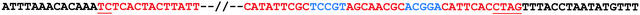Abstract
The maize mutation sh2-7527 was isolated in a conventional maize breeding program in the 1970s. Although the mutant contains foreign sequences within the gene, the mutation is not attributable to an interchromosomal exchange or to a chromosomal inversion. Hence, the mutation was caused by an insertion. Sequences at the two Sh2 borders have not been scrambled or mutated, suggesting that the insertion is not caused by a catastrophic reshuffling of the maize genome. The insertion is large, at least 12 kb, and is highly repetitive in maize. As judged by hybridization, sorghum contains only one or a few copies of the element, whereas no hybridization was seen to the Arabidopsis genome. The insertion acts from a distance to alter the splicing of the sh2 pre-mRNA. Three distinct intron-bearing maize genes were found in the insertion. Of most significance, the insertion bears striking similarity to the recently described DNA helicase–bearing transposable elements termed Helitrons. Like Helitrons, the inserted sequence of sh2-7527 is large, lacks terminal repeats, does not duplicate host sequences, and was inserted between a host dinucleotide AT. Like Helitrons, the maize element contains 5′ TC and 3′ CTRR termini as well as two short palindromic sequences near the 3′ terminus that potentially can form a 20-bp hairpin. Although the maize element lacks sequence information for a DNA helicase, it does contain four exons with similarity to a plant DEAD box RNA helicase. A second Helitron insertion was found in the maize genomic database. These data strongly suggest an active Helitron in the present-day maize genome.
INTRODUCTION
Transposable elements are entities that move within the genome (McClintock, 1949, 1965). Phenotypically, they are recognized when they integrate into functional genes. Mutations associated with such insertions often are unstable, and the excision of the element can restore gene function. Historically, transposable elements were classified into two groups based on their structure and mechanistic mode of propagation. One group, represented by the classic maize transposable elements, have terminal inverted repeats, duplicate host sequences upon integration, and transpose via DNA (Engels, 1983; Fedoroff, 1989). These elements usually leave characteristic footprints after excision (Doring and Starlinger, 1986; Nevers et al., 1986; Jin and Bennetzen, 1989; Singer et al., 1993; Giroux et al., 1996; Kunze et al., 1997). The second group, the retroelements, move through an RNA copy, are related to retroviruses, and include long-terminal repeat and non-long-terminal repeat retrotransposons (Wessler et al., 1995). Long-terminal repeat retrotransposons possess long terminal repeats and encode all of the proteins required for transposition (Boeke and Corces, 1989; Grandbastien, 1992). Despite their diversity, these two classes of transposable elements share two common features. Both make duplications of host sequences upon entry, and the 3′ end of the insertion is either a duplicate of the 5′ terminus or a copy of a poly(A) tail.
Recently, a fundamentally different class of transposable elements, termed Helitrons, was proposed in eukaryotes (Feschotte and Wessler, 2001; Kapitonov and Jurka, 2001). These sequences were identified by computer analysis of the Arabidopsis, rice, and Caenorhabditis elegans genomes. Rather than by cut and paste, these DNA elements are proposed to move through replication and strand replacement. This mode of propagation, called rolling circle transposition, has been reported previously in some insertional sequence groups of transposable elements in prokaryotes (Tavakoli et al., 2000), bacterial plasmids (Khan, 2000), and the circular single-stranded DNA of plant geminiviruses (Rigden et al., 1996).
Helitrons are quite large, ranging from ∼5 to ∼15 kb, lack terminal repeats, do not duplicate host insertion sites, insert precisely between the nucleotides A and T, and constitute ∼2% of the Arabidopsis and C. elegans genomes. In addition, the majority of these elements are nonautonomous, differing from autonomous counterparts by large internal deletions. The only invariant features of Helitrons are the 5′ TC and 3′ CTRR termini as well as palindromes of variable sequences capable of forming a 16- to 20-bp hairpin loop structure 10 to 12 bp upstream of their 3′ termini.
Like other transposable elements, Helitrons may be powerful tools for gene and genome expansion. They frequently capture cellular genes and multiply them during their journey through the genome. Furthermore, it is proposed that the transduced cellular genes that do not contribute to the replication of the Helitrons are destroyed by multiple mutations, whereas genes that provide selective advantage to the transposon are retained. These would exhibit obvious sequence similarity to their genes of origin (Kapitonov and Jurka, 2001).
Many important questions remain to be answered concerning Helitron elements. How widespread are Helitrons, and do they all use the same cellular machinery for transposition? Most importantly, direct proof that these sequences are transposable elements has yet to be provided. Helitrons reported to date have been identified via computer-assisted reconstruction and are thought to have been “active in recent evolutionary history” (Kapitonov and Jurka, 2001). Because the time of transposition of the recently described elements is not known, if they are transposable, it is possible that Helitrons no longer move in present-day genomes. If this is the case, genes that were important for transposition may not be functional or even recognizable in the few genomes shown to harbor Helitrons.
Here, we report a maize mutation that likely occurred in very recent times that has virtually all of the hallmarks of a Helitron. This mutation, sh2-7527, arose spontaneously in a conventional maize breeding program within the shrunken2 (Sh2) gene of maize. The locus encodes the large subunit of ADP-Glc pyrophosphorylase, a key regulatory enzyme in the starch biosynthetic pathway (Giroux et al., 1996; Smidansky et al., 2002). Like Helitrons, this insertion is large, has no sequence or structural similarity to previously described cut-and-paste or copy-and-paste transposable elements, lacks terminal repeats, did not duplicate host sequences upon insertion, was inserted within a host dinucleotide AT, is flanked by a TC 5′ terminus and a CTRR 3′ terminus, and contains two short palindromic sequences near the 3′ terminus that potentially could form a 20-bp hairpin. Although the maize element lacks sequence information for a DNA helicase, it does contain four exons derived from a plant DEAD box RNA helicase (Aubourg et al., 1999). In addition, the element contains two other intron-bearing genes: one shares strong sequence similarity to a rice gene, and the other is >99% identical to a maize EST of unknown function. Finally, this element, like certain other mutations within the sh2 locus, functions from a distance to alter the splicing of the Sh2 transcript.
