Abstract
The serine-threonine kinase Dun1 contains a forkhead-associated (FHA) domain and functions in the DNA damage checkpoint pathway of Saccharomyces cerevisiae. It belongs to the Chk2 family of checkpoint kinases, which includes S. cerevisiae Rad53 and Mek1, Schizosaccharomyces pombe Cds1, and human Chk2. Dun1 is required for DNA damage-induced transcription of certain target genes, transient G2/M arrest after DNA damage, and DNA damage-induced phosphorylation of the DNA repair protein Rad55. Here we report that the FHA phosphoprotein recognition domain of Dun1 is required for direct phosphorylation of Dun1 by Rad53 kinase in vitro and in vivo. trans phosphorylation by Rad53 does not require the Dun1 kinase activity and is likely to involve only a transient interaction between the two kinases. The checkpoint functions of Dun1 kinase in DNA damage-induced transcription, G2/M cell cycle arrest, and Rad55 phosphorylation are severely compromised in an FHA domain mutant of Dun1. As a consequence, the Dun1 FHA domain mutant displays enhanced sensitivity to genotoxic stress induced by UV, methyl methanesulfonate, and the replication inhibitor hydroxyurea. We show that the Dun1 FHA domain is critical for direct kinase-to-kinase signaling from Rad53 to Dun1 in the DNA damage checkpoint pathway.
DNA damage checkpoints coordinate the cellular responses to genotoxic stress and ensure genomic integrity (31, 40, 55, 60). Besides cell cycle transitions, DNA damage checkpoints in the yeast Saccharomyces cerevisiae control damage-induced transcription; DNA replication; DNA repair and genomic stability; deoxynucleoside triphosphate metabolism; the relocalization of the Sir3/4, Ku80, and Rap1 proteins; and possibly other physiological responses to genotoxic stress (5, 20, 35, 55, 58, 60, 61).
Central to the DNA damage checkpoints in S. cerevisiae is a branched kinase cascade consisting of five protein kinases (Mec1, Tel1, Rad53, Chk1, and Dun1) (55, 60). Mec1 and Tel1 are both high-molecular-weight phosphoinositide 3-kinase-related protein kinases that are activated by unknown mechanisms. Their human counterparts, ATM and ATR, are also essential for the human DNA damage checkpoints. Rad53 and Dun1 are related forkhead-associated (FHA) domain kinases (see Fig. 1) and have counterparts in other organisms, including fission yeast Cds1 and human Chk2 (40, 60). Finally, Chk1 kinase, as well as its fission yeast and human homologs, is critical for the G2 cell cycle arrest in response to DNA damage (41). Genetic analysis of S. cerevisiae established that Mec1 controls the activities of the three downstream kinases Rad53, Dun1, and Chk1 (4, 38, 41, 42, 61). Under certain conditions, Tel1 controls the activation of Rad53 kinase in a Mec1-independent fashion (52). The exact mechanisms of how DNA damage checkpoints are activated and how the checkpoint kinases transmit and possibly amplify the signal, as well as control the effector pathways, are only beginning to be understood.
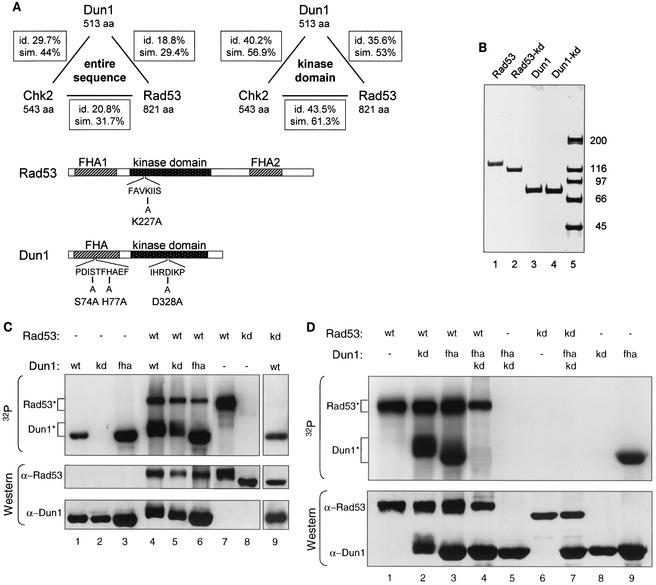
FIG. 1.
Rad53 kinase directly phosphorylates Dun1 kinase in vitro, dependent on the FHA domain of Dun1. (A) Schematic representation of the relatedness of Rad53, Dun1, and Chk2 kinases and of the Rad53 and Dun1 mutants used in this study. Top panel, Dun1, Rad53, and human Chk2 are related FHA domain kinases. Their entire sequences or their kinase domains were aligned by using the algorithm of Needleman-Wunsch, and the overall sequence identities (id.) and similarities (sim.) are indicated. aa, amino acids. Bottom panel, the rad53-kd allele changes an invariant lysine residue (K227) in subdomain II, which is directly involved in phosphotransfer (22). Rad53-K227A was shown to be nonfunctional and kinase deficient (4, 59). The dun1-kd allele changes an invariant aspartic acid residue (D328) in subdomain VI, which has been implicated in the catalytic mechanism (22). Dun1-D328A was previously shown to be nonfunctional and kinase deficient (25, 61). The Dun1 FHA domain mutant was created by changing serine 74 and histidine 77, two of the four invariable residues in FHA domains (24), to alanine. Previous work with the FHA domain of fission yeast Cds1 showed that such a double mutation abolishes its function (7). The dun1-H77A mutation alone was previously found to have little phenotypic consequence (25). (B) Partial purification of Rad53 and Dun1 kinases. One microgram of purified GST-tagged kinase was analyzed by SDS-4 to 12% PAGE and visualized with Coomassie brilliant blue R250 staining. Wild-type Rad53 (lane 1) and Dun1 (lane 3) proteins were isolated after cell exposure to 0.1% MMS for 2 h, which resulted in an electrophoretic mobility shift caused by phosphorylation (see also panel C and Fig. 2A). The Rad53 (lane 2) and Dun1 (lane 4) kinase-deficient proteins were isolated from cells not exposed to MMS for technical reasons related to protein stability. Control experiments showed no difference between Rad53-kd and Dun1-kd isolated after overexpression from induced or uninduced cells in SDS-PAGE or in in vitro kinase experiments (data not shown). The full-length kinases constituted more than 95% of the preparations as quantified in gel scans. Lane 5, molecular mass standards (in kilodaltons). (C) In vitro phosphorylation of Dun1 kinase by Rad53 kinase. In vitro kinase assays with GST-affinity purified wild-type and mutant proteins (see panel B) were performed. GST-Dun1 fusions were overexpressed in WDHY1413 (dun1-Δ), while all GST-Rad53 fusions were in DES453 (rad53-Δ). Proteins were resolved by SDS-8% PAGE and transferred to nitrocellulose filters. Upper panel, autoradiogram to assess incorporation of 32P into the proteins during the reaction. Rad53* and Dun1* show the positions of the respective phosphorylated protein species. Lower panel, immunoblot analysis of the same filter with anti-Rad53 and anti-Dun1 rat antibodies. wt, wild type; kd, kinase-deficient mutants; fha, FHA domain mutant. Lanes 1 to 8 are from one gel with intervening lanes spliced out; lane 9 is from a different gel. (D) In vitro phosphorylation depends on the Dun1 FHA domain. An in vitro kinase assay was performed with purified GST-fusion proteins overexpressed as described for panel C. Upper panel, 32P incorporation; lower panel, immunoblot analysis with anti-Rad53 and anti-Dun1 rat antibodies. All samples were analyzed on the same gel, but some intervening lanes were spliced out.
Dun1 kinase functions in DNA damage-induced transcription of a subset of damage-inducible genes, including the RNR genes, by controlling the inactivating phosphorylation of the Crt1 transcriptional repressor (26, 27, 61). Mutations in DUN1 cause sensitivity to DNA damaging-agents and the replication inhibitor hydroxyurea (HU). This sensitivity can be partly suppressed by elevating the deoxynucleoside triphosphate pools through deletion of the ribonucleotide reductase inhibitor Sml1 or by overexpression of RNR1, the gene encoding the large subunit of ribonucleotide reductase (58, 61). In addition, Dun1 functions in one pathway with Rad53 kinase to cause a G2/M arrest in response to DNA damage by negatively regulating mitotic exit. A second G2/M pathway, controlled by Chk1 regulating anaphase entry, acts in parallel (16, 41). Dun1 kinase is controlled by Rad53 kinase (4, 43), but it is unclear whether Rad53 kinase controls Dun1 activation directly or indirectly.
