Abstract
Adult mesenchymal stem cells (MSCs) derived from bone marrow contribute to the regeneration of multiple types of mesenchymal tissues. Here we describe the functional role of a novel form of cross-talk between the transforming growth factor β1 (TGF-β1) and Wnt signaling pathways in regulating the activities of human MSCs. We show that TGF-β1 induces rapid nuclear translocation of β-catenin in MSCs in a Smad3-dependent manner. Functionally, this pathway is required for the stimulation of MSC proliferation and the inhibition of MSC osteogenic differetiation by TGF-β1, likely through the regulation of specific downstream target genes. These results provide evidence for a new mode of cooperation between the TGF-β and Wnt signaling pathways in this specific cellular context and suggest a potentially important role for this distinct signaling pathway in the control of self-renewal and differentiation of a specific type of MSCs.
Keywords: Human mesenchymal stem cells, TGF-β1, β-catenin
Multipotent stem cell populations found in adult tissues have been of great interest because they serve as reservoirs for tissue renewal after trauma, disease, and aging. One important type of adult stem cell derived from bone marrow is the mesenchymal stem cell (MSC), which contributes to the regeneration of mesenchymal tissues such as bone, cartilage, muscle, tendon, and adipose (Pittenger et al. 1999). However, lack of knowledge at the molecular level on the regulatory mechanisms underlying the self-renewal and differentiation of MSCs has limited the potential use of MSCs in practical applications such as tissue engineering and gene therapy (Caplan 2000).
Transforming growth factor β (TGF-β) and Wnt superfamily proteins play important roles in the regulation of many developmental processes and maintenance of normal tissue homeostasis (Cadigan and Nusse 1997; Massague 1998). TGF-β initiates signaling by binding and activating the membrane-anchored type II and type I receptor Ser/Thr kinases, which subsequently phosphorylate the effectors Smad2/Smad3. Phosphorylated Smad2/Smad3 can then form complexes with the common Smad, Smad4, and translocate into the nucleus. Accumulation of Smads in the nucleus results in the activation or repression of downstream target genes by recruiting various transcriptional coactivators or corepressors (Derynck et al. 1998; Massague 1998). Canonical Wnt signaling originates from binding of Wnt molecules to its membrane-associated receptor, Frizzled, and coreceptor LRP5/6. In the absence of Wnt ligand, cytoplasmic β-catenin levels are kept low by a continued process of phosphorylation-dependent ubiquitination and degradation. Upon Wnt stimulation, the kinases that phosphorylate and destabilize β-catenin are inhibited. As a result, β-catenin accumulates in the nucleus, where it can associate with transcription factors such as TCF/LEF to activate expression of Wnt-responsive genes (He et al. 2004; Logan and Nusse 2004).
In addition to their distinct functions, TGF-β and Wnt signaling pathways act cooperatively in certain biological contexts. Several studies have shown that cross-talk between TGF-β superfamily members and Wnt/wingless signaling pathways plays important roles in regulation of certain developmental events in Xenopus and Drosophila. This cooperative regulation is mediated by the association between Smads and TCF/LEF in the nucleus, and results in the synergistic activation of specific target genes (Labbe et al. 2000; Nishita et al. 2000). In this report, we demonstrate a novel level of cross-talk between TGF-β and Wnt signaling pathways in MSCs derived from adult human bone marrow, and this cross-talk may play an important role in regulating self-renewal and differentiation programs of those MSCs.