RESULTS
Transcripts of sh2-7527 Differ by Alternative Splicing within Sh2 and Contain Non-Sh2 Sequences
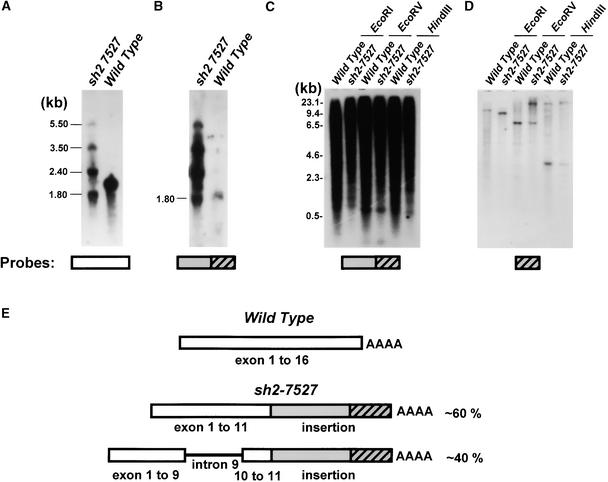
Previous RNA gel blot analysis demonstrated that the mutant sh2-7527 produced at least two abundant transcripts: one larger and one smaller than the wild-type transcript (Giroux and Hannah, 1994). DNA gel blot analysis of the mutant genomic DNA suggested an insertion or rearrangement at the sh2 locus (data not shown). To further characterize sh2-7527, total RNA from mutant and wild-type developing endosperms was electrophoresed, blotted, and probed with different segments of the Sh2 cDNA. The full-length Sh2 cDNA hybridized to four transcripts of ∼1.8, 2.4, 3.5, and 5.5 kb in the mutant and to a single 2.2-kb transcript in the wild type (Figure 1A). A Sh2 cDNA probe containing exons 12 to 16 did not hybridize to any mutant transcripts (data not shown).
Figure 1.
Insertion of sh2-7527.
(A) RNA gel blot analysis of wild-type and sh2-7527 RNA. Total endosperm RNA, 20 days after pollination, was subjected to electrophoresis, blotted, and probed with a full-length Sh2 cDNA probe. MW, molecular mass.
(B) The blot from (A) was stripped and probed with the PCR-derived, 1.2-kb, non-Sh2 portion of the sh2-7527 cDNA.
(C) Genomic DNA from wild-type and sh2-7527 plants was digested with the indicated enzymes, electrophoresed, blotted, and hybridized with the PCR-derived, 1.2-kb, non-Sh2 portion of the sh2-7527 cDNA.
(D) The blot from (C) was stripped and probed with a 3′ portion of the insertion of sh2-7527.
(E) Scheme of wild-type and sh2-7527 cDNAs. The PCR-amplified sh2-7527 regions used to generate the probes are shown.
To elucidate the nature of the sh2-7527 mutation and resulting transcripts, cDNA and genomic libraries were constructed from the mutant and screened with a full-length Sh2 cDNA. Sequence analysis of 28 resulting cDNA clones identified two classes of Sh2 transcripts (Figure 1E). The two classes, consisting of 12 and 16 members, differed only by the presence or absence of Sh2 intron 9. In addition, an extraneous 1.2-kb sequence, not of Sh2 origin, was positioned exactly at the 3′ terminus of exon 11 in all 28 clones. This sequence then terminated with a poly(A) tail. Based on the wild-type Sh2 sequence and the sh2-7527 foreign cDNA insertion, the calculated lengths of these clones (2372 and 2440 bp) likely correspond to that of the 2.4-kb mutant transcript doublet shown in Figure 1A. When the 3′ non-Sh2 sequence was used as a probe on RNA gel blots, a single transcript of low abundance was detected in wild-type RNA (Figure 1B).
Some but not all of the non-Sh2 sequences isolated from the mutant cDNAs are highly repetitive in the maize genome, as judged by genomic DNA gel blot analysis (Figure 1C). When the 3′ segment of the insertion isolated from a genomic clone was used as a probe, only one or two hybridizing fragments were detected in both mutant and wild-type genomic DNA (Figure 1D).
The Extraneous Sequence Is Spliced into the sh2-7527 Transcript from an Insertion Located within Intron 11 of Sh2
Identification of the extraneous sequence immediately at the terminus of exon 11 in the mature transcript raised the possibility that the insertion might lie in a distal portion of Sh2 or distal to Sh2 and that RNA splicing conjoins the foreign sequence to exon 11. To explore this possibility, the sh2-7527 genomic λ library was screened with a full-length Sh2 cDNA clone. Although clones containing the two Sh2-insertion borders were isolated, we were unable to isolate single or overlapping clones that contained the entire insertion.
Sequencing of 6.87 kb from the 5′ end and 3.88 kb from the 3′ terminus of the insertion identified the placement of the insertion within Sh2. The insertion lies within intron 11; hence, the covalent attachment of the inserted sequences to Sh2 exon 11 of the mutant transcript comes about by pre-mRNA splicing. The insertion lies within the dinucleotide AT 139 bp from the 5′ terminus of Sh2 intron 11, starts with 5′ TC, ends with 3′ CTAG, and contains two short palindromic sequences near its 3′ terminus. These features give it the potential of forming a 20-bp hairpin (Figure 2).
Figure 2.
Borders of the sh2-7527 Insertion.
The sequences of the insertion and the flanking Sh2 intron 11 are shown in red and black letters, respectively. Conserved sequences at the termini of the element are underlined. Palindromic sequences capable of forming a hairpin are shown in blue letters.
The Removal of Introns from Mosaic Sh2-Insertion Pre-mRNA Follows Conventional Rules of RNA Splicing
Comparison of the insertion's genomic sequence with the processed Sh2 mosaic transcript identified 12 exons. Splicing follows conventional rules for nuclear genes (Simpson and Filipowicz, 1996). With one exception, all introns begin with GT and end with AG. The last intron begins with GC. A GC donor site occurs in 0.5% of Arabidopsis introns (Simpson et al., 1993; Korning et al., 1996).
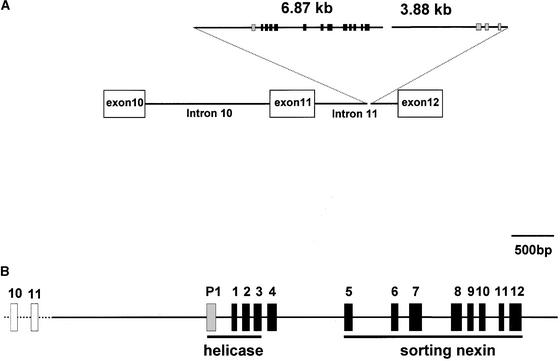
Some introns of the insertion are large, and the 1.2-kb mature sequence in the chimeric Sh2 transcript is spread over a 6.5-kb genomic fragment (Figure 3A). The first intron, separating exon 11 of Sh2 and the first exon of the insertion, is 2388 bp and contains an ∼1.0-kb region that is 67% GC rich.
Figure 3.
Scheme of sh2-7527.
(A) The location of the sh2-7527 insertion within intron 11 of Sh2 is shown as a triangle. Exons are boxed. Exons of the ORF identified in the 3′ portion are listed from right to left.
