ribosome, 1a occasionally interacts with the nascent 2a peptide. Association of 1a with the cytoplasmic face of ER membrane, which might occur before or after interaction with 2a, directs the membrane association of the translated RNA. The RNA is then sequestered into the membrane-bound spherular replication complex formed by multiple 1a proteins (37), and if the RNA possesses the 3′ tRNA-like end that directs initiation, it serves as a template for negative-strand RNA synthesis by 1a and 2a.
FIG. 7.
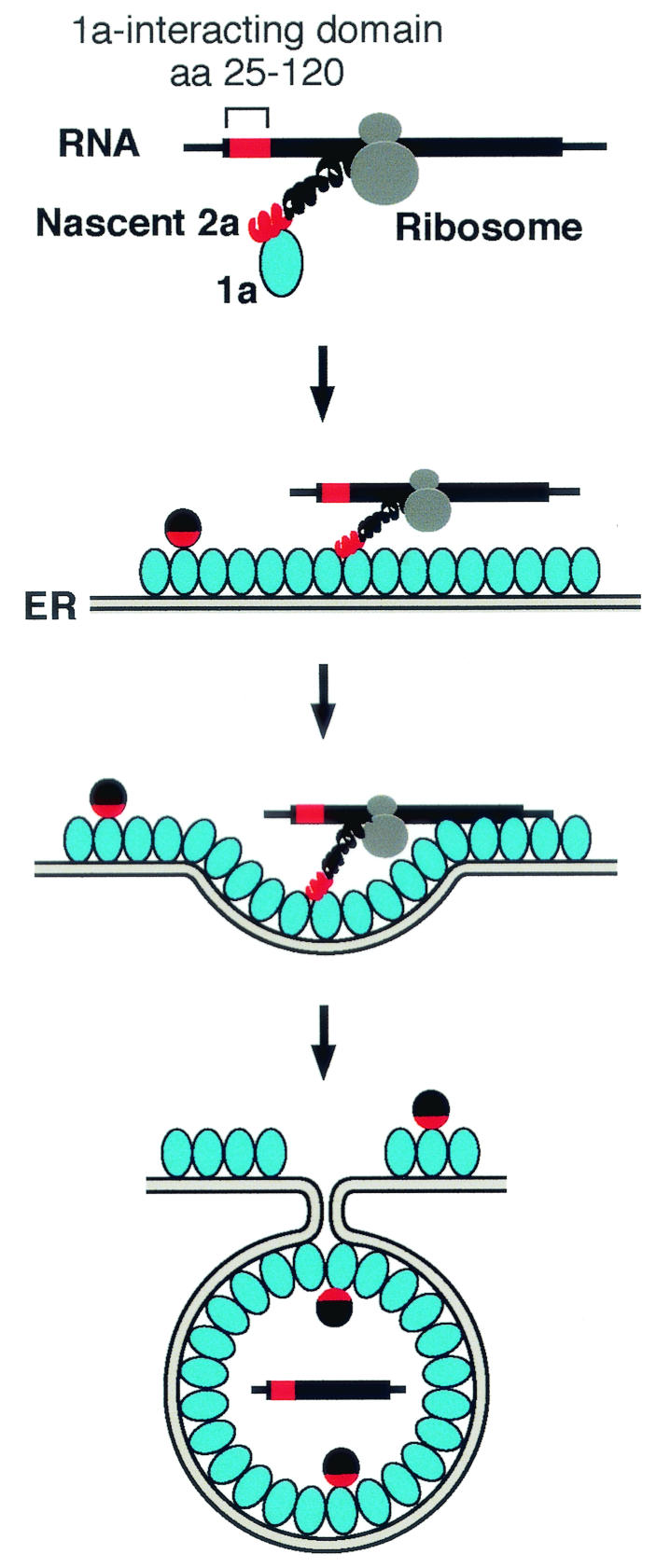
Model for the 2a ORF-dependent 1a responsiveness of highly translated 2a mRNAs. The successive panels illustrate initiation of interaction between 1a and the N terminus of the nascent 2a peptide still associated with its mRNA through the ribosome, localization of the translating 2a peptide and RNA to the ER membrane by 1a, and sequestration of 2a and the RNA into the capsid-like, membrane-bound spherules formed by multiple 1a proteins (37). The red and black spheres represent mature 2a protein, with the N-proximal 1a-interacting domain of 2a indicated by red as in the nascent 2a protein.
Alternate pathway for RNA2 recruitment.