Results
TGF-β1 induces nuclear translocation of β-catenin without affecting the steady-state protein level of β-catenin and independent of canonical Wnt signaling pathway
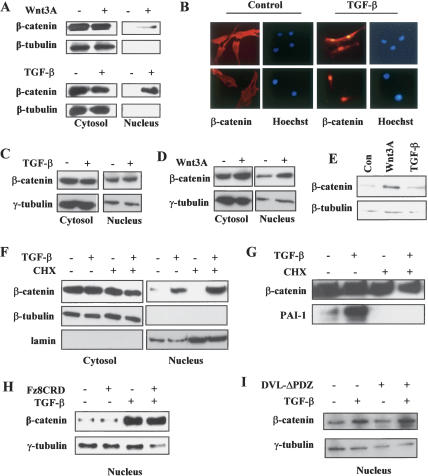
In an attempt to explore the regulatory mechanisms that govern the proliferation and differentiation programs of human MSCs, we investigated the cross-talk between TGF-β and Wnt signaling pathways in this specific cellular context. To do this, we stimulated primary MSCs that were derived from adult human bone marrow with either Wnt3A or TGF-β1. As shown in Figure 1A, a significant amount of β-catenin appeared in the nucleus of MSCs after 2 h incubation with Wnt3A-conditioned medium as determined by nuclear/cytoplasmic fractionation. To our surprise, we found that TGF-β1 was also capable of inducing the nuclear translocation of β-catenin in a manner similar to Wnt3A treatment, since an increasing amount of β-catenin was detected in the nuclear fraction 1–2 h after the cells were treated with TGF-β1 (Fig. 1A). To verify this highly intriguing result, we used immunofluorescence imaging to directly visualize the localization of β-catenin. As shown in Figure 1B, the nuclear staining of endogenous β-catenin was significantly increased in MSCs 1 h after treatment with TGF-β1, confirming the data from the fractionation experiments. When MSCs were plated at a low cell density to maintain their undifferentiated state, strong staining of β-catenin in the nucleus was detected in >90% of the cells following treatment with TGF-β1. Importantly, β-catenin accumulation in the nucleus was rapid in response to TGF-β1 treatment, suggesting that the TGF-β1-induced β-catenin nuclear translocation in MSCs is likely to be mechanistically distinct from that of the slow accumulation of β-catenin in the nucleus in response to TGF-β1 as previously reported in the context of chondrogenesis of MSCs (Tuli et al. 2003; Zhou et al. 2004). To determine whether the ability of TGF-β1 to induce β-catenin nuclear translocation was cell-type specific, we examined β-catenin localization upon TGF-β1 treatment in Madin-Darby canine kidney (MDCK) epithelial cells. Although Wnt3A treatment increased nuclear β-catenin levels in MDCK cells, TGF-β1 treatment did not (Fig. 1C,D). Taken together with the observations that TGF-β1 failed to induce rapid nuclear accumulation of β-catenin in HaCaT human keratinocytes and BJ human fibroblasts (data not shown), these results suggest that β-catenin nuclear translocation in response to TGF-β1 may be associated specifically with certain cellular contexts such as MSCs.
1.
TGF-β1 induces nuclear translocation of β-catenin without affecting the steady-state protein level of β-catenin and independent of canonical Wnt signaling pathway. (A) Cytosolic and nuclear fractions of protein lysates were isolated from human MSCs treated or untreated with TGF-β1 for 2 h or Wnt3A for 6 h. Western blot was probed with an anti-β-catenin monoclonal antibody. (B) β-catenin localization was detected by immunofluorescence with the anti-β-catenin antibody after cells were treated or untreated with TGF-β1 for 1 h. (C) Cytosolic and nuclear fraction of protein lysates were isolated from MDCK cells treated or untreated with TGF-β1 for 2 h. Western blots were probed with anti-β-catenin antibody. (D) Cytosolic and nuclear fraction of protein lysates were isolated from MDCK cells treated or untreated with Wnt3A. Western blots were probed with anti-β-catenin antibody. (E) Total cell lysates were prepared from MSCs after treatment with TGF-β1 or Wnt3A for 24 h. Western blots were probed with anti-β-catenin antibody. (F) MSCs were pretreated with protein translation inhibitor cyclohexmide for 1 h before TGF-β1 treatment for 2 h. Cytosolic and nuclear fractions of β-catenin were detected by anti-β-catenin antibody. (G) MSCs were pretreated with CHX for 1 h before TGF-β1 treatment for 6 h. Protein levels of β-catenin and PAI-1 in whole-cell lysates were determined by Western blot with anti-β-catenin and PAI-1 antibodies. (H) MSCs were pretreated with Fz8CRD containing conditioned medium for 6 h before TGF-β1 treatment for 2 h. Nuclear fractions of β-catenin were detected by anti-β-catenin antibody. (I) The levels of nuclear β-catenin in vector control and DVL-ΔPDZ adenovirus-infected MSCs treated with TGF-β1 for 2 h were determined by Western blot analysis.
