Abstract
The latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus can associate with mitotic chromosomes and promote latent episome maintenance and segregation. Here we report that LANA also mediates the replication of plasmid DNAs bearing viral terminal repeats. The predicted secondary structure of LANA's C terminus reveals striking similarity to the known structure of the DNA-binding domain of Epstein-Barr virus EBNA1, despite the absence of primary sequence homology between these proteins, suggesting conservation of the key mechanistic features of latent gammaherpesvirus DNA replication.
Kaposi's sarcoma-associated herpesvirus (KSHV) is a lymphotropic gammaherpesvirus that is strongly associated with neoplasms of lymphoid (primary effusion lymphoma, multicentric Castleman's disease) and endothelial (Kaposi's sarcoma) origin (6, 22-24). The majority of tumor cells in these malignancies harbor latent KSHV episomes (20, 21, 23), and latent infection is thought to play a major role in the development of KSHV-associated neoplasms. To establish latent infection in dividing cells, herpesviruses have to ensure that viral episomes are (i) replicated before the cell divides and (ii) segregated to the daughter cells during mitosis. Several different observations indicate that the latency-associated nuclear antigen (LANA) encoded by ORF73 of KSHV is responsible for episomal maintenance during latency. First, LANA is expressed in all latently infected cells and binds specifically to a sequence within the viral terminal repeats (TRs) (3, 8, 11). Second, when plasmids containing two units of the TRs are introduced into LANA-expressing B cells, single-cell clones which appear to harbor stable episomes can be selected (2, 3). Third, LANA is able to bind to mitotic host chromosomes (7, 17). Together, these observations have led to a model in which LANA tethers viral episomes to host chromosomes, thereby ensuring faithful segregation of the viral genome during mitosis.
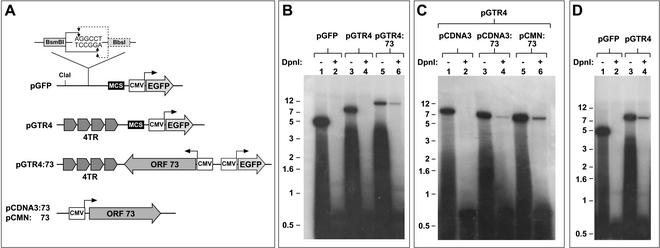
The association of LANA expression with episome stability in selected cell lines (2, 3) suggests that LANA might also be able to promote latent DNA replication. To address this issue directly, we constructed a series of reporter constructs containing the functional elements depicted in Fig. 1A. To generate the vector backbone (pGFP) for the reporters, the polylinker of the green fluorescent protein (GFP) expression vector pEGFP-N1 (Clontech) was removed and an alternative polylinker was inserted upstream of the cytomegalovirus (CMV) promoter. Another linker was inserted into a unique BsaI site to permit the cloning of TR units. A single TR was excised from pML1 (15) with NotI (which cuts once per TR unit) and inserted into the BsmBI site of pGFP. From the resulting construct, pGTR1, a fragment containing the TR unit was excised with ClaI and BbsI and reintroduced into the same donor construct digested with ClaI and BsmBI, resulting in a construct with two contiguous TRs (pGTR2). Another round of excision and reintroduction yielded construct pGTR4, which contains four TR units in authentic head-to-tail orientation. To generate pGTR4:73, the CMV/ORF73 expression cassette from pCDNA3:ORF73 (19) was excised by using the MfeI/PvuII sites and introduced into the polylinker upstream of the CMV promoter in pGTR4.
FIG. 1.