As noted in the introduction, cis-acting signals necessary and sufficient for normal RNA template recruitment by 1a have been mapped in BMV genomic RNA3 and RNA2. In RNA3, the 1a-responsive signal consists of an approximately 150-nt intergenic sequence, while in RNA2 the 1a-responsive signal consists of the 5′ UTR plus adjacent downstream sequences. Structure probing shows that the RNA3 1a-responsive signal folds into an extended stem-loop that presents the conserved box B motif at the apex and mimics a tRNA TΨC stem-loop (7). Like the RNA3 signal, the RNA2 5′ end signal is predicted to form an extended stem-loop similarly presenting the box B motif at its apex (Fig. 1A). Moreover, 5′-proximal box B sequences with the potential to be presented in similar stem-loops are conserved in RNA1 and RNA2 of bromoviruses and cucumoviruses (10).
The role of the 5′ UTR in 1a recruitment of wt RNA2 templates to the replication complex is well established by many results, such as the effect of deleting the subdomain A stem-loop (Fig. 1) (10) and the ability of the RNA2 5′ UTR to function as a self-contained signal sufficient to confer box B-dependent 1a responsiveness on a nonviral RNA (10). Thus, it was unexpected to find that RNA2 derivatives with the yeast GAL1 5′ UTR, and therefore completely lacking the box B-containing 5′-terminal stem-loop, were highly 1a responsive (Fig. 1, G2aA or G2aT). Moreover, simultaneous deletion of the box B-containing 5′-terminal stem-loop (subdomain A) and subdomain B (Fig. 1) from the wt RNA2 5′ UTR also yielded wt 1a responsiveness, although deletion of the 5′ stem-loop alone or small deletions and base substitutions within it largely abolished RNA2 1a responsiveness (Fig. 1) (10). These results revealed an alternate 1a-responsive pathway that is independent of the 5′ stem-loop and box B sequences. Furthermore, the tRNA-like 3′ end was dispensable for both the 5′ box B-dependent and -independent pathways of 1a responsiveness (Fig. 1) (10). Therefore, the 5′ box B-independent 1a responsiveness must be mediated by sequences inside the 2a ORF.
As previously found for BMV RNA3 (31, 40), 3′ tRNA-like sequences containing the negative-strand initiation site were essential but insufficient to direct negative-strand synthesis from RNA2 derivatives in vivo (Fig. 2). In addition, negative-strand RNA2 synthesis required sequences mediating RNA2 recruitment into viral RNA replication complexes, as reflected by 1a-induced membrane association. Just as for wt RNA2 recruited by 1a through the box B-dependent pathway, RNA2 derivatives recruited through the 2a ORF-dependent pathway (Fig. 1) were assembled into functional replication complexes and served as templates for wt levels of negative-strand RNA synthesis (Fig. 2).
Dependence on efficient translation for 2a ORF-mediated 1a responsiveness.
Why are most RNA2 mutants defective in 1a responsiveness when 5′ sequences are mutated to inactivate the box B-dependent pathway, if the 2a ORF can mediate an alternate 1a-responsive pathway in some RNA2 derivatives? Comparison of Fig. 1 and 3 revealed a strong correlation between 2a ORF-mediated 1a responsiveness and the translation efficiency of RNA2 derivatives. Thus, 2a ORF-mediated 1a responsiveness required not only the 2a ORF but also its efficient translation. Efficient translation is likely required to provide more opportunities for 1a to interact with the nascent 2a peptide, possibly compensating for the short time available for interaction during 2a translation. Alternatively, multiple 1a-2a interactions per RNA might be required. wt RNA2 and RNA2ΔA, which showed essentially no box B-independent 1a responsiveness (Fig. 1), were translated at only 7 to 15% of the rate of chimeric 2a mRNAs with the yeast GAL1 5′ UTR (Fig. 3). wt RNA2 translation is selectively attenuated through the RNA2-specific, 5′ UTR subdomain B sequences (28), possibly through secondary structure (Fig. 1A). This down-regulation helps to maintain the ∼25:1 ratio of 1a to 2a in BMV RNA replication complexes (37). Like the similar excess of Gag to Pol in retrovirus cores, this ratio may be important for replication complex assembly (37). In contrast to the wt RNA2 5′ UTR, the 3′ tRNA-like structure of RNA2 only slightly reduced translation compared to a 3′ poly(A) tail (Fig. 3).
1a interaction with the N terminus of nascent 2a peptide.
Deletions overlapping 2a ORF codons 50 to 113, but not other deletions, simultaneously inhibited 1a-induced membrane association of 2a protein and 2a mRNA (Fig. 6). This agrees with prior findings that sequences within 2a aa 25 to 140 interact directly with the C-terminal helicase-like domain of 1a (24) and that sequences within the first 120 aa are necessary and sufficient for 1a-dependent localization of GFP-2a to ER membrane (9). Further insight into this strong linkage between 1a interaction with the N terminus of 2a and the box B-independent 1a responsiveness of some 2a mRNAs comes from the finding that expressing 2a in trans did not restore the 1a responsiveness of frameshifted RNAs (Fig. 5). All of these results support the hypothesis that 1a recruits efficiently translated 2a mRNAs through interaction with the N terminus of nascent 2a proteins.