Wnt-induced nuclear accumulation of β-catenin has been established in multiple cellular systems as the consequence of β-catenin stabilization (Orford et al. 1997). To determine whether TGF-β1 induces β-catenin nuclear translocation via a similar mechanism, we measured the steady-state protein levels of β-catenin in MSCs in the presence or absence of TGF-β1 proteasome inhibitors. Interestingly, no change in the levels of β-catenin was observed after the MSCs were treated with TGF-β1 for 24 h (Fig. 1E) or three different types of proteasome inhibitors (Supplementary Fig. 1), suggesting that β-catenin nuclear translocation in response to TGF-β1 is not mediated by a significant change in the stability of β-catenin in MSCs. As a control, Wnt3A treatment still induced an increase in β-catenin protein levels in this cellular context (Fig. 1E).
Since the expression of several members of the Wnt family is known to be regulated by TGF-β1 (Zhou et al. 2004), β-catenin nuclear translocation in response to TGF-β1 could be a consequence of TGF-β1-induced Wnt production and action through an autocrine mechanism. To test this possibility, we pretreated MSCs with the protein translation inhibitor cycloheximide (CHX) before the addition of TGF-β1. As shown in Figure 1F, the presence of CHX did not have an effect on the ability of TGF-β1 to induce β-catenin nuclear accumulation, even though the induction of a TGF-β1 target gene plasminogen activator inhibitor-1 (PAI-1) was completely blocked (Fig. 1G), suggesting that this novel activity of TGF-β1 is not mediated by an increase in the production of Wnt proteins. Again, no significant changes in the levels of total β-catenin protein were detected under the same conditions of CHX treatment (Fig. 1G). This result, together with those derived from treatment of proteasome inhibitors (Supplementary Fig. 1), indicates that β-catenin is highly stable in MSCs. To explore whether TGF-β1 induced nuclear translocation of β-catenin required Wnt signaling, we used a competitive inhibitor of the Wnt receptor Frizzled, Fz8CRD (Hsieh et al. 1999). As shown in Figure 1H, addition of Fz8CRD to the culture medium of MSCs had no effect on TGF-β1-stimulated β-catenin nuclear translocation, even though the same treatment blocked the nuclear accumulation of β-catenin induced by Wnt3A (Supplementary Fig. 2). This result suggests that the observed β-catenin nuclear translocation in response to TGF-β1 is unlikely a Wnt ligand-dependent process. To further address this question, we used a dominant-negative form of disheveled (DVL-ΔPDZ), whose ectopic expression was shown to disrupt Wnt signaling (Hino et al. 2003). As a positive control, the adenovirus-mediated expression of DVL-ΔPDZ was able to inhibit the ability of Wnt3A to stabilize β-catenin in C57MG cells (Supplementary Fig. 2). However, DVL-ΔPDZ expression did not block the translocation of β-catenin in response to TGF-β1 (Fig. 1I). These results indicate that TGF-β1-induced β-catenin nuclear translocation does not require the canonical Wnt signaling pathway involving β-catenin dephosphorylation and stabilization.
β-catenin nuclear translocation is mediated by the TGF-β signaling pathway
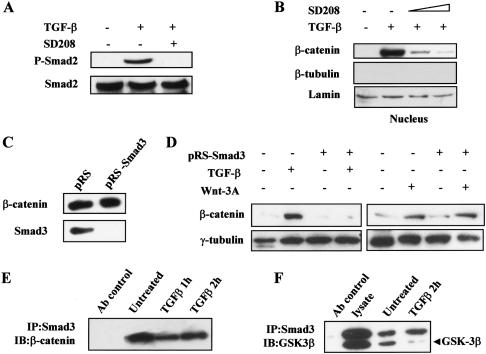
To explore the mechanism by which TGF-β1 induces β-catenin nuclear translocation, we next examined whether nuclear translocation of β-catenin is dependent on the signaling activity of the TGF-β type I receptor. To probe this, we pretreated MSCs with an inhibitor of the TGF-β type I receptor kinase, SD208 (Uhl et al. 2004), for 30 min before applying TGF-β1 treatment for another hour. Not only did SD208 block the phosphorylation of Smad2 (Fig. 2A), demonstrating that SD208 effectively inhibited TGF-β1 signaling, it also inhibited the nuclear translocation of β-catenin (Fig. 2B). These data indicate that TGF-β1-induced nuclear translocation of β-catenin was mediated by the bona fide TGF-β signaling pathway via the activation of the type I receptor kinase.