The FHA domain is a phosphoprotein recognition domain with a 55- to 75-amino-acid homology region that mediates specific phosphorylation-dependent protein-protein interactions (11, 24, 29). In eukaryotes, many FHA domain-containing proteins reside in the nucleus and function in DNA metabolism. The S. cerevisiae genome encodes 13 FHA domain-containing proteins, three of which (Rad53, Dun1, and Mek1) are related protein kinases known to act in DNA checkpoints (http://smart.embl-heidelberg.de). Structural and biochemical analysis identified the FHA domain as a unique phosphothreonine-specific phosphoprotein recognition fold consisting of 11 β-strands, which form two large, twisted antiparallel β-sheets folding into a β-sandwich (reviewed in reference 51). Structural analysis of phosphorylated peptides bound to the Rad53 FHA1 and -2 domains revealed that the four invariant amino acid residues of the FHA domain are located close to the peptide binding site (12, 54). Despite the considerable amino acid sequence divergence between the individual FHA domains, the secondary structures of four FHA domains whose structures have been analyzed (Rad53 FHA1 and -2, KAPP, and Chfr) are remarkably similar (reviewed in reference 51). The different binding specificities of individual FHA domains to phosphothreonine peptides are likely a reflection of the underlying amino acid sequence variation in FHA domains (12).
The specific biological function of individual FHA domains is poorly understood. Few in vivo data on FHA domain mutants are available, and only a limited number of bona fide physiological binding substrates of FHA domains have been identified. These include substrates for the FHA domains of Arabidopsis KAPP, human NIPP1, human KI-67 antigen, fission yeast Cds1, and the FHA2 domain of S. cerevisiae Rad53 (7, 29). The Rad53 kinase binds specifically to phosphorylated Rad9 protein via its FHA2 domain, and it has been proposed that this binding leads to Rad53 activation (13, 18, 45, 49, 53). Phosphorylation of Rad9 protein, presumably by Mec1 (or Tel1) kinase, may lead to the recruitment of Rad53 to Mec1 kinase for direct phosphorylation of Rad53 by Mec1 kinase. Experiments with fission yeast and mammalian cells support the idea that Cds1/Chk2 (S. cerevisiae Rad53) is directly phosphorylated by Rad3/ATM (S. cerevisiae Mec1/Tel1) (34, 50). Alternatively, it was suggested that Rad9 phosphorylation leads to autophosphorylation and activation of Rad53 kinase in the absence of direct phosphorylation of Rad53 kinase by Mec1 (18). FHA domain mutations in the human CHK2 gene were found to be associated with a rare familial multicancer syndrome, Li-Fraumeni syndrome (6). The binding substrate for the Chk2 FHA domain has not been identified yet. The Dun1 FHA domain binds to phosphothreonine-containing peptides (39), and its importance for Dun1 function has been demonstrated by mutational analysis (25). Based on two-hybrid data, an interaction between the Dun1 FHA domain and the poly(A) nuclease has been suggested to regulate DNA repair (21).
While the general function of the FHA domain as a phosphoprotein-specific protein-protein interaction domain is well established, the specific functions and interaction partners of individual FHA domains are poorly understood. Here we report that the FHA domain of the DNA damage checkpoint kinase Dun1 mediates specific protein-protein contacts with Rad53 kinase leading to direct kinase-to-kinase signaling between Rad53 and Dun1 kinases. Dun1 FHA domain mutants were defective in trans phosphorylation by Rad53 kinase in vivo and in vitro. As a consequence of their compromised checkpoint function, Dun1 FHA domain mutants exhibited increased sensitivity to genotoxic stress.
MATERIALS AND METHODS
Strains, plasmids, and oligonucleotides.
The strains used and their full genotypes are described in Table 1. Except for the strain used to purify the kinases, all strains were isogenic derivatives of W303. All plasmid constructions employed PCR cloning with Pfu polymerase. Plasmid pJN58 was used as a yeast overexpression vector of glutathione S-transferase (GST) fusion proteins under the control of the GAL promoter. pGAL-GST-DUN1 and pGAL-GST-RAD53 were constructed by in-frame cloning of the corresponding PCR-generated coding sequences into BamHI and HindIII vector sites by using oligonucleotide pairs olWDH131-olWDH134 and olWDH130-olWDH135, respectively. For the yeast two-hybrid analysis, plasmids pEG202 and pJG4-5 were employed (19). The coding sequence of DUN1 (olWDH248-olWDH249) was cloned into EcoRI- and XhoI-cleaved vectors. The coding sequences of RAD53 generated with primer pairs olWDH148-olWDH152 and olWDH148-olWDH151 were cloned into EcoRI- and NcoI-cleaved pEG202 or EcoRI-cleaved pJG4-5. The Dun1-fha coding sequence was amplified by PCR with olWDH131-olWDH134 and cloned in the BamHI site of pYES-TRP (a kind gift of S. Elledge) to come under the control of the GAL1 promoter.
TABLE 1.
S. cerevisiae strains used in this studya
| Strain | Relevant genotype | Source |
|---|---|---|
| CRY1 | Wild type | S. Elledge |
| DES453 | rad53-Δ::HIS3 TRP1::GAP-RNR1 | S. Elledge |
| DES460 | TRP1::GAP-RNR1 | S. Elledge |
| MHY26 | dun1-Δ100::HIS3 | S. Elledge |
| WDHY1413 | dun1-Δ::KanMX | This study |
| WDHY1619 | dun1-fha TRP1::GAP-RNR1 | This study |
| WDHY1620 | dun1-kd TRP1::GAP-RNR1 | This study |
| WDHY1748 | dun1-kd | This study |
| WDHY1751 | dun1-fha | This study |
| WDHY1757 | dun1-Δ100::HIS3 TRP1::GAP-RNR1 | This study |
| WDHY1759 | cdc13-1 TRP1::GAP-RNR1 | This study |
| WDHY1769 | dun1-fha cdc13-1 TRP1::GAP-RNR1 | This study |
| WDHY1781 | dun1-Δ100::HIS3 cdc13-1 TRP1::GAP-RNR1 | This study |
| WDHY1782 | dun1-kd cdc13-1 TRP1::GAP-RNR1 | This study |
| WDHY1887 | mec1-Δ::HIS3 cdc13-1 TRP1::GAP-RNR1 | This study |
| WDHY1934 | DUN1-myc18::TRP1 | This study |
| WDHY1935 | dun1-kd-myc18::TRP1 | This study |
| WDHY1936 | dun1-fha-myc18::TRP1 | This study |
Strain WDHY1413 has the additional genotype MATa leu2-3,112 trp1-289 ura3-52 his7-2 lys1-I. All other strains are isogenic W303 derivatives with the common genotype MATa can1-100 ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 rad5-535.
Integration vector YIp5-DUN1 for the allele replacement was constructed by cloning of the DUN1 coding sequence together with 1 kb of upstream and 136 bp of downstream sequences (olWDH246-olWDH247) between the NheI and NruI sites. For overexpression of His6-Dun1 protein in Escherichia coli, the DUN1 coding sequence (olWDH131-olWDH250) was cloned between the NdeI and BamHI sites of pET14-b (Novagen). DUN1 wild-type (CRY1) and mutant (WDHY1748 and WDHY1751) genes were tagged by PCR product-mediated integrative transformation, using olWDH298-olWDH299 for amplification of a myc18-TRP1 cassette. All oligonucleotide sequences are available upon request.
Generation of chromosomal dun1-kd (D328A) and dun1-fha (S74A,H77A) alleles.
To generate chromosomal point mutations in the FHA and kinase domains of DUN1, the pop-in, pop-out method was used (44). Briefly, yeast integration plasmid YIp5-dun1-kd or YIp5-dun1-fha was cut with EspI, and strain DES460 was transformed with the linearized plasmids. Ura+ transformants were grown under nonselective conditions and plated on plates containing 5-fluoroorotic acid and uracil to select “pop-out” of the URA3 marker of YIp5. Clones with dun1-kd and dun1-fha alleles were identified among 5-fluoroorotic acid-resistant colonies by sensitivity to methyl methanesulfonate (MMS) after replica plating on the corresponding plates. The presence of the desired mutations in the genome was confirmed by genomic sequencing.
DNA manipulations.
Site-specific mutagenesis of the DUN1 and RAD53 genes was performed by using the QuickChange system (Stratagene). Sets of mutagenic primers were used: olWDH170-olWDH171 to generate the dun1-D328A (kinase-deficient [kd]) mutation in DUN1, olWDH240-olWDH241 to generate the dun1-S74A,H77A mutation in the Dun1 FHA domain, and olWDH153-olWDH154 to produce rad53-K227A (kinase deficient). All mutations were confirmed by sequencing. The RNeasy kit (Qiagen) was used to prepare the total RNA from yeast cells. Northern blot analysis was performed with PCR-generated RNR2 (olWDH50 and olWDH51) and ACT1 (olWDH238 and olWDH239) probes labeled with 32P.