(B) Expanded version of the 5′ portion. Exons of Sh2 and the sh2-7527 insertion are boxed. Sh2-7527 insertion sequences are shown in bold. Exon P1 refers to a sequence with strong similarity to RNA helicase that is not found in the mature chimeric transcript. Exons with similarity to helicase and the sorting nexin are underlined.
The Insertion in sh2-7527 Alters Sh2 Splicing from a Distance
The complexity of the pre-mRNA splicing process is made evident by the splicing pattern of sh2-7527. Although the insertion lies in intron 11 and sequences from exons 7 to 11 are wild type, intron 9 is excised from only approximately half of the mature transcripts, even though the closer intron 10 is removed faithfully in these transcripts. Whether other mutant transcripts noted on RNA gel blots represent additional distortions in splicing is unknown, because clones that correspond to them were not isolated. Although not common, mutations that affect the splicing of distant introns have been described in vertebrates and plants (McNellis et al., 1994; Marillonnet and Wessler, 1997; Lal et al., 1999b).
The parameters underlying this complexity are unknown. Possibly, the insertion-induced change in the environment surrounding intron 9 is causal, as is the case with the splicing of the transposable element Ds (Lal and Hannah, 1999). The relatively small size (68 bp) of intron 9 and its poor fit to a consensus donor site may render its splicing more vulnerable to local sequence context (Goodall and Filipowicz, 1990).
The sh2-7527 Insertion Contains Putative Pseudogenes
The non-Sh2 portion of the mutant transcript contains two long open reading frames (ORFs). The first ORF contains 131 codons and covers exons 1 through 3 (Figure 3B). The putative translation product shows significant partial similarity to RNA helicases. The second ORF encompasses 179 codons and covers exons 5 through 12. The derived amino acid sequence shows significant partial similarity to the Arabidopsis sorting nexin putative protein (At5g06140).
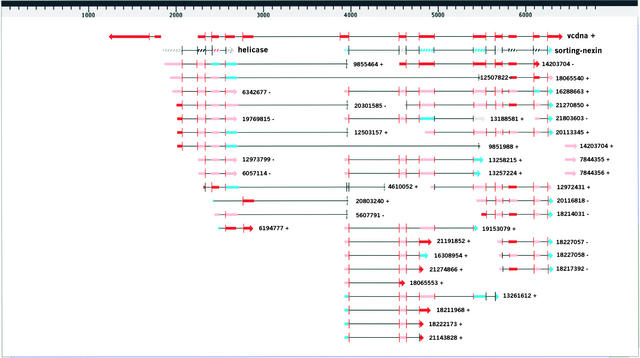
The concatenation of apparently unrelated ORFs and the lack of similarity over the entire lengths of the corresponding proteins suggest that the sh2-7527 insertion encodes either highly diverged proteins or pseudogenes. To investigate these possibilities more closely, we determined significantly matching ESTs by spliced alignment using the GeneSeqer World Wide Web service (http://bioinformatics.iastate.edu/cgi-bin/gs.cgi) (Usuka et al., 2000; Brendel and Zhu, 2002). The results are shown schematically in Figure 4; the detailed alignments are displayed at http://gremlin1.zool.iastate.edu/~volker/v5html/. None of the ESTs match strongly enough to determine the sh2-7527 insertion as their cognate locus. However, 48 ESTs displayed high-quality matches that are largely consistent with the observed exon-intron structure in the two parts containing the long ORFs. One maize EST (GenBank identification number gi:14203704) matched with >90% identity spanning 578 bases from exons 6 to 11. This EST presumably corresponds to a native maize homolog of the Arabidopsis sorting nexin protein. It is plausible that transcripts from that gene gave rise to the single band detected with the non-Sh2 probe in wild-type RNA (Figure 1B).
Figure 4.
Spliced Alignment Results for the 5′ End of the sh2-7527 Insertion.
The figure was produced with the MyGV viewer (Zhu and Brendel, 2002) for GeneSeqer (Usuka and Brendel, 2000; Usuka et al., 2000). Predicted exons are represented by boxes, with colors indicating various similarity scores (red, >0.9; pink, >0.8; cyan, >0.7; and gray, >0.6). vcdna+ indicates the sequenced transcript (cf. Figure 3B), helicase indicates Arabidopsis RNA helicase At1g00660, and sorting-nexin indicates Arabidopsis protein At5g06140. The other gene structures are spliced alignments with ESTs identified by their GenBank gi numbers followed by + or − to indicate the aligned strand. Vertical bars delineate introns and are of proportional length to their predicted splice site strengths. Spliced alignment to predict gene structure using the same tools is reviewed by Brendel and Zhu (2002).
The EST spliced alignment revealed an additional potential exon 5′ to exon 1 in the insertion (Figure 3B). Spliced alignment with the Arabidopsis RNA helicase encoded by At4g00660 using the SplicePredictor World Wide Web service (http://bioinformatics.iastate.edu/cgi-bin/sp.cgi) (Usuka and Brendel, 2000) suggests that this exon is part of a predicted pseudogene with similarity to a maize RNA helicase. The alignment is shown in Figure 5. Although the similarity at the amino acid level is highly significant, multiple nonsense mutations and frameshift-inducing small insertions clearly identify this segment as a remnant of an RNA helicase–encoding gene that no longer encodes a full-length protein. Alignment of the sorting nexin protein to the distal exons displays a similar pattern of nonsense mutations and insertions/deletions (data not shown).
Figure 5.
Alignment of the sh2-7527 Genomic Insertion Sequence with the Arabidopsis DEAD Box RNA Helicase (Aubourg et al., 1999).
The alignment was produced by the program GeneSeqer (Usuka and Brendel, 2000). In the alignment, codons and amino acids representing conserved substitutions are marked +, and neutral substitutions are marked with dots between the amino acids. Predicted introns are not translated.
In contrast to vertebrates and yeast, the U content of plant introns is much higher than that of their flanking exons (Brendel et al., 1998). This difference has been implicated in defining splice sites (Goodall and Filipowicz, 1989; Simpson and Brown, 1993; Luehrsen and Walbot, 1994; Brown and Simpson, 1998). As noted, the first exon of the RNA helicase gene predicted by EST and protein evidence is retained in the mutant intron and is not spliced from the primary transcript. We postulate that the change in the strategic location of this exon in the context of the Sh2 sequence interferes with its recognition, leading to exon skipping. Exon skipping was demonstrated recently in two mutants of maize and several mutants of Arabidopsis (Brown, 1996; Brown and Simpson, 1998; Simpson et al., 1998; Lal et al., 1999a, 1999b).