2.
β-catenin nuclear translocation is mediated by the TGF-β signaling pathway. (A) Whole-cell lysates of MSCs untreated or treated with TGF-β1 and the type I TGF-β receptor inhibitor SD208 for 30 min were blotted with a polyclonal antiphospho-Smad2 antibody. (B) Nuclear fractions of MSCs untreated or treated with TGF-β1 and SD208 were detected by anti-β-catenin antibody. The concentrations of SD208 used in this assay were 50 and 100 nM. (C) MSCs were infected with control (pRS) or Smad3 siRNA (pRS-Smad3) expressing retrovirus and selected by puromycin. Whole-cell lysates from stable cell populations were blotted with an anti-Smad3 rabbit polyclonal antibody. Equal loading was confirmed by the presence of β-catenin. (D) Nuclear fractions from pRS or pRS-Smad3 retroviruse-infected MSCs untreated or treated with TGF-β1 for 2 h or Wnt3A for 6 h were blotted with the anti-β-catenin antibody. (E) Whole-cell lysates from MSCs untreated or treated with TGF-β1 for 1 or 2 h were subjected to immunoprecipitation with an anti-Smad3 polyclonal antibody. An anti-Flag antibody was used as the Ab control for the immunopre-cipitation. The Western blots were carried out using antibodies against β-catenin. (IP) Immunoprecipitation; (IB) immunoblotting. (F) Immunoprecipitation was performed as in E with lysates from MSCs untreated or treated with TGF-β for 2 h. GSK-3β was detected by a mouse monoclonal antibody.
To further explore this mechanism, we investigated whether the primary effectors of TGF-β signaling, the Smads, are directly involved in the process of β-catenin nuclear translocation. To do this, we evaluated the effect of Smad3 knockdown on the ability of TGF-β1 to induce β-catenin nuclear translocation. By introducing a Smad3-specific small interfering RNA (siRNA) construct into MSCs through retroviral transfer, the expression of Smad3 protein was reduced by >90% (Fig. 2C). Subsequently, we examined β-catenin nuclear translocation in response to TGF-β1 in MSCs stably expressing the Smad3-siRNA in comparison with those infected with a retroviral vector control. As shown in Figure 2D, the cell fractionation results clearly indicate that Smad3 is required for the TGF-β1-induced nuclear translocation of β-catenin, since the amount of β-catenin in the nuclear fraction was barely detectable in MSCs with reduced expression of Smad3. This result also indicates that Smad2 may not be involved in the mediation of TGF-β1-induced nuclear translocation of β-catenin. Importantly, under the same conditions, Wnt3A treatment could still increase the level of β-catenin in the nucleus (Fig. 2D), suggesting that the mechanism of Wnt3A-induced β-catenin nuclear accumulation in MSCs is distinct from that of TGF-β1-induced β-catenin nuclear translocation.
The requirement of Smad3 for β-catenin nuclear translocation suggests the possibility that Smad3 could actively transport β-catenin into the nucleus. Previous reports have shown that Smad3 can interact with β-catenin and its binding partners Axin and CKIε (Labbe et al. 2000; Furuhashi et al. 2001; Waddell et al. 2004). Thus, it is possible that Smad3 and β-catenin coexist in a complex in the cytoplasm, then translocate into the nucleus simultaneously upon TGF-β1 stimulation. Consistent with this postulation, we found that Smad3 and β-catenin were indeed in the same complex in MSCs, since endogenous β-catenin and Smad3 could be coimmunoprecipitated by an anti-Smad3 antibody, and this interaction was minimally affected by TGF-β1 (Fig. 2E). Furthermore, endogenous Smad3 was found to interact with GSK-3β (Fig. 2F), another protein known to be associated with β-catenin. Interestingly, the association between Smad3 and GSK-3β decreases in response to TGF-β (Fig. 2F). Attempts to detect association between Smad3 and Axin or CKIε were unsuccessful, possibly because of the low levels of endogenous Axin or CKIε. Nevertheless, taken together with previous findings that both Smad3 and β-catenin could interact with Axin/CKIε and the association between Smad3 and Axin/CKIε decreases in response to TGF-β in cell types other than MSCs (Furuhashi et al. 2001; Waddell et al. 2004), these results support a model in which the nuclear translocation of β-catenin in response to TGF-β1 can be directly linked to changes in the composition and dynamics of a protein complex possibly containing β-catenin, Smad3, GSK-3β, Axin, and CKIε.