Replication of TR-containing plasmids in B cells. (A) Functional elements of reporter and expression constructs. The reporter constructs used in this study are based on the vector backbone pGFP. The vector contains a GFP expression cassette driven by the CMV promoter and a multiple-cloning site (MCS) upstream of the CMV promoter. The linker inserted into the BsaI site of pGFP and the cleavage sites of BsmBI and BbsI are shown enlarged. Cleavage with either enzyme generates identical NotI-compatible overhangs. These sites together with the indicated ClaI site were used to assemble the four head-to-tail-oriented TR units of pGTR4 (see text for details). The reporter construct pGTR4:73 contains the CMV-driven ORF73 expression cassette of pCDNA3:73 inserted into the MCS of pGTR4. The LANA expression constructs pCDNA3:73 and pCMN:73 were used to provide LANA in trans. EGFP, enhanced GFP. (B to D) DpnI resistance assays. The indicated constructs were introduced into either BJAB cells (B and C) or the stable LANA-expressing cell line BJAB:73 (D) as described in the text. Episomal plasmids were recovered 72 h later by Hirt extraction and subjected to digestion with either XhoI (which cuts once in the reporters) (−) or XhoI and DpnI (+). Standards were analyzed by Southern blotting, and reporter constructs were detected with a radioactively marked GFP probe. The positions of size markers (in kilobase pairs) are shown. Quantitation of linearized versus DpnI-digested plasmids was carried out with a phosphorimager (Molecular Dynamics Storm860 and ImageQuant software). A relatively large proportion of cells undergo cell death following the electroporation procedure; the smears below the linearized bands result from degraded DNA released from dead cells as well as degraded, adherent extracellular plasmid DNA and thus were not included in the quantitation.
The ability of the constructs to replicate in mammalian cells was evaluated by DpnI resistance assays (12). For this purpose, the reporters were amplified in dam+ bacteria and 20 μg of DNA was electroporated into 1.5 × 107 BJAB B cells. Cells were harvested 72 h later, passed through a 40-μm-pore-size cell strainer, and washed three times with cold phosphate-buffered saline. Episomal DNA was subsequently isolated from 1 × 107 (in single-transfection experiments) or 2 × 107 (in cotransfection experiments) cells by a modified Hirt procedure (1). DNA was eluted in 100 μl of water, and 40 μl (corresponding to 4 × 106 or 8 × 106 cells) was digested overnight with either 20 U of XhoI to linearize the reporter constructs (input control) or 20 U of XhoI and 40 U of DpnI to digest unreplicated episomes. Samples were subsequently analyzed by Southern blotting, and reporter constructs were detected with a 32P-labeled GFP probe. As shown in Fig. 1B, no replicated DNA was recovered when the pGFP or pGTR4 reporters were introduced into BJAB cells (lanes 1 to 4), indicating that the presence of four TRs alone does not confer the ability to detectably replicate in this assay. However, when a LANA expression cassette is provided in cis in addition to TRs, DpnI-resistant episomes become easily detectable (lanes 5 and 6). Quantitation of the full-length linear DNA in the XhoI lane (lane 5) versus that in the XhoI-plus-DpnI lane (lane 6) reveals that 21% of the total episomal DNA consists of replicated plasmids.
Can LANA also mediate replication in trans? To address this question, the pGTR4 reporter was cotransfected into BJAB cells along with either an empty expression vector (Fig. 1C, lanes 1 and 2) or the LANA expression construct pCDNA3:73 (lanes 3 and 4) or pCMN:73 (lanes 5 and 6). While pCDNA3:73 and pCMN:73 contain the same functional elements (Fig. 1A), pCMN:73 differs from pCDNA3:73 in the absence of noncoding sequences upstream of ORF73 and the presence of a Kozak-optimized start codon for ORF73, resulting in at least 10-fold-higher expression levels for LANA, as judged by Western blot analysis (data not shown). Both LANA expression constructs enabled replication of the cotransfected pGTR4 reporter plasmid, with efficiencies of 4 (pCDNA3:73) and 8% (pCMN:73). The lower percentage of replicated plasmids obtained when LANA is provided in trans is likely to be a result of reduced levels of LANA compared to those generated by plasmids in which the TRs are linked to LANA in cis. (Unlike pGTR4:73, pCDNA3:73 and pCMN:73 do not contain TR units and are therefore not replicated, resulting in declining plasmid levels over the 72-h period.) In another set of experiments, either pGFP or pGTR4 was transfected into BJAB:73 cells, a cell line stably expressing LANA. The BJAB:73 cell line was generated by stable transduction of BJAB cells with a retrovirus expressing ORF73 as well as a puromycin resistance gene. Approximately 60% of these puromycin-resistant mass cultures expressed LANA, as judged by immunofluorescence analysis (data not shown). As shown in Fig. 1D, only pGTR4, not pGFP, is replicated in BJAB:73 cells, indicating that replication is strictly dependent on the presence of the TRs. Fourteen percent of pGTR4 episomes recovered from BJAB:73 cells were DpnI resistant. This percentage is comparable to the values obtained with pGTR4:73 in BJAB cells when the proportion of LANA-negative cells (40%) in the BJAB:73 cultures is considered.