Protein methods.
Production of polyclonal antibodies against His-tagged Dun1 (overexpressed in E. coli) and against GST-tagged Rad53 (overexpressed in yeast) in rats and rabbits and their affinity purification were performed as described previously (5). The anti-Myc (9E10) and the goat anti-Rad53 antibodies were purchased from Santa Cruz Biotechnology. Immunoprecipitation and immunodetection of proteins were done as described previously (5). Large-scale purification of the wild-type and mutated GST-tagged Dun1 and Rad53 proteins was performed as described previously (46), except that galactose induction was for 3 h. When desired, cell cultures were treated with 0.1% MMS for an additional 2 h. Small-scale (250-ml cultures) affinity purification of overexpressed proteins by using glutathione-Sepharose beads was performed under conditions identical to those for the immunoprecipitation experiments.
Kinase assays.
Kinase assays were performed in 25 μl of reaction mix containing 50 mM Tris HCl (pH 7.5), 10 mM MgCl2, 10 mM MnCl2, 1 mM dithiothreitol, 0.25 mM ATP, and 1.5 μCi of [γ-32P]ATP (>7,000 Ci/mmol). When kinase bound to glutathione-Sepharose 4B beads was used, the beads were washed twice prior to the reaction in 500 μl of buffer containing 50 mM Tris HCl (pH 7.5), 10 mM MgCl2, and 10 mM MnCl2, and the reaction mixtures were supplemented with 10 mM glutathione (reduced form). Reaction mixtures were incubated for 30 min at 30°C. Proteins were resolved by sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis (SDS-8% PAGE), transferred to nitrocellulose filters, and exposed to X-ray film. We noted that kinase overexpression led to some induction of the DNA damage checkpoint as measured by Rad53 autophosphorylation, resulting in an electrophoretic shift (data not shown).
In vivo assays. (i) HU sensitivity.
Four-microliter-portions of 10-fold serial dilutions of late-log-phase cultures grown in yeast extract-peptone-dextrose (YPD) were spotted on YPD plates with or without 100 mM HU.
(ii) MMS sensitivity.
Cell survival after acute exposure to MMS was assayed by treating exponentially growing cells for 0 to 50 min with 0.5% MMS. Prior to plating on YPD to determine the surviving fraction, the MMS was inactivated by the addition of an equal volume of 10% sodium thiosulfate, and cells were washed with water on a 0.22-μm-pore-size filter. The surviving fraction for each strain was determined after incubating plates for 3 days. Experiments were performed in triplicate.
(iii) UV sensitivity.
Exponentially growing cells were plated on YPD plates and exposed to 0 to 200 J of UVC light/m2. The surviving fraction was determined after incubating plates for 3 days. Experiments were repeated at least three times.
(iv) Petite colony formation.
Single colonies grown on YPD were inoculated in YPD and grown for 2 days, and appropriate dilutions were plated on YPD plates. After 3 days of growth, the frequency of petite colonies was determined as the frequency of smaller, white colonies. It was confirmed that such colonies were indeed petite by the absence of growth on nonfermentable carbon sources. For each strain, three determinations were performed. At least 300 cells were counted for each strain in each experiment.
(v) Two-hybrid assay.
The LexA DNA binding domain fusion plasmids pEG202:DUN1, pEG202:Dun1-fha, and pEG202:RAD53 and activation domain fusion plasmids pJG4-5:DUN1, pJG4-5:Dun1-fha, and pJG4-5:RAD53 were used as described previously (19).
(vi) G2/M arrest assay.
Cell cycle arrest in response to cdc13-induced DNA damage was quantitatively assayed as described previously (16, 41). Cells were grown to early log phase at 23°C (permissive for cdc13), arrested in G1 with α-factor (5 μg/ml) for 2 h, washed with water by filtration, and released in YPD at 36°C (restrictive for cdc13). Samples were taken every 30 min after release, fixed in 70% ethanol, and stained with DAPI (4′,6′-diamidino-2-phenylindole). Cell morphology and nuclear status in at least 100 cells from each sample were scored microscopically.
RESULTS
In vitro phosphorylation of Dun1 by Rad53 kinase depends on the Dun1 FHA domain.
S. cerevisiae Dun1 is in length, overall structure, overall sequence identity, and overall sequence similarity more closely related to human Chk2, which contains also a single N-terminal FHA domain, than to S. cerevisiae Rad53 (two FHA domains) or than Rad53 is to human Chk2 (Fig. 1A). Sequence alignments of only the kinase domains of the three kinases confirm that both yeast kinases are more closely related to human Chk2 than to each other. The kinase domain of Rad53 appears to be slightly more related to that of Chk2 than to the Dun1 kinase domain (Fig. 1A). DNA damage-induced hyperphosphorylation of Dun1 kinase was shown to be genetically dependent on Rad53 kinase (4, 43). We tested the simplest hypothesis, i.e., that Rad53 kinase directly phosphorylates Dun1 kinase. Furthermore, we suspected that the Dun1 FHA domain mediates the specific interaction between the two kinases. Rad53 is known to undergo extensive genotoxic stress-induced phosphorylation, providing possible FHA domain target sites (38, 43, 48). To test this hypothesis, we performed in vitro kinase assays with wild-type and mutant Rad53 and Dun1 kinases. The kinases were expressed in S. cerevisiae as GST fusion proteins that complemented the MMS sensitivity of the respective deletion mutants in their wild-type but not mutant form (data not shown). Expression in yeast allowed the activation of the kinases by the application of DNA damage in vivo before purification. The purified kinases were substantially free of contaminating proteins (Fig. 1B). The Dun1 and Rad53 kinase preparations exhibited the autophosphorylation patterns expected from previous immunoprecipitation experiments (43). Dun1 kinase displayed a small electrophoretic mobility shift due to phosphorylation after DNA damage (see Fig. 2A). Wild-type Rad53 kinase showed a typical electrophoretic shift indicative of its extensive autophosphorylation after DNA damage induction (18, 38, 43), which is absent in the kinase-deficient Rad53 mutant protein (Fig. 1C and data not shown). From these data we conclude that the GST-Rad53 and GST-Dun1 fusion proteins retained a significant amount of biological function and are useful tools for further analysis.
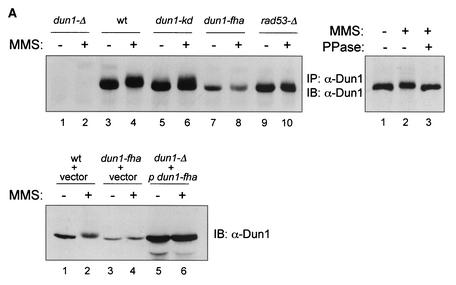
FIG. 2.
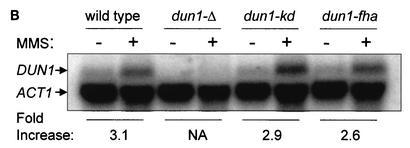
In vivo phosphorylation of Dun1 kinase depends on the Dun1 FHA domain. (A) Dun1 hyperphosphorylation depends on its FHA domain and Rad53 kinase. Upper left panel, immunoprecipitation (IP)-immunoblotting (IB) analysis of Dun1 protein level and phosphorylation status in wild-type (DES460) (lanes 3 and 4), dun1-Δ (MHY26) (lanes 1 and 2), dun1-kd (WDHY1620) (lanes 5 and 6), dun1-fha (WDHY1619) (lanes 7 and 8), and rad53-Δ (DES453) (lanes 9 and 10) strains before and after DNA damage (2 h in 0.1% MMS). Anti-Dun1 rabbit antibodies were used for IP. Immunodetection was performed with rat anti-Dun1 antibodies and the ECL system (Amersham Pharmacia Biotech). Upper right panel, IP mixtures from wild-type cells (DES460) exposed to 0.1% MMS for 2 h (lanes 2 and 3) or left without MMS (lane 1) were either incubated with 1,000 U of λ phosphatase (PPase) (New England Biolabs) at 30°C for 30 min (lane 3) or left untreated (lanes 1 and 2). Dun1 protein was detected as described above. Lower panel, immunoblot of extracts from wild-type (CRY1 with empty vector pYES-TRP1) (lanes 1 and 2), dun1-fha (WDHY1751 with empty vector) (lanes 3 and 4), and dun1-Δ (MHY26 containing plasmid pYES-TRP1-Dun1-fha where Dun1-fha is overexpressed from the GAL1 promoter) (lanes 5 and 6) cells, using rat anti-GST-Dun1 antibodies. Cells were induced for 3 h with 2% galactose and subsequently exposed to 0.1% MMS for 2 h or left without MMS. (B) dun1-fha has no effect on the steady-state and DNA damage-induced DUN1 mRNA levels. Northern blot analysis of DUN1 mRNA levels in wild-type (DES460), dun1-Δ (WDHY1757), dun1-kd (WDHY1620), and dun1-fha (WDHY1619) strains is shown. DUN1 transcript levels in wild-type and mutant strains were determined before and after DNA damage (0.1% MMS for 1 h) and normalized against the ACT1 transcript level. The DNA damage induction of the DUN1 transcript is expressed as fold increase. NA, not applicable.