The 3′ sequenced part of the sh2-7527 insertion has similarity to several ESTs (http://gremlin1.zool.iastate.edu/~volker/v3html/). A three-exon structure close to the 3′ end (Figure 3A) shows significant similarity to the Arabidopsis protein At1g04010 of unknown function.
The Foreign Sequence within Sh2 Is Attributable to an Insertion and Not to a Chromosomal Rearrangement
Two fundamentally different types of rearrangements—interchromosomal and intrachromosomal—were considered as causal for the sh2-7527 mutation. To determine whether the sh2-7527 mutation was caused by an interchromosomal breakage and reunion, we crossed sh2-7527 to the wild type and then crossed the resulting progeny as female to the mutant sh2-R. Translocation heterozygotes usually are associated with 50% female sterility (Brink and Burnham, 1929); hence, semisterile ears were expected from the second cross if sh2-7527 was caused by a translocation. No evidence of semisterility was found. In rare cases, duplication-deficient gametes arising from translocation heterozygotes are viable and semisterility is not observed. These gametes lack one chromosomal arm but contain two copies of another chromosomal segment. If this is the case, at least some of the aneuploid and viable megaspores would transmit only one of the two translocated chromosomes containing Sh2. That portion of sh2-7527 contained on the deficient chromosome would be missing in the resulting progeny. If duplication-deficient gametes produced 50% of the mutant seed in the cross described above, then the probability of not finding a plant lacking, by chance alone, one of the two Sh2 borders in a population of 16 plants arising from mutant seeds is (1–0.5)16 or 1.5 × 10−5.
PCR primers for each of the two Sh2–foreign insertion borders were used to analyze genomic DNA from each of 16 plants derived from mutant seeds of the cross described above. Both borders were found in all plants. We conclude that viable duplication-deficient gametes do not occur and that the sh2-7527 mutant is not the result of an interchromosomal rearrangement.
Next, we determined whether an intrachromosomal rearrangement gave rise to sh2-7527. Intrachromosomal exchanges could have occurred in either the direct or the inverted orientation. In the first case, the centromere distal portion of Sh2 would be placed on an acentric chromosomal ring, whereas the centromere proximal portion of Sh2 would remain on chromosome 3. Chromosomal rings are mitotically unstable and are maintained only by selection (McClintock, 1938). We believe that this possibility is quite unlikely, because we had propagated this mutation through at least seven generations of heterozygosity with a wild-type chromosome before analysis. Any selection would have been relaxed in heterozygotes.
The hypothesis that an intrachromosomal rearrangement involving a nonhomologous exchange in the inverted orientation gave rise to sh2-7527 also was tested. In this case, all of chromosome 3 remains attached to the centromere. The sequence between the two breakpoints would be inverted, and the distal portion of Sh2 would not be lost. In this scenario, the two sequences abutting Sh2 would lie adjacent to each other before the rearrangement event occurred. As a direct test, primers complementary to the two borders were used in PCR experiments with wild-type DNA. If an inversion were causal, a PCR product of 532 bp would be predicted. An amplified product was not produced. We conclude that the foreign sequence within Sh2 is attributable to an insertion and not to a chromosomal rearrangement.
Finally, we note that the wild-type Sh2 sequences, albeit divided, remained unaltered in this mutant. Chromosomal rearrangements involving the rejoining of nonhomologous breaks in plant DNA normally are associated with the loss of sequences at the breakpoints or with the addition of filler DNA (Gorbunova and Levy, 1997). Junctions with no sequence alterations, as found in sh2-7527, are rare for bona fide chromosomal rearrangements. All of the accumulated data strongly suggest an insertion as the genesis of sh2-7527.
The sh2-7527 Insertion Is Not a Repetitive Sequence in Closely Related Sorghum
In previous work, it was noted that at least some of the highly repeated sequences of maize hybridize to only a few fragments in sorghum DNA (Hulbert et al., 1990). These observations suggest that the repetitive sequences of maize may have evolved at a higher rate than low-copy-number genomic sequences or that they have a different evolutionary origin. To detect possible orthologs of the sh2-7527 insertion in other plant species and to determine whether this insertion follows the same pattern of evolution as other repetitive maize sequences, digested and blotted genomic DNAs of maize, sorghum, and Arabidopsis were probed with this insertion. As expected, the highly repetitive nature of this insertion in maize was made evident by the smearing pattern of the maize DNA (Figure 6). By contrast, only two sorghum fragments hybridized to the insertion, whereas no hybridization was seen with Arabidopsis DNA. Hence, this insertion may not be found in all plants and, like the other repetitive maize sequences described, is not found in high copy numbers in closely related sorghum.
Figure 6.
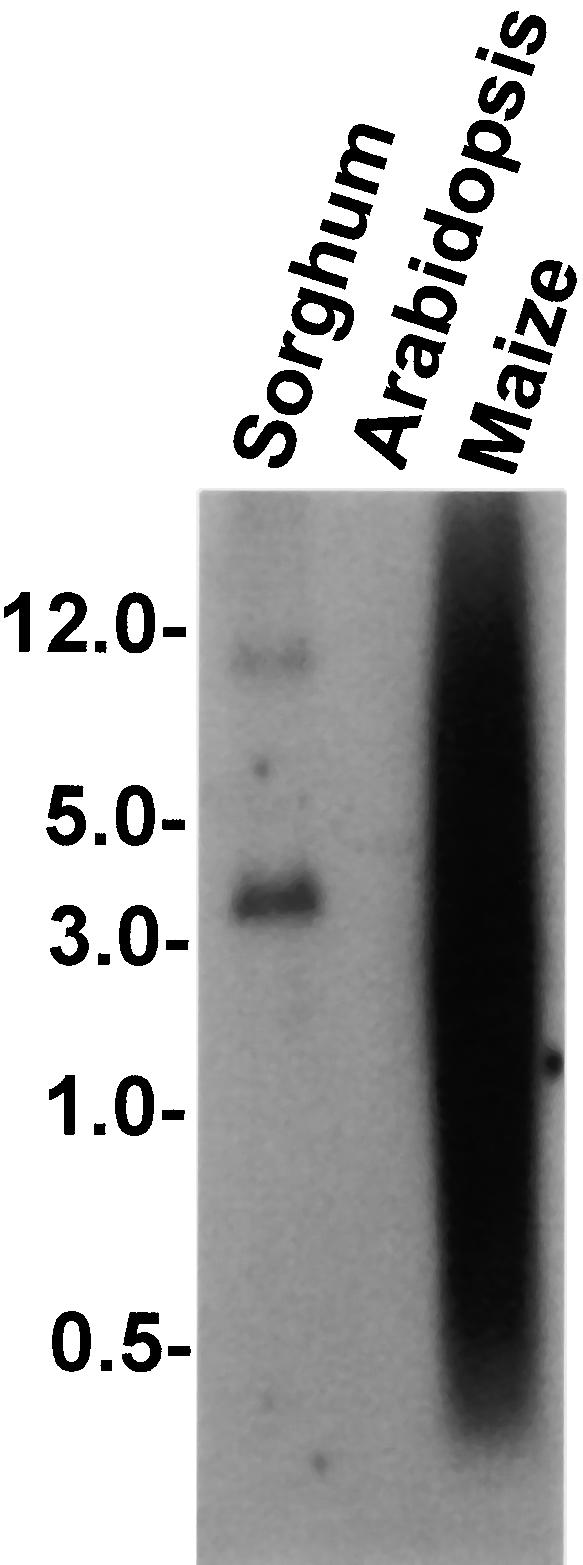
Presence of the sh2-7527 Insertion in Maize, Sorghum, and Arabidopsis.