In an additional experiment, reporters containing one, two, or four TRs were found to have comparable replication efficiencies (data not shown; available as supplemental information on http://itsa.ucsf.edu/∼micro/Faculty/ganem_folder/data/supp Fig. 1.html). We speculate that the high number of TR copies in the viral genome (15) exist principally to provide additional attachment sites for the tethering of episomes to host chromosomes during metaphase rather than to provide a multitude of viral replication origins. However, we cannot exclude the possibility that added TRs might lead to more-efficient replication at the very high TR numbers (30 to 40) found in the viral genome.
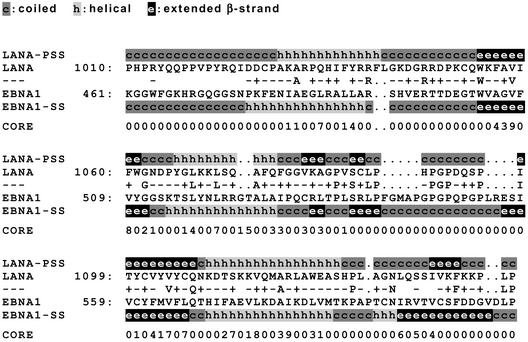
The EBNA1 protein, encoded by Epstein-Barr virus (EBV), like LANA mediates segregation and replication of episomes containing a viral origin of replication (designated oriP in EBV) (25). Despite this functional conservation, the two proteins do not exhibit significant amino acid homology. However, when we used the 3D-PSSM program (9, 14) to search for proteins with homology to the predicted secondary structure of LANA, a striking similarity between the C-terminal domains of LANA and EBNA1 was observed. The 3D-PSSM algorithm uses primary sequence information to predict the secondary structures of proteins; these structures are then used to search a database of proteins for which the crystal structures have been solved. The DNA-binding domain of EBNA1 (fold library entry d1b3ta, amino acids [aa] 461 to 607 of EBNA1) was returned as the top match in this search, with an E value of 0.002 (certainty, 95%). This result is highly significant, since the next-best match (a representative structure of polypyrimidine tract-binding riboucleoproteins) had an E value of only 0.731. An alignment of the predicted secondary structure of the relevant regions of LANA and EBNA-1 is shown in Fig. 2. Since only aa 461 to 607 of EBNA1 have been structurally examined (4, 5), it is unclear whether LANA also shows structural homology to regions outside of EBNA1's DNA-binding domain.
FIG. 2.
3D-PSSM alignment of EBNA1 and LANA. The carboxy-terminal domain of LANA (aa 932 to 1162; GenBank accession no. AAC57158) was used to search the 3D-PSSM database (program version 2.6.0; Imperial College of Science, Technology and Medicine [http://www.sbg.bio.ic.ac.uk/∼3dpssm/]). Shown is a modified version of the alignment of the relevant regions in LANA (aa 1010 to 1144) and EBNA1 (aa 461 to 607) generated by 3D-PSSM. The primary sequences of LANA and EBNA1 (accession no. NP_039875) are given. Dots, gaps introduced into the aligned sequences. Identities and positive (+) or negative (−) scores for aligned residues as assigned by 3D-PSSM are shown between the primary sequence data for LANA and EBNA1. The secondary structure of LANA (LANA-PSS) as predicted by the 3D-PSSM algorithm and the known secondary structure of EBNA1 (EBNA1-SS) are shown above or below the primary sequences of LANA and EBNA1, respectively. The CORE values shown below the aligned sequences provide an index for the contribution of residues to hydrophobic interactions (on a scale from 0 to 9) within the resolved EBNA1 structure. High values indicate buried residues important for the core structure, whereas low values indicate residues on the surface of the protein.