Additional support for this hypothesis was provided by the inverse correlation between 1a responsiveness and the length of translated 2a peptide (Fig. 4). Interestingly, although partial deletions collectively encompassing the region from aa 113 to the end of 2a (Fig. 6, G2aAΔ4-Δ8) exhibited no dramatic effects on 1a responsiveness, frameshifts at aa 122 or 261 (Fig. 4A, G2aA-fs3 and -fs4) abolished or seriously inhibited 1a responsiveness. Why was deletion of this region allowed while frameshift within it was not allowed? Effective interaction of 1a with the nascent 2a peptide may require translation to continue beyond the N-terminal 2a domain for multiple reasons: to extrude the 1a-interacting 2a domain from the translating ribosome and to allow time for protein folding, 1a interaction, and possibly movement to the membrane, all while still maintaining linkage to the RNA through the translating ribosome and nascent 2a protein.
While 1a interaction with the nascent 2a peptide is a critical step in these processes (Fig. 7), 2a ORF-dependent recruitment of RNA to the replication complex may also involve a later step in which 1a interacts directly with the RNA. This interaction could be mediated by the well-established, normally low-level, nonspecific interaction of 1a with RNAs (10, 40), possibly stimulated by the RNA proximity provided by 1a interaction with nascent 2a.
Biological significance of the 2a ORF-dependent pathway of RNA2 recruitment.
Since inactivating the box B-dependent pathway largely abolished the 1a responsiveness of wt RNA2 (Fig. 1) (10), it appears that the contribution of the 1a-2a interaction-dependent pathway to wt RNA2 recruitment normally is small relative to that of the dominant box B pathway. This fits the low translation efficiency of wt RNA2 and the requirement for high-level translation for RNA recruitment via the 1a-2a interaction-dependent pathway (Fig. 3) (28). However, since the translation efficiency of wt RNA2 could vary between yeast and the natural plant hosts of BMV, the relative contribution of the 1a-2a interaction pathway to template recruitment might also vary in plant cells. Nevertheless, even if its relative contribution in plant cells is low, as is that in yeast, the 1a-2a interaction pathway offers significant insights into RNA recruitment, as outlined below.
First, RNA2 derivatives recruited through the 1a-2a interaction-dependent pathway were assembled into functional replication complexes and served as templates for wt levels of negative-strand RNA synthesis, just as for RNA2 recruited through the box B-dependent pathway (Fig. 2). Thus, although independent of box B, at least the latter steps of the 2a ORF-dependent RNA2 recruitment pathway (Fig. 7) must overlap those of the normal, box B-dependent pathway (10, 37, 40).
As noted above, the box B-dependent pathways that normally dominate 1a-mediated recruitment of wt RNA2 and RNA3 require cis-acting signals in which box B mimics the exact sequence and structure of the conserved TΨC stem-loop of tRNAs (7, 10, 40). Since essentially no mutation of this tRNA sequence or structural mimicry is tolerated, the box B-dependent pathway of 1a-mediated RNA recruitment appears likely to involve interaction with host factors that normally interact with tRNAs (7, 10). Such factors may be involved either in the 1a- and box B-dependent inhibition of BMV RNA translation to allow RNA replication, in communicating with the tRNA-like 3′ end to facilitate later initiation, or in both (7, 22). Nevertheless, the discovery of the box B-independent, 1a-2a interaction-dependent pathway reported here shows that alternate mechanisms of interacting with 1a can allow effective RNA recruitment. Further study and comparison of the 1a-2a interaction-dependent and box B-dependent pathways should reveal the shared, essential features of 1a-mediated RNA recruitment and provide further insights into the particular roles of tRNA-like, TΨC stem-loop mimicry in facilitating the normal pathways of RNA template recruitment and initiation of RNA synthesis.
In addition, the results presented here (Fig. 4 to 6) strongly imply that 1a interacts in vivo with nascent 2a protein. Such early interaction with 1a may be important in recruiting 2a into the replication complex while avoiding or minimizing the aggregation found in 2a derivatives lacking 1a interaction sequences (9).
Acknowledgments
We thank Yuriko Tomita and Masayuki Ishikawa for generously providing pB1YT2 and members of our laboratory for helpful discussions throughout these experiments.