Genomic DNA from leaves was digested with EcoRI, electrophoresed, blotted, and hybridized with cDNA derived from the sh2-7527 insertion. Fragment sizes (kb) are shown at left.
DISCUSSION
The Maize Mutation sh2-7527 Most Likely Was Caused by a Helitron
Recently, a novel family of eukaryotic transposable elements, termed Helitrons, was discovered by computer-assisted genome analysis (Kapitonov and Jurka, 2001). Helitrons presumably transpose through a mechanism that is fundamentally different from those of other eukaryotic transposable elements and involves catalysis by a DNA helicase. Because the structures of these recently identified elements are based on inactive copies, it is unknown whether an active autonomous element exists in a modern genome. If Helitrons are active in present-day genomes, then one would expect to observe movement of a Helitron into sequences known to lack the element. Analysis of the maize mutant sh2-7527 strongly suggests that Helitrons are present and active in the modern maize genome. Like Helitrons, the insert of sh2-7527 is large, lacks terminal repeats, does not duplicate host sequences, was inserted within a host AT dinucleotide, contains 5′ TC and 3′ CTRR termini, and contains two palindromic sequences at its 3′ terminus. We suggest that Helitrons are functional in the modern maize genome.
A search of maize sequences identified the presence of another Helitron element. This element resides in BAC clone gi:(gi|13606087|gb| AF090447.2| AF090447) and is 17,749 bp in length, starting at position 4408 and ending at position 22,158. It bears sequence similarity to the sh2-7527 element only in the last 20 bp of each terminus. Twelve pseudogenes are located randomly throughout its length. Five of the 12 genes are composed of more than one exon. Based on a comparison of the insertion site with maize ESTs, this Helitron insertion lies in an active gene within a host AT dinucleotide.
The Helitron insertion of sh2-7527 contains many features typical of transposable elements. It is highly repetitive in its host genome but has only single/low copy numbers in the close relative sorghum. Second, it apparently collects host sequences, as was noted with the second maize Helitron element described above. Sequence analysis of the 5′ and 3′ regions of the sh2-7527 insertion revealed similarity to three distinct intron-bearing maize genes. The capture and movement of cellular genes by retroelements are well documented in vertebrates and to some extent in plants. For instance, ∼1% of the human genome apparently has been transduced by L1 retroposon (Pickeral et al., 2000). The movement of cellular plant genes by transposable elements also has been reported in maize. Talbert and Chandler (1988) first noted that a sequence found within a Mu element termed MRS-A has striking sequence similarity to an expressed gene found in maize and its relatives. A version of the maize retroposon Bs1 contains sequences similar to a portion of the spliced transcript of a plasma membrane H+-ATPase (Bureau et al., 1994; Jin and Bennetzen, 1994; Palmgren, 1994). The recent discovery of two additional genes transduced by Bs1 suggests that capture of multiple genes by retroelements may facilitate gene evolution by the formation of new hybrid genes (Elrouby and Bureau, 2001). Similarly, the origin of ORF23 of the maize retroposon Grande1 has been postulated to be caused by a transduction event (Martinez-Izquierdo et al., 1997). Recently, Takahashi et al. (1999) reported the capture of a genomic sequence homologous with sequences that encode a domain of the high-mobility-group DNA binding proteins by the element Tpn1 of Japanese morning glory. Tpn1 bears sequence similarity to the En/Spm maize element. Similarly, capture of a portion of genomic sequence similar in sequence to the homeobox gene Athb-1 by a MULE-1 element has been reported in Arabidopsis (Le et al., 2000).
The presence of sequences similar to part of an RNA helicase rather than a DNA helicase gene in this maize element raises a number of interesting possibilities. At present, we do not know whether it is a remnant of a cellular gene captured by the element or whether the complete RNA helicase gene in an autonomous Helitron element plays a role in transposition. RNA helicase activity has been invoked in many functions, including RNA unwinding, RNA splicing, RNA editing, translation initiation, and rRNA processing (Luking et al., 1998). Intriguingly, some RNA helicases also recognize DNA/RNA hybrids and even DNA/DNA duplexes (Linder et al., 1989). If the RNA helicase captured in sh2-7527 can recognize DNA/DNA hybrids, then it could function like the DNA helicase, as proposed by Kapitonov and Jurka (2001) for Helitrons. Targeting to DNA/RNA hybrids via recognition by the RNA helicase raises the intriguing possibility that transposition could be directed to genes undergoing transcription. We also note that a disruption of an RNA helicase in C. elegans can lead to both the activation of retroelements and the inhibition of RNA interference (Ketting et al., 1999). This led to the proposal that RNA interference serves as a “genome immune system” against transposable elements (Plasterk, 2002). Thus, an RNA helicase–encoding transposon, when activated, would be equipped to suppress the host genome defense mechanism and hence to facilitate transposition.
As noted, the sh2-7527 Helitron element contains sequences similar to coding sequences of a sorting nexin. This family of proteins is involved in intracellular vesicle protein trafficking (Haft et al., 1998) and is expressed widely in many tissues, at least in animals. Although plant genes bearing sequence similarity to human sorting nexins can be found in plant genome databases, specific roles for these plant proteins have not been elucidated. At present, we do not know whether the high level of sequence similarity of the sh2-7527 element to sorting nexins reflects a role of this protein in Helitron function or whether it simply represents a recent incorporation of this gene into this particular Helitron element.
We also note that some of the structural features of the sh2-7527 insertion resemble those of the Y′ family of repeated DNA elements in yeast. Like the sh2-7527 insertion, these elements lack direct repeats, and unlike known class 1 and 2 transposable elements, they appear to move via recombination. Of most significance, a Y′ ORF encodes an RNA helicase that presumably is required for its maintenance (Louis and Haber, 1992).