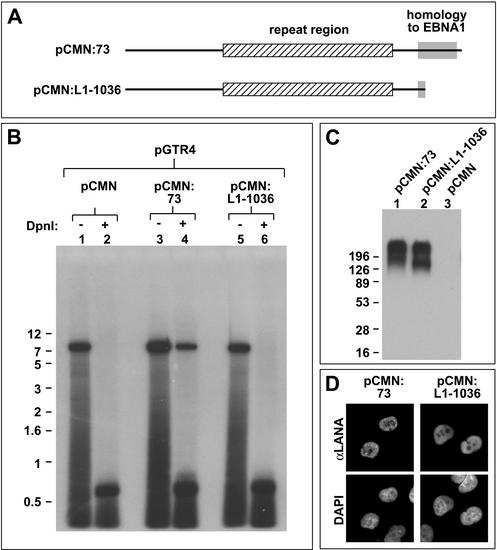
Our computational analysis predicts that disruption of the C-terminal domain of LANA should abolish DNA replication activity. We therefore generated a LANA mutant truncated after aa 1036, thus lacking most of the domain homologous to EBNA1 (Fig. 3A). Western and immunofluorescence analyses revealed that the mutant localized to the nucleus and was expressed at levels similar to those for wild-type LANA (Fig. 3D and C, respectively). As shown in Fig. 3B, cotransfection of the mutant together with pGTR4 did not yield any replicated reporter plasmids, indicating that aa 1037 to 1164 of LANA are absolutely essential for replication of TR-containing episomes. An additional mutant comprising only the C terminus of LANA (aa 925 to 1162) fused to a nuclear localization sequence was inactive in our replication assays; however, immunofluorescence analysis revealed that this mutant formed large aggregates in the cytoplasm of transfected cells (data not shown). It is therefore unclear whether the C-terminal region alone is able to support DNA replication.
FIG. 3.
The carboxy-terminal region of LANA is required for replication activity. (A) Cartoon showing the proteins encoded by the expression vectors pCMN:73 (full-length LANA) and pCMN:L1-1036 (lacking the carboxy-terminal 126 aa of LANA). Mutant L1-1036 is truncated within the region showing structural homology to EBNA1. All amino acid positions conform to the LANA sequence with GenBank accession no. AAC57158. (B) DpnI resistance assays. Expression construct pCMN:73 or pCMN:L1-1036 or the empty expression vector pCMN was transfected into BJAB cells together with the reporter construct pGTR4. Episomes were recovered 72 h later by Hirt extraction and analyzed as described for Fig. 1. (C) Western blot analysis of LANA expression. Aliquots of the transfected BJAB cells (4 × 10 6 cells) described above were harvested 48 h posttransfection, and extracts containing 40 μg of total protein were analyzed by Western blotting using a polyclonal antiserum against LANA (18). The positions of size markers (in kilodaltons) are shown. Note that LANA shows multiple bands and runs higher than expected from its calculated molecular mass (135 kDa). The protein shows similar behavior in KSHV-positive BCBL1 cells (data not shown). (D) Immunofluorescence analysis of endothelial SLK cells transfected with the expression construct pCMN:73 (left) or pCMN:L1-1036 (right). LANA was detected with polyclonal antibodies, and DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole).
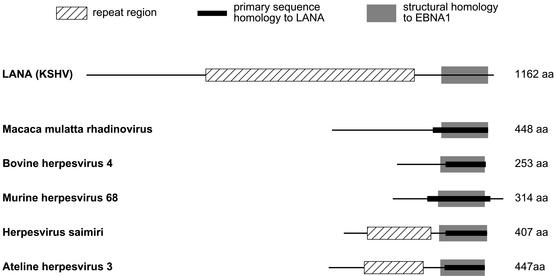
KSHV LANA consists of an N-terminal unique region, a central domain of repeated acidic residues, and a C-terminal region that, as just noted, is related to the EBV DNA-binding domain. ORF73 and LANA are positionally conserved in other members of the rhadinovirus genus. Although quite different in size and sequence organization, some of these proteins (most of whose functions are unknown) exhibit limited primary sequence homology to LANA. When present, such sequence homology is always restricted to a region located within aa 963 to 1162 of LANA (Fig. 4 and Table 1, center columns), the region of LANA with structural similarity to EBNA1's C terminus. Computational analysis reveals that, in all ORF73-encoded proteins, regions of primary sequence homology to LANA are also similar to EBNA1 in their predicted secondary structures (Fig. 4 and Table 1, right columns). The E values (between 6.5 × 10−2 and 1.8) and ranks (between 1 and 9) with which the DNA-binding domain of EBNA1 was returned as a match in the initial searches (employing the full-length sequences) varied. However, in a refined search in which only the regions showing primary sequence homology to LANA were analyzed, EBNA1's DNA-binding domain ranked first for all ORF73-encoded homologues, with highly significant E values between 4 × 10−3 and 5.7 × 10−2 (Table 1, right columns). These data indicate the presence of a structurally conserved domain likely devoted to DNA binding and suggest that the respective positional ORF73-encoded homologues have functions similar to those of LANA during viral replication. (The two herpesviruses whose ORF73 products lack homology to KSHV LANA [alcelaphine herpesvirus 2 and ovine herpesvirus 2] showed no structural relationship to EBNA1's C-terminal domain.)