TGF-β1 and nuclear β-catenin exert similar biological effects on MSCs
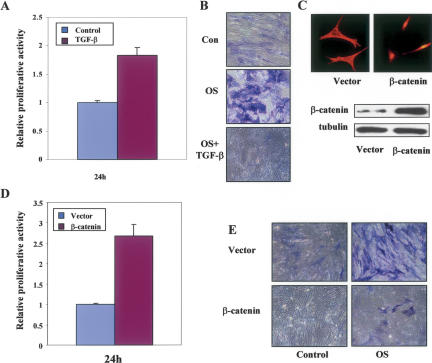
To investigate whether activation of this novel Smad3/β-catenin-mediated TGF-β1 signaling pathway is associated with a specific biological response in MSCs, we examined the effects of TGF-β1 on the regulation of proliferation and osteogenic differentiation in MSCs. As shown in Figure 3A and Supplementary Figure 3, TGF-β1 stimulated the proliferation of MSCs, an activity that contrasts the potent antiproliferative effect of TGF-β1 on many other cell types (Massague 1998). To determine whether TGF-β1 could also affect the differentiation program of these MSCs, an osteogenic assay was performed to measure the production of alkaline phosphatase (ALP) by culturing the MSCs in the osteogenic supplemental (OS) medium in the presence or absence of TGF-β1. In the absence of TGF-β1, an expected enhancement in the staining of ALP was observed in MSCs driven toward osteogenic differentiation by the culturing of those cells in OS medium (Fig. 3B). In contrast, the presence of TGF-β1 in the OS medium resulted in a much lower level of ALP staining (Fig. 3B), suggesting that TGF-β1 inhibits the osteogenic effect of the OS medium on MSCs.
3.
TGF-β1 and nuclear β-catenin have similar biological effects on MSCs. (A) Proliferation of human MSCs was examined by 3H-thymidine incorporation after the cells were untreated or treated with TGF-β1 for 24 h and relative proliferation activities are shown. Error bars were calculated from three duplicate experiments. (B) ALP activity was probed for osteogenic differentiation after MSCs were cultured in OS medium in the absence or presence of TGF-β1 for 10 d. (C) MSCs were infected with MSCV-mutant β-catenin or vector control retroviruses. Localization and ectopic expression of the mutant β-catenin were shown. (D) Proliferation of MSCs infected with the mutant β-catenin or vector control retroviruses was examined by 3H-thymidine incorporation. Relative proliferation activities are shown and error bars were calculated for standard deviation from three duplicate experiments. (E) ALP activity was probed for osteogenic differentiation after MSCs were infected with the mutant β-catenin or vector control retroviruses and cultured in OS medium for 10 d.
We then tested the postulation that nuclear translocation of β-catenin was directly linked to the TGF-β1-mediated regulation of proliferation and osteogenic differentiation of MSCs. To begin, we evaluated the effects of nuclear accumulation of β-catenin on the activities of MSCs by the introduction of a mutant form of β-catenin into those cells via retroviral infection. This β-catenin mutant retains full transcriptional activity but contains alanine substitutions at the four serine phosphorylation sites to prevent it from ubiquitination-mediated degradation (Barth et al. 1999). Interestingly, the ectopically expressed mutant form of β-catenin was almost completely localized in the nucleus of MSCs (Fig. 3C), in contrast to the previously reported predominant cell–cell junction localization of this same mutant at the plasma membrane (Barth et al. 1999). Similar to the proliferative effect of TGF-β1, expression of this β-catenin mutant in MSCs led to a significant increase in proliferation when compared with control cells (Fig. 3D). Consistent with the antiosteogenic effect of TGF-β1 in the MSCs, expression of this β-catenin mutant caused a substantial decrease in ALP staining (Fig. 3E). These results suggest that nuclear-localized β-catenin could exert similar biological effects on MSCs as those of TGF-β1, strongly supporting the postulation that there is a direct correlation between the activation of the Smad3/β-catenin-mediated TGF-β1 signaling pathway and the elicitation of unique biological responses in MSCs.