The Capture of Genes by Transposable Elements Likely Plays an Important Role in Evolution
Transposable elements likely play an important role in the organization and evolution of plant genomes (Lonnig and Saedler, 1997; White and Doebley, 1998). The transduced genes can be moved from their original positions and inserted into different regions of the genome. This effect has two noteworthy consequences. First, it can result in gene duplication without duplication of the neighboring chromosomal segment. This may contribute to the recently noted lack of micro-colinearity in closely related genomes (Tikhonov et al., 1999) and even within a species, as shown recently in maize (Fu and Dooner, 2002). A second significant outcome is exon shuffling. As reported here, exons of Sh2 and the two different genes within the sh2-7527 insertion were fused to produce a novel transcript that may evolve to encode a novel protein.
METHODS
Plant Material
Maize (Zea mays) plants were grown in the field or in the greenhouse at the University of Florida. The mutant sh2-7527, kindly provided by the late Oliver Nelson (University of Wisconsin, Madison), was described previously (Giroux and Hannah, 1994). The wild-type Sh2 allele used in this study was isolated originally from McClintock's a1-m3 stock (Giroux et al., 1996).
Construction and Screening of Genomic and cDNA Libraries
Genomic DNA isolated from sh2-7527 leaves was partially digested with Sau3A1 and size-fractionated by centrifugation at 25,000g at 4°C for 24 h through a 10 to 40% linear Suc gradient in sterile TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8.0). Selected DNA fractions were ligated into the cloning vector λDASH II and packaged according to the manufacturer (Stratagene). This library was screened with a full-length Sh2 cDNA probe. Eight clones were identified after screening of ∼1,000,000 plaques. These were purified, blotted, and probed with PCR-amplified 5′ (exons 1 to 10) and 3′ (exons 12 to 16) ends of Sh2 cDNA. Three plaques hybridized only to the 5′ end, and five plaques hybridized only to the 3′ end of Sh2 cDNA. DNA from these clones was restriction mapped, and the longest inserts were sequenced directly with synthesized primers. The first sequencing reaction was performed using primers SH211F (5′-TACACAAGGGTT-GGTCGTTCT-3′) and SH212R (5′-GGGTGCAGTGAAGAAAGGTG-3′). These primers are complementary to Sh2 exon 11 and 12 sequences, respectively. The inserts were fully sequenced in both directions. The PCR-amplified terminal regions of the insert were used as a probe to screen additional λ clones for overlapping 5′ and 3′ ends of the insert.
A cDNA library was constructed from RNA extracted from sh2-7527 endosperms at 20 days after pollination using a cDNA synthesis kit (λZAP; Stratagene) according to the instructions of the manufacturer. Screening of ∼20,000 plaques from this library identified 28 positive clones. These were purified and excised in vivo to produce pBSKS using the Exassist/SOLR system (Stratagene). Sequencing was performed at the University of Florida Interdisciplinary Center for Biotechnology Research DNA Sequencing Core Laboratory using the ABI Prism Dye Terminating system developed by Applied Biosystems (Foster City, CA).
RNA Isolation and RNA Gel Blot Analysis
Total RNA from maize kernels at 20 days after pollination was isolated as described previously (McCarty, 1986). A total of 10 μg of sh2-7527 and wild-type RNA was subjected to electrophoresis on a 1.3% agarose gel and blotted onto a Hybond N+ membrane (Amersham) according to the protocol of the manufacturer. Hybridization was performed according to Church and Gilbert (1984) in a buffer containing 0.5 M Na2HPO4, pH 7.2, 7% SDS, and 1% BSA. Blots were first probed with 32P-dCTP–labeled full-length Sh2 cDNA and then reprobed with the sh2-7527 cDNA insertion after two washes with 0.1 × SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 1% SDS for 15 min at 90°C and overnight exposure on film to ensure first probe removal.
Genomic DNA Isolation and DNA Gel Blot Analysis
Genomic DNA from mutant and wild-type leaves was digested with restriction endonucleases (Life Technologies, Rockville, MD) in 100 μL containing 4% spermidine (Dellaporta et al., 1983). After electrophoresis on 0.7% agarose gels and blotting, the digested DNA was first probed with full-length non-Sh2 cDNA from sh2-7527. The probe was synthesized via PCR using primers ALUp1 (5′-TACACA-GGGTTGGTCGTTCT-3′) and ALLo1 (5′-GTGTCGTGATCATCCTTA-GC-3′). These primers span the entirety of the non-Sh2 cDNA. The blot was stripped and reprobed with the 3′ region of this cDNA. This probe was synthesized using PCR primers ALGSUp1 (5′-GCAACGGAAACATATCCTAC-3′) and ALGSLo1 (5′-GGTCCGTGCGTCTCC-AG-3′) and a genomic sh2-7527 clone as a template.
PCR Amplification of the sh2-7527 Sequence
A total of 1 μg of sh2-7527 genomic DNA was subjected to PCR amplification using primers Sh27F (5′-GGGCTAGTGAAGATTGATCA-3′) and Sh211R (5′-TACACAGGGTTGGTCGTTCT-3′), which are complementary to Sh2 exons 7 and 11, respectively. The resulting 651-bp product was cloned into pBluescript KS+ (Stratagene) and sequenced in both directions. Similarly, the 5′ and 3′ ends of the non-Sh2 inserted DNA were amplified using primers Sh2E11F (5′-ACGGGCTATTGGGAGGATGT-3′) and 75275′R (5′-CATGCCTGCTACAGA-GAAAG-3′), which are complementary to Sh2 exon 11 and the 5′ insertion sequence, respectively, and primers Sh2E12R (5′-GGGTGC-AGTGAAGAAAGGTG-3′) and 75273′F (5′-CTGCAGTCACAGAAGGAAAC-3′), which are complementary to Sh2 exon 12 and the 3′ insertion sequence, respectively. DNA sequences derived from the mutant were compared with the Sh2 sequence (Shaw and Hannah, 1992) using DNA analysis software (Lasergene; DNASTAR, Madison, WI).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The accession numbers for the sequences mentioned in this article are M81603 (Z. mays ADP-glucose pyrophosphorylase [shrunken-2] gene; wild-type Sh2), AF162682 (Z. mays mutant sh2-7527 alien mRNA sequence; sh2-7527 foreign cDNA insertion), AF293457 (Z. mays truncated insertion sequence mutant shrunken-2 [sh2]; 6.87 kb from the 5′ terminus), AF293458 (Z. mays truncated insertion sequence mutant shrunken-2 [sh2]; 3.88 kb from the 3′ terminus), and g3775993 (Arabidopsis DEAD box RNA helicase).