Rad53 kinase caused hyperphosphorylation of Dun1 kinase in vitro (Fig. 1C, lane 4) as indicated by the increased incorporation of 32P and by the electrophoretic shift of Dun1 protein. The hyperphosphorylation depended on the kinase activity of Rad53 (lane 9). A kinase-deficient allele of Rad53, Rad53-K227A, exhibited no detectable autophosphorylation and no detectable trans phosphorylation of the Dun1 substrate (Fig. 1C, lanes 8 and 9). trans phosphorylation of Dun1 kinase by Rad53 was independent of the kinase activity of Dun1 (Fig. 1C, lane 5). A Dun1 kinase-deficient allele, Dun1-D328A (Fig. 1A), that exhibited no detectable autophosphorylation (lane 2), was phosphorylated efficiently by Rad53 kinase. The difference in 32P incorporation between the wild-type and Dun1-kd substrates in lanes 4 and 5 is due to the smaller amount of Rad53 kinase present in the reaction mixture loaded in lane 5. The extents of the electrophoretic mobility shifts of both substrates are very similar (Fig. 1C, upper panel, lanes 4 and 5).
To test whether the FHA domain of the Dun1 kinase was involved in mediating protein-protein interactions necessary for in vitro phosphorylation, we mutated the FHA domain by replacing two of the four invariant residues with alanine (Fig. 1A). Importantly, the Dun1-fha mutant substrate failed to be hyperphosphorylated by wild-type Rad53 kinase, as indicated by the absence of the electrophoretic shift (Fig. 1C, lane 6). We refer to Dun1 hyperphosphorylation as the Rad53-dependent phosphorylation that leads to an electrophoretic shift which occurs in addition to the level of Dun1 autophosphorylation which does not lead to an appreciable electrophoretic shift. Note that the Dun1-fha mutant protein is kinase proficient and able to autophosphorylate (Fig. 1D, lane 9). To further analyze the effect of the Dun1-fha mutant in in vitro kinase assays, we created a double mutant, the dun1-kd,fha mutant, that eliminated the Dun1 autophosphorylation in the background of the FHA domain mutant. The Dun1-kd,fha double mutant protein no longer served as a substrate for Rad53 kinase, as indicated by the virtual absence of 32P incorporation and by the severe reduction of the electrophoretic shift (Fig. 1D, lane 4). Control immunoblotting experiments confirmed that nearly identical amounts of Dun1-fha and Dun1-kd,fha substrates were present in the reaction mixtures (Fig. 1D, lower panel, lanes 2 to 4).
Taken together, these data show that Rad53 kinase can directly phosphorylate Dun1 kinase in vitro and that trans-phosphorylation by Rad53 kinase depended on the FHA domain but not on the kinase activity of the Dun1 substrate.
Rad53 kinase phosphorylates Dun1 kinase in vivo.
The dependence of the in vitro phosphorylation of Dun1 by Rad53 kinase on FHA domain-mediated protein-protein contacts provided an indication that Rad53 might phosphorylate Dun1 in vivo. To provide further arguments that Rad53 kinase directly phosphorylates Dun1 kinase in vivo, we analyzed the Dun1 phosphorylation status in vivo in the presence and absence of DNA damage by an immunoprecipitation-immunoblotting assay with anti-Dun1 antibodies. For this purpose, the DUN1 mutations were introduced into the genome so that the mutant proteins are expressed from the normal chromosomal locus. In response to DNA damage, Dun1 undergoes an electrophoretic shift (Fig. 2A, upper left panel, lane 4) that is caused by phosphorylation, as the electrophoretic shift can be reversed by phosphatase treatment (Fig. 2A, upper right panel). This is consistent with the previous observation showing DNA damage-induced hyperphosphorylation of Dun1 kinase in metabolic labeling experiments (4) and the in vitro data shown in Fig. 1C and D. The electrophoretic mobility shift was partly dependent on the Dun1 kinase activity, as the kinase-deficient Dun1 protein underwent a slightly diminished shift (compare Fig. 2A, lanes 4 and 6). Hyperphosphorylation was abolished in cells lacking Rad53 kinase (lanes 9 and 10), as shown before (4). Importantly, the dun1-fha mutation abolished the DNA damage-induced hyperphosphorylation of Dun1 kinase (lanes 7 and 8). Longer exposures failed to detect electrophoretically shifted Dun1-fha protein (data not shown). These data demonstrate that also in vivo, Dun1 hyperphosphorylation after DNA damage is dependent on the Dun1 FHA domain.
The Dun1-fha mutant protein consistently exhibited lower steady-state levels than the wild-type or kinase-deficient variants. The diminished Dun1-fha protein level is unlikely to be the direct cause for the inability of Rad53 kinase to phosphorylate this substrate in vivo, as overexpression of the Dun1-fha mutant protein did not restore the electrophoretic shift after MMS treatment (Fig. 2A, lower panel, lanes 5 and 6). Northern blot experiments showed that the steady-state level of the RNA was not significantly affected (Fig. 2B), suggesting that the reduced steady-state protein level is due to posttranscriptional events. It had been previously shown that Dun1 is transcriptionally induced after DNA damage (1), and the data in Fig. 2B suggest that this induction is not an autoregulatory loop.
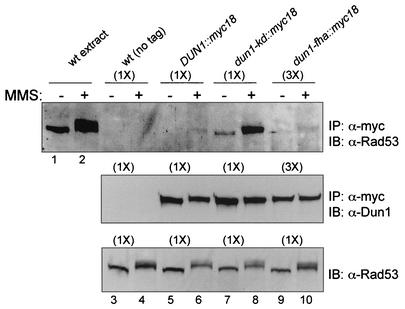
To more firmly establish that Dun1 kinase is a direct in vivo substrate of Rad53 kinase, we sought evidence that the proteins interacted in vivo. To this end, we monitored interaction between the two proteins by the sensitive two-hybrid assay (Table 2). A weak, but significant and reproducible, interaction between the two proteins could be identified. This interaction partly depended on the Dun1 FHA domain, as the interaction signal was significantly reduced, but not abolished, when the dun1-fha mutant was used. The induction of DNA damage by addition of MMS did not alter the interaction monitored in the two-hybrid system (data not shown). Overexpression from the vectors of the two-hybrid system appeared to result in a low-level, constitutive induction of the DNA damage response, as indicated by Rad53 autophosphorylation (data not shown). To substantiate the two-hybrid data, a highly efficient and sensitive tag (myc18) was introduced to the DUN1 chromosomal wild-type and mutant genes. This allowed us to demonstrate by immunoprecipitation an association of the Dun1 and Rad53 kinases in vivo at their native protein level (Fig. 3). The interaction appeared to preferentially involve activated Rad53 kinase, as the complex appeared to be more abundant after DNA damage and contained electrophoretically shifted (activated) Rad53 kinase (Fig. 3, upper panel, lanes 5 and 6). This is consistent with previous data (23) that showed that Dun1 preferentially interacts with the activated form of Rad53 kinase. Interestingly, the Dun1-kd mutant protein resulted in a more stable association with Rad53, which was still controlled by activation of the DNA damage checkpoint (Fig. 3, upper panel, lanes 7 and 8). Although the interaction signal was at the detection limit, it appeared that the Dun1-fha mutant protein was still proficient to form a complex with activated Rad53 kinase (Fig. 3, upper panel, compare lanes 6 and 10). Note that three times as much extract was used in the analysis of the Dun1-fha protein (Fig. 3), since its protein level was found to be reduced by about threefold (Fig. 2A).
TABLE 2.
Two-hybrid interaction between Rad53 and Dun1 is partly dependent on the Dun1 FHA domain
| DNA binding domain fusion | Activation domain fusion | β-Galactosidase activitya | Fold increase |
|---|---|---|---|
| Dun1 | —b | 1.4 ± 0.1 | 1 |
| Rad53 | 10.2 ± 1.2 | 7.3 | |
| Dun1-fha | Rad53 | 3.5 ± 0.6 | 2.5 |
| Rad53 | — | 0.9 ± 0.2 | 1 |
| Dun1 | 2.5 ± 1.6 | 2.8 | |
| Dun1-fha | 1.4 ± 0.2 | 1.6 |
Results are means and standard deviations of three determinations.