FIG. 4.
ORF73-encoded proteins of rhadinoviruses: homology to LANA and EBNA1. Shown is a graphical representation of the results presented in Table 1. Hatched boxes, repeat regions within LANA and ORF73-encoded proteins of other rhadinoviruses. Primary amino acid homology of the ORF73-encoded proteins to LANA was evaluated by BLASTP pairwise sequence alignment. Black bars, regions showing significant homology to LANA; grey boxes, regions which show predicted structural homology to EBNA1, as judged by 3D-PSSM analysis (see Table 1 for details). The ORF73-encoded proteins of alcelaphine herpesvirus 2 and ovine herpesvirus 2 are not depicted since they do not show homology to either LANA or EBNA1.
TABLE 1.
Secondary structure similarity between rhadinovirus ORF73-encoded homologues and EBNA1a
| Virus carrying ORF73 | Expression pattern | Protein size (aa) | Homology to:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LANA (BLASTP)
|
EBNA1 (3D-PSSM)
|
|||||||||
| % Identity/similarity | Region of homology on:
|
Initial search
|
Refined search
|
|||||||
| Subject | LANA | Region of homology | Rank | E value | Rank | E value | ||||
| KSHV | Latent | 1,162 | 1010-1144 | 1 | 0.002 | |||||
| M. mulatta rhadinovirus | ? | 448 | 34/48 | 285-446 | 1025-1144 | 311-445 | 3 | 0.288 | 1 | 0.004 |
| Bovine herpesvirus 4 | ? | 253 | 33/52 | 137-253 | 1028-1147 | 123-250 | 2 | 0.391 | 1 | 0.057 |
| Murine herpesvirus 68 | ? | 314 | 29/40 | 95-274 | 963-1162 | 125-259 | 1 | 0.065 | 1 | 0.012 |
| Herpesvirus saimiri | Latent | 407 | 28/47 | 288-406 | 1025-1145 | 269-405 | 7 | 1.51 | 1 | 0.040 |
| Ateline herpesvirus 3 | ? | 447 | 27/50 | 328-446 | 1025-1145 | 316-445 | 9 | 1.81 | 1 | 0.027 |
| Alcelaphine herpesvirus 2 | ? | 1,300 | — | — | — | — | — | — | ||
| Ovine herpesvirus 2 | ? | 495 | — | — | — | — | — | — | ||
In the left columns the viral strains encoding the homologues are sorted from top to bottom according to the level of primary sequence identity of the respective ORF73-encoded homologue to LANA. The expression patterns, if known, and total lengths in amino acids are shown. In the center columns homology of each ORF73-encoded protein to LANA was evaluated by BLASTP pairwise sequence alignment. —, no detectable primary sequence homology. The positions of homologous regions in the ORF73-encoded proteins and LANA are indicated by their amino acid positions. In the right columns results of the 3D-PSSM search with respect to structural similarity to EBNA1 are shown. A 3D-PSSM search was carried out with the complete sequence of each ORF73-encoded homologue (the program accepts only sequences up to 800 aa; the homologues of KSHV and alcelaphine herpesvirus 2 where therefore submitted in two N- and C-terminal portions). The rank with which EBNA1 appeared as a match in this initial search, the E value for the match, and the matched region (in amino acids) within each ORF73-encoded homologue are shown. A refined search was carried out by using the subregions showing primary sequence homology to LANA, and the ranks and E values with which EBNA1 appeared in this search are also shown. The ORF73-encoded proteins of alcelaphine herpesvirus 2 and ovine herpesvirus 2 do not show primary sequence homology to LANA or structural homology to EBNA1. None of the ORF73-encoded homologues showed primary sequence homology to EBNA1 (BLASTP pairwise sequence alignment).