Acknowledgments
We thank Tom Peterson, Don McCarty, and Rob Ferl for helpful comments. We thank Maureen Clancy for assistance with manuscript preparation and Gabrielle Stryker for her help with the figures. This research was supported by National Science Foundation Grants IBN-9316887 and MCB-9420422 and U.S. Department of Agriculture Competitive Grants 94-37300-453, 97-36306-4461, 95-37301-2080, and 98-01006. This is Florida Agricultural Experiment Station Journal Series No. R-07881.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.008375.
References
- Aubourg, S., Kreis, M., and Lecharny, A. (1999). The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res. 27, 628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J., and Corces, V. (1989). Transcription and reverse transcription of retrotransposons. Annu. Rev. Microbiol. 43, 403–434. [DOI] [PubMed] [Google Scholar]
- Brendel, V., Carle-Urioste, J.C., and Walbot, V. (1998). Intron recognition in plants. In A Look beyond Transcription: Mechanisms Determining mRNA Stability and Translation in Plants, J. Bailey-Serres and D.R. Gallie, eds (Rockville, MD: American Society of Plant Physiologists), pp. 20–28.
- Brendel, V., and Zhu, W. (2002). Computational modeling of gene structure in Arabidopsis thaliana. Plant Mol. Biol. 48, 49–58. [PubMed] [Google Scholar]
- Brink, R.A., and Burnham, C.R. (1929). Inheritance of semi sterility in maize. Am. Nat. 63, 301–316. [Google Scholar]
- Brown, J.W.S. (1996). Arabidopsis intron mutations and pre-mRNA splicing. Plant J. 10, 771–780. [DOI] [PubMed] [Google Scholar]
- Brown, J.W.S., and Simpson, C.G. (1998). Splice site selection in plant pre-mRNA splicing. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 77–95. [DOI] [PubMed] [Google Scholar]
- Bureau, T.E., White, S.E., and Wessler, S.R. (1994). Transduction of a cellular gene by a plant retroelement. Cell 77, 479–480. [DOI] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation: Version 2. Plant Mol. Biol. Rep. 1, 19–21. [Google Scholar]
- Doring, H.-P., and Starlinger, P. (1986). Molecular genetics of transposable elements in plants. Annu. Rev. Genet. 20, 175–200. [DOI] [PubMed] [Google Scholar]
- Elrouby, N., and Bureau, T.E. (2001). A novel hybrid open reading frame formed by multiple cellular gene transductions by a plant long terminal repeat retroelement. J. Biol. Chem. 276, 41963–41968. [DOI] [PubMed] [Google Scholar]
- Engels, W.R. (1983). The P family of transposable elements in Drosophila. Annu. Rev. Genet. 17, 315–344. [DOI] [PubMed] [Google Scholar]
- Fedoroff, N.V. (1989). About maize transposable elements and development. Cell 56, 181–191. [DOI] [PubMed] [Google Scholar]
- Feschotte, C., and Wessler, S.R. (2001). Treasures in the attic: Rolling circle transposons discovered in eucaryotic genomes. Proc. Natl. Acad. Sci. USA 98, 8923–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., and Dooner, H.K. (2002). Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99, 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux, M.J., and Hannah, L.C. (1994). ADP-glucose pyrophosphorylase in shrunken-2 and brittle-2 mutants of maize. Mol. Gen. Genet. 243, 400–408. [DOI] [PubMed] [Google Scholar]
- Giroux, M.J., Shaw, J., Barry, G., Cobb, G.B., Greene, T., Okita, T., and Hannah, L.C. (1996). A single mutation that increases maize seed weight. Proc. Natl. Acad. Sci. USA 93, 5824–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall, G.J., and Filipowicz, W. (1989). The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58, 473–483. [DOI] [PubMed] [Google Scholar]
- Goodall, G.J., and Filipowicz, W. (1990). The minimum functional length of pre-mRNA introns in monocots and dicots. Plant Mol. Biol. 14, 727–733. [DOI] [PubMed] [Google Scholar]
- Gorbunova, V., and Levy, A. (1997). Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 25, 4650–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien, M.-A. (1992). Retroelement in higher plants. Trends Genet. 8, 103–108. [DOI] [PubMed] [Google Scholar]
- Haft, C.R., Sierra, M.L., Barr, V.A., Haft, D.H., and Taylor, S.I. (1998). Identification of a family of sorting nexin molecules and characterization of their association with receptors. Mol. Cell. Biol. 18, 7278–7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert, S.H., Richter, T.E., Axtell, J.D., and Bennetzen, J.L. (1990). Genetic mapping and characterization of sorghum and related crops by means of maize ADH probes. Proc. Natl. Acad. Sci. USA 87, 4252–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y.-K., and Bennetzen, J.L. (1989). Structure and coding properties of Bs1, a maize retrovirus-like transposable element. Proc. Natl. Acad. Sci. USA 86, 6235–6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y.-K., and Bennetzen, J.L. (1994). Integration and nonrandom mutation of a plasma membrane proton ATPase gene fragment within the Bs1 retroelement of maize. Plant Cell 6, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov, V.V., and Jurka, J. (2001). Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. USA 98, 8714–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting, R.F., Haverkamp, T.H., van Luenen, H.G., and Plasterk, R.H. (1999). Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99, 133–141. [DOI] [PubMed] [Google Scholar]
- Khan, S.A. (2000). Plasmid rolling circle replication: Recent development. Mol. Microbiol. 37, 477–484. [DOI] [PubMed] [Google Scholar]
- Korning, P.G., Hebsgaard, S.M., Rouzé, P., and Brunak, S. (1996). Cleaning the GenBank Arabidopsis thaliana data set. Nucleic Acids Res. 24, 316–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, R., Saedler, H., and Lonnig, W.-E. (1997). Plant transposable elements. Adv. Bot. Res. 27, 331–470. [Google Scholar]
- Lal, S., Choi, J.H., and Hannah, L.C. (1999. a). The AG dinucleotide terminating introns is important but not always required for pre-mRNA splicing in the maize endosperm. Plant Physiol 120, 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal, S., Choi, J.H., Shaw, J., and Hannah, L.C. (1999. b). A splice site mutant of maize activates cryptic splice sites, elicits intron inclusion and exon exclusion, and permits branch point elucidation. Plant Physiol. 121, 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal, S., and Hannah, L.C. (1999). Maize transposable element Ds is differentially spliced in endosperm and suspension cells. Biochem. Biophys. Res. Commun. 261, 798–801. [DOI] [PubMed] [Google Scholar]