—, empty vector control.
FIG. 3.
Rad53 and Dun1 kinases form a complex in vivo. Immunoprecipitation (IP)-immunoblotting (IB) analysis of Rad53- and Dun1-containing complexes is shown. Upper panel, immunoprecipitation of Dun1-myc18 proteins with 9E10 anti-Myc antibodies from the wild type (wt) with untagged DUN1 (CRY1) (lanes 3 and 4), the wild type with DUN1::myc18 (WDHY1934) (lanes 5 and 6), the dun1-kd::myc18 mutant (WDHY1935) (lanes 7 and 8), and the dun1-fha::myc18 mutant (WDHY1936) (lanes 9 and 10), followed by immunoblotting with goat anti-Rad53 antibodies. In lanes 1 and 2, extract from 0.5 optical density unit of wild-type cells (CRY1) was loaded as size standards for activated and nonactivated Rad53. Cells were exposed to 0.1% MMS for 2 h prior to harvesting or not exposed. Note that 250 optical density units of cells was used in lanes 3 to 8 (1×), whereas 750 optical density units was used in lanes 9 and 10 (3×), because the cellular level of the Dun1-fha protein is about threefold lower (see Fig. 2A). Middle panel, loading control for Dun1 levels. The procedure was as for the upper panel, but immunoblotting was with rat anti-Dun antibodies. Lower panel, extract control for Rad53 level. Extracts from 1.5 optical density units of the same cell cultures used for the immunoprecipitation experiment shown in the upper and middle panels were blotted directly with goat anti-Rad53 antibodies. Note that the same amount was used in all lanes.
The combination of the in vitro kinase data (Fig. 1) and in vivo phosphorylation and interaction data (Fig. 2 and 3; Table 2) strongly suggests that Dun1 interacts, probably in a transient fashion after DNA damage checkpoint activation, with Rad53 kinase, which leads to hyperphosphorylation and activation of Dun1 kinase. The Dun1-fha mutant had some effect on the Dun1-Rad53 interaction as measured in the two-hybrid system, but it did not abolish the formation of Dun1-Rad53-containing complexes.
Checkpoint functions of Dun1 kinase depend on its FHA domain.
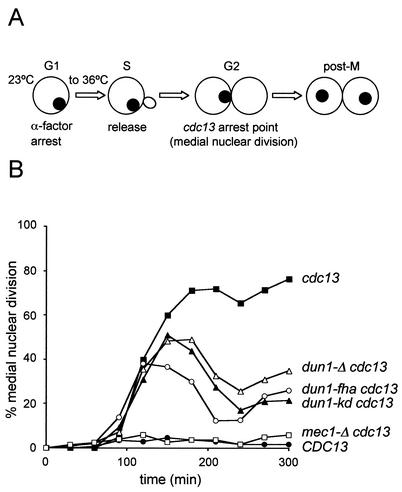
To explore the molecular defects caused by the FHA domain mutations in Dun1 kinase, we analyzed several DNA damage checkpoint responses (G2/M cell cycle arrest, DNA damage-induced transcription, and Rad55 phosphorylation) that are known to depend on Dun1 kinase. Transient arrest at the G2/M border is the major cell cycle response controlled by the DNA damage checkpoints in S. cerevisiae (55, 60). The arrest is mediated by two pathways, one dependent on both Rad53 and Dun1 kinases controlling primarily mitotic exit and the other dependent on Chk1 kinase inhibiting anaphase entry (16, 41). A quantitative G2/M arrest assay (Fig. 4A) which is based on using the cdc13 mutation was employed (16). The CDC13 gene product binds to the ends of chromosomes, and cdc13 mutant cells accumulate single-stranded DNA near their chromosome ends, triggering a pronounced cell cycle arrest (17). Logarithmically grown cells were synchronized in G1 and released at the restrictive temperature for cdc13. During S phase, DNA damage is generated, leading to a DNA damage checkpoint-dependent arrest at the G2/M border. cdc13 cells arrest and exhibit medial nuclear division (Fig. 4A) which can easily be quantified (see Materials and Methods). Checkpoint mutant cells fail to arrest and enter the second cycle. This protocol allows quantification of the kinetics of establishing arrest, the extent of the arrest, and the maintenance of the arrest. cdc13 cells with an intact DNA damage checkpoint arrested in G2/M and maintained this arrest throughout the experiment at the restrictive temperature (Fig. 4B). This arrest is completely eliminated in mec1-Δ cdc13 cells (Fig. 4B), demonstrating that it is entirely checkpoint-mediated. CDC13 cells (i.e., in the absence of DNA damage) with an intact DNA damage checkpoint traversed G2/M in a normal cell cycle and did not arrest (Fig. 4B). dun1-Δ cells exhibit a partial G2/M arrest defect, as previously reported (16). The G2/M arrest mediated by Dun1 kinase was dependent on its kinase activity, as the dun1-kd allele exhibited a nearly identical defect as the gene deletion. Importantly, the dun1-fha mutant exhibited a G2/M arrest defect that was at least as strong as that in the kinase-deficient or deletion mutants. It was previously shown that the G2/M arrest defect of rad53 and dun1 deletion is epistatic (16). Therefore, these results suggest that signaling of the G2/M arrest via Rad53 kinase is routed entirely through the Dun1 FHA domain interaction to the downstream effectors. This result also demonstrates that the FHA domain mutant is not leaky.
FIG. 4.
The FHA domain is important for the checkpoint function of Dun1 kinase. (A) Scheme for the G2/M arrest assay. G1 phase synchronized cells were released into the cell cycle at the restrictive temperature for cdc13. After S-phase traversal, DNA damage accumulates, leading to a checkpoint-mediated G2/M arrest with medial nuclear division morphology (see text) (modified from reference 16). (B) dun1-fha is defective for the G2/M cell cycle arrest after DNA damage. The cdc13 DUN1 (WDHY1759), cdc13 DUN1 mec1 (WDHY1887), CDC13 DUN1 (DES460), dun1-Δ cdc13 (WDHY1781), dun1-kd cdc13 (WDHY1782), and dun1-fha cdc13 (WDHY1769) strains were used to determine the kinetics and maintenance of the G2/M arrest. (C) dun1-fha is defective in DNA damage-induced gene expression. Northern blot analysis of the RNR2 transcript levels in wild-type (DES460), dun1-Δ (WDHY1757), dun1-kd (WDHY1620), and dun1-fha (WDHY1619) strains is shown. The transcript level of RNR2 mRNA before and after DNA damage (0.1% MMS for 1 h) was normalized against the ACT1 transcript level and is expressed as fold increase after DNA damage. (D) dun1-fha is defective in DNA damage-induced phosphorylation of Rad55 protein. The Rad55 phosphorylation status in wild-type (wt) (DES460) (lanes 1 and 2), dun1-Δ (WDHY1757) (lanes 3 and 4), dun1-fha (WDHY1619) (lanes 5 and 6), and dun1-kd (WDHY1620) (lanes 7 and 8) strains was analyzed by immunoprecipitation (IP) and immunoblotting (IB). Cells were grown either in the absence or in the presence of 0.1% MMS for 2 h.
DNA damage-induced transcription is another checkpoint-controlled response to genotoxic stress. Dun1 kinase was first isolated in a screen for genes that are essential for damage-induced transcription of RNR genes (61). To assess the effect of the various DUN1 mutations on damage-inducible transcription, we quantified the steady-state transcript levels of the DNA damage-inducible gene RNR2, encoding a ribonucleotide reductase subunit, in comparison to the noninducible ACT1 gene, encoding actin (Fig. 4C). In wild-type cells, RNR2 transcription was induced almost sixfold after MMS treatment, which is largely dependent on Dun1 kinase. This is consistent with previous observations after UV irradiation (1) and after exposure to MMS and HU (61). The Dun1-dependent component of the RNR2 induction was entirely dependent on the kinase activity of Dun1, as the kinase-deficient allele exhibited as strong a defect as the gene deletion. The dun1-fha mutant significantly decreased the transcriptional induction of RNR2, almost to the level of the kinase-deficient and deletion alleles. We conclude that the Dun1 FHA domain is important for full transcriptional induction after DNA damage. It is unclear whether the small difference between the FHA domain and kinase-deficient and deletion alleles is significant.
It has been speculated that the DNA damage checkpoint directly instructs DNA repair pathways by phosphorylating DNA repair proteins (31, 55, 60). One such protein is the repair protein Rad55, which is specifically phosphorylated after genotoxic stress in a DNA damage checkpoint-dependent manner (5). Rad55 phosphorylation can be visualized as an electrophoretic shift in immunoprecipitation-immunoblotting experiments. DNA damage-induced Rad55 phosphorylation depends partially on Dun1 kinase (5). The dun1-kd and the dun1-fha mutants showed a very similar, partial defect in DNA damage-induced Rad55 phosphorylation that was similar, if not identical, to the effect of the deletion mutant (Fig. 4D).