This work was supported by the National Institutes of Health through grant GM35072. P.A. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ahlquist, P. 1992. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2:71-76. [DOI] [PubMed] [Google Scholar]
- 2.Ahola, T., and P. Ahlquist. 1999. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J. Virol. 73:10061-10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahola, T., J. A. den Boon, and P. Ahlquist. 2000. Helicase and capping enzyme active-site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J. Virol. 74:8803-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allison, R., C. Thompson, and P. Ahlquist. 1990. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc. Natl. Acad. Sci. USA 87:1820-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argos, P. 1988. A sequence motif in many polymerases. Nucleic Acids Res. 16:9909-9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 7.Baumstark, T., and P. Ahlquist. 2001. Brome mosaic virus RNA3 intergenic replication enhancer folds to mimic a tRNA TΨC stemloop and is modified in vivo. RNA 7:1652-1670. [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman, M. R., and C. C. Kao. 1999. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 286:709-720. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J., A. Noueiry, and P. Ahlquist. 2001. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 75:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.den Boon, J., J. Chen, and P. Ahlquist. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75:12370-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreher, T. W., A. L. Rao, and T. C. Hall. 1989. Replication in vivo of mutant brome mosaic virus RNAs defective in aminoacylation. J. Mol. Biol. 206:425-438. [DOI] [PubMed] [Google Scholar]
- 13.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.French, R., and P. Ahlquist. 1988. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J. Virol. 62:2411-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.French, R., and P. Ahlquist. 1987. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J. Virol. 61:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.French, R., M. Janda, and P. Ahlquist. 1986. Bacterial gene inserted in an engineered RNA virus: efficient expression in monocotyledonous plant cells. Science 231:1294-1297. [DOI] [PubMed] [Google Scholar]
- 17.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restrictions sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 19.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Cell Biol. 3:419-429. [Google Scholar]
- 20.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haseloff, J., P. Goelet, D. Zimmern, P. Ahlquist, R. Dasgupta, and P. Kaesberg. 1984. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc. Natl. Acad. Sci. USA 81:4358-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janda, M., and P. Ahlquist. 1998. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. USA 95:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janda, M., and P. Ahlquist. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961-970. [DOI] [PubMed] [Google Scholar]
- 24.Kao, C. C., and P. Ahlquist. 1992. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J. Virol. 66:7293-7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kao, C. C., R. Quadt, R. P. Hershberger, and P. Ahlquist. 1992. Brome mosaic virus RNA replication proteins 1a and 2a form a complex in vitro. J. Virol. 66:6322-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong, F., K. Sivakumaran, and C. Kao. 1999. The N-terminal half of the brome mosaic virus 1a protein has RNA capping-associated activities: specificity for GTP and S-adenosylmethionine. Virology 259:200-210. [DOI] [PubMed] [Google Scholar]
- 27.Mise, K., R. F. Allison, M. Janda, and P. Ahlquist. 1993. Bromovirus movement protein genes play a crucial role in host specificity. J. Virol. 67:2815-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noueiry, A. O., J. Chen, and P. Ahlquist. 2000. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc. Natl. Acad. Sci. USA 97:12985-12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novak, J. E., and K. Kirkegaard. 1991. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J. Virol. 65:3384-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pogue, G. P., L. E. Marsh, J. P. Connell, and T. C. Hall. 1992. Requirements for ICR-like sequences in the replication of brome mosaic virus genomic RNA. Virology 188:742-753. [DOI] [PubMed] [Google Scholar]
- 31.Quadt, R., M. Ishikawa, M. Janda, and P. Ahlquist. 1995. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc. Natl. Acad. Sci. USA 92:4892-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Restrepo-Hartwig, M., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Restrepo-Hartwig, M., and P. Ahlquist. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 73:10303-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Restrepo-Hartwig, M. A., and J. C. Carrington. 1994. The tobacco etch potyvirus 6-kilodalton protein is membrane associated and involved in viral replication. J. Virol. 68:2388-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sacher, R., and P. Ahlquist. 1989. Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection. J. Virol. 63:4545-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 38.Sivakumaran, K., C. H. Kim, R. J. Tayon, and C. C. Kao. 1999. RNA sequence and secondary structural determinants in a minimal viral promoter that directs replicase recognition and initiation of genomic plus-strand RNA synthesis. J. Mol. Biol. 294:667-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smirnyagina, E., Y. H. Hsu, N. Chua, and P. Ahlquist. 1994. Second-site mutations in the brome mosaic virus RNA3 intercistronic region partially suppress a defect in coat protein mRNA transcription. Virology 198:427-436. [DOI] [PubMed] [Google Scholar]
- 40.Sullivan, M., and P. Ahlquist. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan, M., and P. Ahlquist. 1997. Cis-acting signals in bromovirus RNA replication and gene expression: networking with viral proteins and host factors. Semin. Virol. 8:221-230. [Google Scholar]