- Le, Q.H., Wright, S., Yu, Z., and Bureau, T. (2000). Transposon diversity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder, P., Lasko, P.F., Ashburner, M., Leroy, P., Nielsen, P.J., Nishi, K., Schnier, J., and Slonimski, P.P.P. (1989). Birth of the D-E-A-D box. Nature 340, 246–249. [DOI] [PubMed] [Google Scholar]
- Lonnig, W.-E., and Saedler, H. (1997). Plant transposons: Contributors to evolution? Gene 205, 245–253. [DOI] [PubMed] [Google Scholar]
- Louis, E.J., and Haber, J.E. (1992). The structure and evolution of subtelomeric Y′ repeats in Saccaromyces cerevisiae. Genetics 1331, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen, K.R., and Walbot, V. (1994). Intron creation and polyadenylation in maize are directed by AU-rich RNA. Genes Dev. 8, 1117–1130. [DOI] [PubMed] [Google Scholar]
- Luking, A., Stahl, U., and Schmidt, U. (1998). The protein family of RNA helicases. Crit. Rev. Biochem. Mol. Biol. 33, 259–296. [DOI] [PubMed] [Google Scholar]
- Marillonnet, S., and Wessler, S.R. (1997). Retroposon insertion into the maize waxy gene results in tissue-specific RNA processing. Plant Cell 9, 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Izquierdo, J.A., Garcia-Martinez, J., and Vicient, C.M. (1997). What makes Grande1 retrotransposon different? Genetica 100, 15–28. [PubMed] [Google Scholar]
- McCarty, D.R. (1986). A simple method for extraction of RNA from maize tissue. Maize Genet. Coop. Newsl. 60, 61. [Google Scholar]
- McClintock, B. (1938). The production of homozygous deficient tissues with mutant characteristics by means of the aberrant behavior of ring shaped chromosome. Genetics 23, 315–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1949). Mutable loci in maize. Carnegie Inst. Washington Year Book 48, 142–154. [PubMed] [Google Scholar]
- McClintock, B. (1965). Components of action of the regulators Spm and Ac. Carnegie Inst. Washington Year Book 65, 568–578. [Google Scholar]
- McNellis, T.W., von Arnim, A.G., Akari, T., Komeda, Y., Misera, S., and Deng, X.-W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for multiple protein domains. Plant Cell 6, 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevers, P., Shepherd, N., and Saedler, H. (1986). Plant transposable elements. Adv. Bot. Res. 12, 102–203. [Google Scholar]
- Palmgren, M.G. (1994). Capturing of host DNA by a plant retroelement: Bs1 encodes plasma membrane H+-ATPase domains. Plant Mol. Biol. 25, 137–140. [DOI] [PubMed] [Google Scholar]
- Pickeral, O.K., Makaoski, W., Boguski, M.S., and Boeke, J.D. (2000). Frequent human genomic DNA transduction driven by LINE-1 retroposition. Genome Res. 10, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk, R.H.A. (2002). RNA silencing: The genome's immune system. Science 296, 1263–1265. [DOI] [PubMed] [Google Scholar]
- Rigden, J.E., Dry, I.B., Krake, L.R., and Rezaian, M.A. (1996). Plant virus DNA replication processes in Agrobacterium: Insight into the origin of geminiviruses? Proc. Natl. Acad. Sci. USA 93, 10280–10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, J.R., and Hannah, L.C. (1992). Genomic sequence of the wild type Shrunken-2 allele of Zea mays. Plant Physiol. 98, 1214–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, C.G., and Brown, J.W.S. (1993). Efficient splicing of AU-rich antisense intron sequence. Plant Mol. Biol. 21, 205–211. [DOI] [PubMed] [Google Scholar]
- Simpson, C.G., and Filipowicz, W. (1996). Splicing of precursors to mRNA in higher plants: Mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol. Biol. 32, 1–41. [DOI] [PubMed] [Google Scholar]
- Simpson, C.G., Leader, D.J., and Brown, J.W.S. (1993). Plant intron sequences. In Plant Molecular Biology Labfax, R.R.D. Croy, ed (Oxford, UK: BIOS Scientific Publishers), pp. 183–251.
- Simpson, C.G., McQuade, C., Lyon, J., and Brown, J.W.S. (1998). Characterization of exon skipping mutants of the COP1 gene from Arabidopsis. Plant J. 15, 125–131. [DOI] [PubMed] [Google Scholar]
- Singer, M.F., Krek, V., McMillan, J.P., Swergold, G.D., and Thayer, R.E. (1993). LINE-1: A human transposable element. Gene 135, 183–188. [DOI] [PubMed] [Google Scholar]
- Smidansky, E.D., Clancy, M., Meyer, F.D., Lanning, S.P., Blake, N.K., Talbert, L.E., and Giroux, M.J. (2002). Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc. Natl. Acad. Sci. USA 99, 1724–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, S., Inagaki, Y., Satoh, H., Hoshino, A., and Iida, S. (1999). Capture of a genomic HMG domain sequence by the En/Spm-related transposable element Tpn1 in the Japanese morning glory. Mol. Gen. Genet. 261, 447–451. [DOI] [PubMed] [Google Scholar]
- Talbert, L.E., and Chandler, V.L. (1988). Characterization of a highly conserved sequence related to mutator transposable elements in maize. Mol. Biol. Evol. 5, 519–529. [DOI] [PubMed] [Google Scholar]
- Tavakoli, N., Comanducci, A., Dodd, H.M., Lett, M.C., Albiger, B., and Bennett, P. (2000). IS1294, a DNA element that transposes by RC transposition. Plasmid 44, 66–84. [DOI] [PubMed] [Google Scholar]
- Tikhonov, A., SanMiguel, P., Nakajima, Y., Gorenstein, N., Bennetzen, J., and Avramova, Z. (1999). Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96, 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuka, J., and Brendel, V. (2000). Gene structure prediction by spliced alignment of genomic DNA with protein sequences: Increased accuracy by differential splice site scoring. J. Mol. Biol. 297, 1075–1085. [DOI] [PubMed] [Google Scholar]
- Usuka, J., Zhu, W., and Brendel, V. (2000). Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics 16, 203–211. [DOI] [PubMed] [Google Scholar]
- Wessler, S.R., Bureau, T.E., and White, S.E. (1995). LTR-retrotransposons and MITEs: Important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 5, 814–821. [DOI] [PubMed] [Google Scholar]
- White, S., and Doebley, J. (1998). Of genes and genomics and the origin of maize. Trends Genet. 14, 327–332. [DOI] [PubMed] [Google Scholar]
- Zhu, W., and Brendel, V. (2002). Gene structure identification with MyGV using cDNA evidence and protein homologs to improve ab initio predictions. Bioinformatics 18, 761–762. [DOI] [PubMed] [Google Scholar]