From monitoring three DNA damage checkpoint-controlled responses, i.e., G2/M cell cycle arrest, DNA damage-induced transcription, and Rad55 protein phosphorylation, we conclude that the Dun1 FHA domain plays a major role in checkpoint signaling to Dun1 kinase.
The FHA domain is important for the biological function of Dun1 kinase.
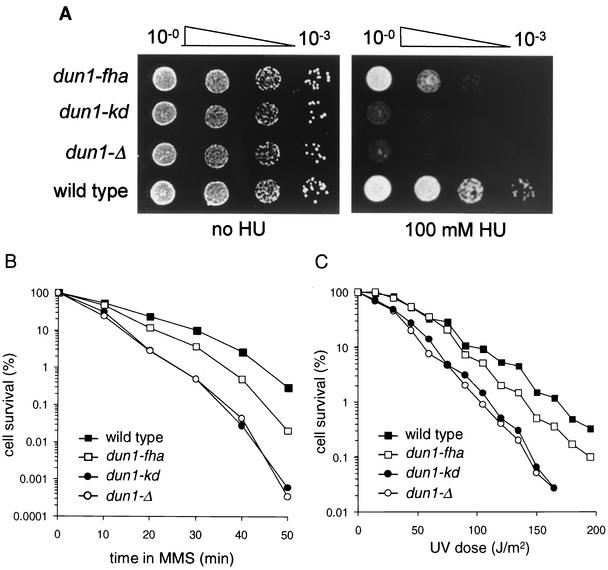
The absence of Dun1 kinase leads to cellular sensitivity to genotoxic stress, such as exposure to the alkylating agent MMS or UV radiation, as well as to HU, a ribonucleotide reductase inhibitor that arrests DNA replication (61). To ascertain that the FHA domain-mediated interaction between Dun1 and Rad53 kinases is biologically important and to relate the molecular checkpoint defects of the dun1-fha mutant to biological function, we analyzed the sensitivity of the dun1-fha mutant strain to genotoxic stress. The deletion of DUN1 or the kinase-deficient allele caused essentially the same level of HU sensitivity, suggesting that kinase activity is essential for Dun1 function in vivo and that the D328A allele is devoid of significant residual activity. The FHA domain mutant was sensitive to HU and exhibited a sensitivity intermediate between those of the wild type and the deletion or kinase-deficient mutants (Fig. 5A). Very similar results were obtained in experiments analyzing the sensitivities of the same strains to acute exposure to MMS (Fig. 5B) and to UV radiation (Fig. 5C).
FIG. 5.
The FHA domain is important for the cellular function of Dun1 kinase. Wild-type (DES460), dun1-Δ (WDHY1757), dun1-kd (WDHY1620), and dun1-fha (WDHY1619) strains were used. (A) The dun1-fha strain is sensitive to HU. Serial dilutions of cultures were spotted on YPD plates with or without HU. Plates were photographed after 2 days. (B) The dun1-fha strain is sensitive to MMS. Survival after acute exposure to MMS was determined. The results of one representative experiment are shown. The differences in sensitivity between the strains were highly reproducible. (C) The dun1-fha strain is sensitive to UV. Survival after UVC exposure was determined. The results of one representative experiment are shown. The differences in sensitivity between the strains were highly reproducible.
In S. cerevisiae, petite colonies are formed by cells that do not contain functional mitochondria. Petite colonies are easily identified as slow-growing white colonies that are unable to grow on nonfermentable carbon sources. Mutations in DUN1 are known to increase the formation of petite colonies (15). Deletion of DUN1 and the kinase-deficient allele led to a fivefold increase in petite colony frequency (from 4.7% ± 0.6% for the wild type to 24.3% ± 4.8% and 23.4% ± 2.8% for dun1-Δ and dun1-kd, respectively [means and standard deviations from three determinations; the same strains as described in the legend to Fig. 5 were used]). Importantly, the dun1-fha cells also generated significantly more petite colonies than isogenic wild-type cells, but the defect was less pronounced than that in the deletion or kinase-deficient mutants (13.3% ± 1.9%).
We conclude that kinase activity is essential for the full function of Dun1 kinase and that the FHA domain mutant partially cripples the function of kinase-competent Dun1 protein. Note that the Dun1 FHA domain mutant, which is competent for kinase activity (Fig. 1C), was used in these experiments.
The strains used in the experiments described here overexpressed RNR1 to alleviate the nucleotide stress that leads to the inviability of rad53 and mec1 null mutants (9, 58). RNR1 overexpression (data not shown) and the deletion of the ribonucleotide reductase inhibitor SML1 lead to partial suppression of the dun1 phenotypes (58). The dun1-fha mutant exhibited phenotypes very similar to those described above also in the absence of RNR1 overexpression (data not shown). The only consistent difference was that the kinase-deficient allele exhibited sensitivities identical to those of as the deletion allele in the RNR1 overexpression background, but it exhibited less severe HU, UV, and MMS sensitivities and petite colony formation phenotype than the DUN1 deletion in the absence of RNR1 overexpression (data not shown). However, the phenotypes of the kinase-deficient allele were still stronger than those of the FHA domain mutation.
From the results of these experiments, we conclude that the FHA domain of Dun1 is important for its full function in DNA metabolism and that the phenotypes caused by the dun1-fha mutation are unlikely to be the sole result of nucleotide stress. The partial phenotypes of the dun1-fha mutation compared to the deletion or kinase-deficient allele suggest that Dun1 carries out some functions that are independent of its FHA domain.
DISCUSSION
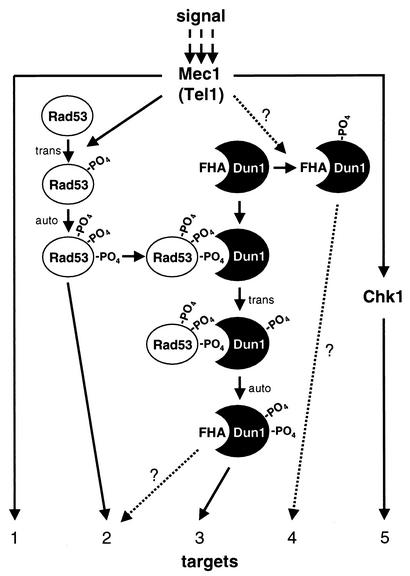
Dun1 kinase is an important signal-transducing kinase in the DNA damage checkpoint system in S. cerevisiae and is related to FHA domain kinases functioning in DNA damage checkpoints in other organisms, including Cds1 in fission yeast and Chk2 in mammals. Here we studied the mechanism by which the DNA damage checkpoint signal is transmitted in the kinase cascade from Rad53 to Dun1. Our data show that Rad53 signals through the Dun1 FHA domain, leading to direct activation of Dun1 kinase by trans phosphorylation. The phenotypes of specific mutations in DUN1 allow us to conclude that Dun1 kinase is controlled not only by Rad53 kinase but also in a Rad53-independent fashion that does not involve the Dun1 FHA domain. This suggests that Rad53 and Dun1 form a signal transduction network that may respond to different input signals, possibly leading to differential outputs (Fig. 6).
FIG. 6.
Model for checkpoint signaling by S. cerevisiae checkpoint kinases. DNA damage activates Mec1 and Tel1 kinases, which directly and indirectly control a web of secondary kinases (Rad53, Dun1, and Chk1) and effector targets (1 to 5). Kinases involving trans-phosphorylation and autophosphorylation are activated (not shown for all kinases). After Rad53 is activated, one or several phosphorylated residues become a recognition motif for the Dun1 FHA domain. The ensuing transient association between the two kinases leads to trans-phosphorylation of Dun1 by Rad53. Activated Dun1 kinase may have specific effectors (target 3), or target overlap with Rad53 kinase may lead to additional signal amplification (target 2). A postulated Rad53-independent pathway of Dun1 activation is indicated as Mec1/Tel1 controlling Dun1 kinase in a direct or indirect fashion. The mechanism of this pathway is not understood, but it may lead to signal-specific targeting of specific effectors (target 4). For details, see text.