In summary, we have shown that LANA, like EBNA-1, can mediate DNA replication of plasmids bearing copies of its DNA-binding site; the two proteins also share secondary structural features. While this manuscript was being readied for submission, Garber et al. (10) reported that plasmids containing KSHV TR sequences can be replicated by LANA in cells of epithelial and endothelial origin; our findings extend these observations to B lymphocytes, a major site of KSHV persistence in vivo. In addition, Garber et al. noted striking organizational similarities between the dyad symmetry element of EBV oriP and the TR sequences bound by LANA. Taken together, these two studies indicate that both the cis- and trans-acting elements of the latent-DNA-replication machinery of EBV and KSHV have common structural features, suggesting that the core mechanistic elements of latent genome replication have been conserved in gammaherpesvirus evolution.
While this manuscript was under review, Lim et al. and Hu et al. (13, 16) reported similar results, showing LANA-dependent replication of TR-containing plasmids.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the ORF73-encoded proteins are as follows: KSHV, AAC57158; Macaca mulatta rhadinovirus, AAF60071; bovine herpesvirus 4, AAK07994; murine herpesvirus 68, NP_044913; herpesvirus saimiri, 1804350B; ateline herpesvirus 3, NP_048045; alcelaphine herpesvirus 2, NP_065570; ovine herpesvirus 2, AAL05844.
REFERENCES
- 1.Arad, U. 1998. Modified Hirt procedure for rapid purification of extrachromosomal DNA from mammalian cells. BioTechniques 24:760-762. [DOI] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, E. Bochkareva, L. Frappier, and A. M. Edwards. 1996. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell 84:791-800. [DOI] [PubMed] [Google Scholar]
- 5.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, W. Furey, Jr., A. M. Edwards, and L. Frappier. 1995. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein EBNA 1. Cell 83:39-46. [DOI] [PubMed] [Google Scholar]
- 6.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 7.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, M. A., II, C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 9.Fischer, D., C. Barret, K. Bryson, A. Elofsson, A. Godzik, D. Jones, K. J. Karplus, L. A. Kelley, R. M. MacCallum, K. Pawowski, B. Rost, L. Rychlewski, and M. Sternberg. 1999. CAFASP-1: critical assessment of fully automated structure prediction methods. Proteins (Suppl. 3):209-217. [DOI] [PubMed]
- 10.Garber, A. C., J. Hu, and R. Renne. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401-27411. [DOI] [PubMed] [Google Scholar]
- 11.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hay, R. T. 1985. The origin of adenovirus DNA replication: minimal DNA sequence requirement in vivo. EMBO J. 4:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, J., A. C. Garber, and R. Renne. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 76:11677-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelley, L. A., R. Maccallum, and M. J. E. Sternberg. 1999. Recognition of remote protein homologies using three-dimensional information to generate a position-specific scoring matrix in the program 3D-PSSM, p. 218-225. In Proceedings of the Third Annual Conference on Computational Molecular Biology. The Association for Computing Machinery, New York, N.Y.
- 15.Lagunoff, M., and D. Ganem. 1997. The structure and coding organization of the genomic termini of Kaposi's sarcoma-associated herpesvirus. Virology 236:147-154. [DOI] [PubMed] [Google Scholar]
- 16.Lim, C., H. Sohn, D. Lee, Y. Gwack, and J. Choe. 2002. Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 76:10320-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polson, A. G., L. Huang, D. M. Lukac, J. D. Blethrow, D. O. Morgan, A. L. Burlingame, and D. Ganem. 2001. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J. Virol. 75:3175-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renne, R., M. Lagunoff, W. Zhong, and D. Ganem. 1996. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J. Virol. 70:8151-8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 22.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 23.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitby, D., and C. Boshoff. 1998. Kaposi's sarcoma herpesvirus as a new paradigm for virus-induced oncogenesis. Curr. Opin. Oncol. 10:405-412. [DOI] [PubMed] [Google Scholar]
- 25.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]