Rad53 kinase controls Dun1 activation in response to DNA damage induced by MMS and replication blocks provoked by HU (4). Genetic analyses suggested that both kinases act in a linear pathway to control the G2/M cell cycle arrest and the transcriptional induction of the Crt1 repressor target genes (16, 26). Here we show that Dun1 kinase is a direct phosphorylation target of Rad53 kinase, forming a transient hetero-oligomeric intermediate in which trans phosphorylation of Dun1 by Rad53 is dependent on the Dun1 FHA domain. The phosphoprotein recognition FHA domain of Dun1 kinase is likely to recognize a particular phosphorylated residue on activated Rad53 kinase (Fig. 6). Activated Rad53 kinase is hyperphosphorylated at many different residues, and the Dun1 FHA domain target residue(s) remains to be determined. It is not known whether the Dun1-Rad53 complex contains or depends on Rad9 protein (18). The strict dependence of Dun1 in vitro phosphorylation by Rad53 on the Dun1 FHA domain with purified kinases argues for a specific and direct interaction between both kinases. However, a contribution of Rad9 in the in vivo interaction cannot be excluded presently. Phosphorylation-dependent transient homo-oligomerization involving its FHA domain has also been suggested to be an intermediate in the activation of Chk2 kinase (2, 57).
While trans phosphorylation of Dun1 by Rad53 was abolished by the Dun1 FHA domain mutant, the association with activated Rad53 kinase appeared to be at least partly intact. The FHA domain mutation changed two invariant residues (S74 and H77) to alanine. The residues equivalent to Dun1 S74 in the Chk2 and Chfr FHA domains are presumed to be in close contact with the phosphorylated target residue (28, 30, 45, 47). It appears that a large number of residues are involved in the interaction between an FHA domain and its target sequence (51). We consider it unlikely that the double mutation of two of the four invariant Dun1 FHA domain residues results in a leaky phenotype, because the cell cycle arrest defect of the dun1-fha mutant was complete and because of previous results with the analogous mutations in Cds1 kinase (7). It is conceivable, but unlikely, that the mutations of the two amino acid residues in the FHA domain to alanine gravely affect the overall structure of the Dun1 protein. This notion is strengthened by the observation that Dun1-fha mutant kinase autophosphorylates to the same extent as the wild-type protein in vitro. Thus, we suggest that although the Dun1 FHA domain mutant might still bind its phosphorylated target sequence on Rad53, it cannot undergo a conformational change that is necessary for Dun1 to be trans phosphorylated by Rad53. In this view, nonactivated Dun1 kinase adopts an active conformation through a series of conformational changes that are triggered by binding to activated Rad53 through its FHA domain and trans phosphorylation by Rad53 (28, 30, 45, 47). The observation that kinase-deficient Dun1 protein forms a more abundant complex with activated Rad53 kinase than wild-type Dun1 indicates that the association is more stable. This suggests that after trans phosphorylation by Rad53 kinase, Dun1 autophosphorylation leads to destabilization of the kinase complex (Fig. 6). It is unclear whether this is achieved by charge repulsion or by another conformational change of Dun1 kinase.
Why would the DNA damage checkpoint employ two kinases in a linear pathway? Little is known about whether the DNA damage checkpoint kinase cascade acts like a switch to provide a threshold distinction, as proposed for some mitogen-activated protein (MAP) kinase cascades (14), or possibly to amplify the initial signal. DNA damage induction by physical or chemical agents is difficult to control, but experiments using the HO endonuclease have demonstrated that a single unrepaired DNA double-strand break triggers a checkpoint response (37). Although the nature of the primary signal for the checkpoint remains elusive, these data suggest a very low threshold for checkpoint induction. Unlike in MAP kinase cascades (10, 14), the kinases in the DNA damage response pathway (Mec1, Rad53, and Dun1) do not associate to form a stable complex (references 33 and 36 and this work). The physical association of the MAP kinases provides signaling specificity (10, 32). In the absence of such stable complex formation, the signaling specificity of the checkpoint appears to be mediated by the FHA phosphoprotein recognition motifs of the individual kinase components. Rad53 kinase has two FHA domains that associate with the adaptor proteins Rad9 and possibly Mrc1. These proteins likely recruit Rad53 kinase to Mec1 kinase after checkpoint induction by DNA damage or replicational stress (3, 13, 49, 53). Likewise, Dun1 kinase activation is mediated by a specific interaction with a phosphorylated residue on activated Rad53 kinase. Such a system of transient specific interactions could provide significant signal amplification, as a single upstream kinase can activate many downstream kinase molecules. Moreover, such a mechanism may be important for checkpoint maintenance and termination.
Several lines of evidence strongly suggest that Dun1 kinase is activated not only in a Rad53-dependent but also in a Rad53-independent pathway. A defect in silencing of telomeric gene expression has been identified in dun1-Δ cells but not in several hypomorphic rad53 alleles. This is likely to be related to an imbalance in the nucleotide pool, as the phenotype can be reversed by RNR1 overexpression or deletion of the SML1-encoded repressor (8). DNA damage-induced transcription of the cross-link repair gene SMN1 was reported to be dependent on Dun1 but independent of Rad53 kinase (56). It is unclear whether this defect in dun1 cells is related to nucleotide pool imbalance. Gross chromosomal rearrangements (GCR) in budding yeast are suppressed by the functions of the DNA damage checkpoint (35). An almost identical, ∼200-fold rate increase of GCR was observed in cells lacking Mec1 kinase, Dun1 kinase, or both, whereas cells lacking Rad53 kinase exhibited only a modest 27-fold increase. Importantly, these effects appear to be independent of the nucleotide pool control of the checkpoint, as the strains were simultaneously deleted for SML1. Thus, it appears that Mec1 and Dun1 kinases form one pathway to repress GCR in budding yeast. These data also imply that Mec1 activation can lead to differential signaling to the downstream kinases Rad53 and Dun1 (Fig. 6).
Presently, it is not clear how much the individual molecular checkpoint responses contribute to survival in genotoxic stress. The Dun1 FHA domain mutation conferred as strong a defect in the G2/M cell cycle arrest assay as the DUN1 deletion and kinase-deficient alleles, suggesting that all Rad53-to-Dun1 signaling involves the Dun1 FHA domain. However, the dun1-fha mutant exhibited less sensitivity to UV, MMS, and HU than null mutants (deletion and kinase deficient), suggesting functions of Dun1 kinase that are independent of its FHA domain but dependent on its kinase activity. Since null mutations in MEC1 abolish all known checkpoint response in budding yeast, the most parsimonious interpretation of our results is that Mec1 also exerts Rad53-independent control on Dun1 kinase (Fig. 6).
Acknowledgments
We thank S. Elledge, F. Fabre, and J. Nelson for strains and plasmids; J. Nunnari for the use of her microscope; and A. Chan for assistance in strain construction. We are very grateful to T. Petes, K. Shiozaki, S. Bärtsch, M. Rolfsmeier, and J. Solinger for comments on the manuscript.
V.I.B. is an International Research Scholar of the Howard Hughes Medical Institute. This work was supported by a grant from the National Institutes of Health (CA-92276) to W.-D.H.
The first two authors contributed equally to this work.
REFERENCES
- 1.Aboussekhra, A., J. E. Vialard, D. E. Morrison, M. A. Delatorreruiz, L. Cernakova, F. Fabre, and N. F. Lowndes. 1996. A novel role for the budding yeast RAD9 checkpoint gene in DNA damage-dependent transcription. EMBO J. 15:3912-3922. [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, J.-Y., X. Li, H. L. Davis, and C. E. Canman. 2002. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J. Biol. Chem. 277:19389-19395. [DOI] [PubMed] [Google Scholar]
- 3.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. H. Hu, P. J. H. Werler, K. Bousset, K. Furuya, J. F. X. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 4.Allen, J. B., Z. Zhou, W. Siede, E. C. Friedberg, and S. J. Elledge. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8:2401-2415. [DOI] [PubMed] [Google Scholar]
- 5.Bashkirov, V. I., J. S. King, E. V. Bashkirova, J. Schmuckli-Maurer, and W. D. Heyer. 2000. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol. Cell. Biol. 20:4393-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell, D. W., J. M. Varley, T. E. Szydlo, D. H. Kang, D. C. R. Wahrer, K. E. Shannon, M. Lubratovich, S. J. Verselis, K. J. Isselbacher, J. F. Fraumeni, J. M. Birch, F. P. Li, J. E. Garber, and D. A. Haber. 1999. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science 286:2528-2531. [DOI] [PubMed] [Google Scholar]
- 7.Boddy, M. N., A. Lopez-Girona, P. Shanahan, H. Interthal, W. D. Heyer, and P. Russell. 2000. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20:8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craven, R. J., and T. D. Petes. 2000. Involvement of the checkpoint protein Mec1p in silencing of gene expression at telomeres in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:2378-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desany, B., A. A. Alcasabas, J. B. Bachant, and S. J. Elledge. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes0 Dev. 12:2956-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dohlman, H. G., and J. W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703-754. [DOI] [PubMed] [Google Scholar]
- 11.Durocher, D., J. Henckel, A. R. Fersht, and S. P. Jackson. 1999. The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell 4:387-394. [DOI] [PubMed] [Google Scholar]
- 12.Durocher, D., I. A. Taylor, D. Sarbassova, L. F. Haire, S. L. Westcott, S. P. Jackson, S. J. Smerdon, and M. B. Yaffe. 2000. The molecular basis of FHA domain-phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell 6:1169-1182. [DOI] [PubMed] [Google Scholar]
- 13.Emili, A. 1998. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell 2:183-189. [DOI] [PubMed] [Google Scholar]
- 14.Ferrell, J. E. 1996. Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem. Sci. 21:460-466. [DOI] [PubMed] [Google Scholar]
- 15.Fikus, M. U., P. A. Mieczkowski, P. Koprowski, J. Rytka, E. Sledziewska-Gojska, and Z. Ciesla. 2000. The product of the DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, specifically functions in mitochondria. Genetics 154:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner, R., C. W. Putnam, and T. Weinert. 1999. RAD53, DUN1, and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J. 11:3173-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garvik, B., M. Carson, and L. Hartwell. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15:6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8:129-136. [DOI] [PubMed] [Google Scholar]
- 19.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1-phase and S-phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 20.Haber, J. E. 1999. Sir-Ku-itous routes to make ends meet. Cell 97:829-832. [DOI] [PubMed] [Google Scholar]
- 21.Hammet, A., B. L. Pike, and J. Heierhorst. 2002. Posttranscriptional regulation of the RAD5 DNA repair gene by the Dun1 kinase and the Pan2-Pan3 poly(A)-nuclease complex contributes to survival of replication blocks. J. Biol. Chem. 277:22469-22474. [DOI] [PubMed] [Google Scholar]
- 22.Hanks, S. K., A. M. Quinn, and T. Hunter. 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241:42-52. [DOI] [PubMed] [Google Scholar]
- 23.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Y. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. V. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann, K., and P. Bucher. 1995. The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends. Biochem. Sci. 20:347-349. [DOI] [PubMed] [Google Scholar]
- 25.Huang, M., and S. J. Elledge. 2000. The FHA domain, a phosphoamino acid binding domain involved in the DNA damage response pathway. CSHSQB 65:413-421. [DOI] [PubMed] [Google Scholar]
- 26.Huang, M., Z. Zhou, and S. J. Elledge. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94:595-605. [DOI] [PubMed] [Google Scholar]
- 27.Huang, M. X., and S. J. Elledge. 1997. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6105-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huse, M., and J. Kuriyan. 2002. The conformational plasticity of protein kinases. Cell 109:275-282. [DOI] [PubMed] [Google Scholar]
- 29.Li, J., G.-I. Lee, S. R. Van Doren, and J. C. Walker. 2000. The FHA domain mediates phosphoprotein interactions. J. Cell Sci. 113:4143-4149. [DOI] [PubMed] [Google Scholar]
- 30.Li, J. J., B. L. Williams, L. F. Haire, M. Goldberg, E. Walker, D. Durocher, M. B. Yaffe, S. P. Jackson, and S. J. Smerdon. 2002. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol. Cell 9:1045-1054. [DOI] [PubMed] [Google Scholar]
- 31.Lowndes, N. F., and J. R. Murguia. 2000. Sensing and responding to DNA damage. Curr. Opin. Genet. Dev. 10:17-25. [DOI] [PubMed] [Google Scholar]
- 32.Madhani, H. D., and G. R. Fink. 1998. The riddle of MAP kinase signaling specificity. Trends Genet. 14:151-155. [DOI] [PubMed] [Google Scholar]
- 33.Mallory, J. C., and T. D. Petes. 2000. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl. Acad. Sci. USA 97:13749-13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuoka, S., G. Rotman, A. Ogawa, Y. Shiloh, K. Tamai, and S. J. Elledge. 2000. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. USA 97:10389-10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myung, K., A. Datta, and R. D. Kolodner. 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104:397-408. [DOI] [PubMed] [Google Scholar]
- 36.Paciotti, V., M. Clerici, M. Scotti, G. Lucchini, and M. P. Longhese. 2001. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol. Cell. Biol. 21:3913-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pellicioli, A., S. B. Lee, C. Lucca, M. Foiani, and J. E. Haber. 2001. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell 7:293-300. [DOI] [PubMed] [Google Scholar]
- 38.Pellicioli, A., C. Lucca, G. Liberi, F. Marini, M. Lopes, P. Plevani, A. Romano, P. P. Di Fiore, and M. Foiani. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18:6561-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pike, B. L., A. Hammet, and J. Heierhorst. 2001. Role of the N-terminal forkhead-associated domain in the cell cycle checkpoint function of the Rad53 kinase. J. Biol. Chem. 276:14019-14026. [DOI] [PubMed] [Google Scholar]
- 40.Rhind, N., and P. Russell. 1998. Mitotic DNA damage and replication checkpoints in yeast. Curr. Opin. Cell Biol. 10:749-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez, Y., J. Bachant, H. Wang, F. H. Hu, D. Liu, M. Tetzlaff, and S. J. Elledge. 1999. Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science 286:1166-1171. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez, Y., B. A. Desany, W. J. Jones, Q. H. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez, Y., Z. Zhou, M. X. Huang, B. E. Kemp, and S. J. Elledge. 1997. Analysis of budding yeast kinases controlled by DNA damage. Methods Enzymol. 283:398-410. [DOI] [PubMed] [Google Scholar]
- 44.Scherer, S., and R. W. Davis. 1979. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl. Acad. Sci. USA 76:4951-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz, M. F., J. K. Duong, Z. X. Sun, J. S. Morrow, D. Pradhan, and D. F. Stern. 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9:1055-1065. [DOI] [PubMed] [Google Scholar]
- 46.Solinger, J. A., G. Lutz, T. Sugiyama, S. C. Kowalczykowski, and W.-D. Heyer. 2001. Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J. Mol. Biol. 307:1207-1221. [DOI] [PubMed] [Google Scholar]
- 47.Stavridi, E. S., Y. Huyen, I. R. Loreto, D. M. Scolnick, T. D. Halazonetis, N. P. Pavletich, and P. D. Jeffrey. 2002. Crystal structure of the FHA domain of the Chfr mitotic checkpoint protein and its complex with tungstate. Structure 10:891-899. [DOI] [PubMed] [Google Scholar]
- 48.Sun, Z. X., D. S. Fay, F. Marini, M. Foiani, and D. F. Stern. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10:395-406. [DOI] [PubMed] [Google Scholar]
- 49.Sun, Z. X., J. Hsiao, D. S. Fay, and D. F. Stern. 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281:272-274. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka, K., M. N. Boddy, X. B. Chen, C. H. McGowan, and P. Russell. 2001. Threonine-11, phosphorylated by Rad3 and ATM in vitro, is required for activation of fission yeast checkpoint kinase Cds1. Mol. Cell. Biol. 21:3398-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai, M.-D. 2002. FHA: a signal transduction domain with diverse specificity and function. Structure 10:887-889. [DOI] [PubMed] [Google Scholar]
- 52.Usui, T., H. Ogawa, and J. H. J. Petrini. 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7:1255-1266. [DOI] [PubMed] [Google Scholar]
- 53.Vialard, J. E., C. S. Gilbert, C. M. Green, and N. F. Lowndes. 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17:5679-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, P., I. J. L. Byeon, H. Liao, K. D. Beebe, S. Yongkiettrakul, D. Pei, and M. D. Tsai. 2000. Structure and specificity of the interaction between the FHA2 domain of Rad53 and phosphotyrosyl peptides. J. Mol. Biol. 302:927-940. [DOI] [PubMed] [Google Scholar]
- 55.Weinert, T. 1998. DNA damage checkpoints update: getting molecular. Curr. Opin. Genet. Dev. 8:185-193. [DOI] [PubMed] [Google Scholar]
- 56.Wolter, R., W. Siede, and M. Brendel. 1996. Regulation of SNM1, an inducible Saccharomyces cerevisiae gene required for repair of DNA cross-links. Mol. Gen. Genet. 250:162-168. [DOI] [PubMed] [Google Scholar]
- 57.Xu, X., L. M. Tsvetkov, and D. F. Stern. 2002. Chk2 activation and phosphorylation-dependent oligomerization. Mol. Cell. Biol. 22:4419-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao, X. L., E. G. D. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]
- 59.Zheng, P., D. S. Fay, J. Burton, H. Xiao, J. L. Pinkham, and D. F. Stern. 1993. SPK1 is an essential S-phase-specific gene of Saccharomyces cerevisiae that encodes a nuclear serine/threonine/tyrosine kinase. Mol. Cell. Biol. 13:5829-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, B. B. S., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 61.Zhou, Z., and S. J. Elledge. 1993. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell 75:1119-1127. [DOI] [PubMed] [Google Scholar]