Nuclear β-catenin is required for the primary effects of TGF-β1 on MSCs through regulation of specific downstream target genes
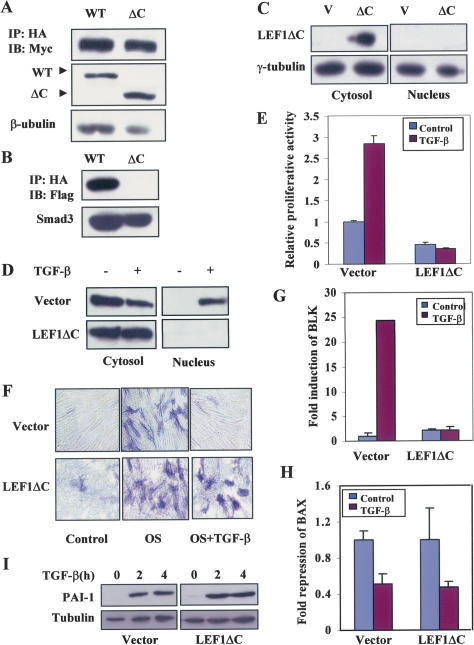
To further test this hypothesis, we next attempted to reduce the expression of β-catenin by using the siRNA strategy that was reported in a previous study (van de Wetering et al. 2003). However, this approach was unsuccessful due to the relatively high stability of β-catenin protein in this specific cell type (Fig. 1G; Supplementary Fig. 1). Consequently, we resorted to the use of a C-terminal truncation mutant of LEF1, LEF1ΔC, which lacks the HMG box and the nuclear localization sequence B box (Prieve et al. 1996). As a transcription factor, the wild-type LEF1 is known to reside on specific promoter sequences of Wnt-responsive genes and form a complex with β-catenin via a domain located in the N-terminalregion of LEF1 to activate transcription after β-catenin translocates into the nucleus (Logan and Nusse 2004). In addition, the HMG box of LEF1 was shown to mediate the interaction between LEF1 and Smad3 in the context of synergistic activation of specific target genes by the two transcription factors (Labbe et al. 2000). Thus, this mutant form of LEF1 is expected to retain β-catenin in the cytoplasm, since its ability to interact with β-catenin through its N-terminal region should be unaltered, consequently acting as a dominant-negative inhibitor that prevents the nuclear translocation of β-catenin without affecting the movement of Smad3. Consistent with this prediction, LEF1ΔC maintained its ability to interact with β-catenin in a similar fashion as that of the wild-type protein, but was unable to associate with Smad3 (Fig. 4A,B). When LEF1ΔC was introduced into the MSCs via an adenoviral delivery system (He et al. 1998), its expression was found to be mainly restricted in the cytoplasm (Fig. 4C). Under this condition, TGF-β was unable to induce β-catenin translocation to the nucleus, in contrast to that in cells infected with the control adenovirus (Fig. 4D). Most importantly, TGF-β-induced cell proliferation was significantly inhibited by the expression of LEF1ΔC in MSCs (Fig. 4E), strongly suggesting that the stimulatory effect of TGF-β1 on MSCs proliferation is mediated by this newly defined signaling pathway via induction of β-catenin translocation into the nucleus. Interestingly, the basal level of cell proliferation of MSCs was also reduced by the expression of LEF1ΔC (Fig. 4E), implicating the possible presence of an autocrine Wnt and/or TGF-β1 signaling pathway that functions to maintain proliferation of MSCs in culture. Additionally, we found that expression of LEF1ΔC eliminated the inhibitory effect of TGF-β1 on osteogenic differentiation of MSCs (Fig. 4F), further supporting the notion that rapid nuclear translocation of β-catenin is required for TGF-β to exert its primary biological effects on MSCs.
4.
Nuclear β-catenin is required for the primary effects of TGF-β1 on MSCs through regulation of specific downstream target genes. (A) 293T cells were cotransfected with pcDNA3-Myc-β-catenin and pcDNA3-HA-LEF1 or pcDNA3-HA-LEF1ΔC, and coimmunoprecipitations were performed with anti-HA and immunoblotted with anti-Myc. (WT) Wild type; (ΔC) LEF1ΔC. (B) 293T cells were cotransfected with pRK5-Flag-Smad3 and pcDNA3-HA-LEF1 or pcDNA3-HA-LEF1ΔC, and coimmunoprecipitations were performed with anti-HA and immunoblotted with anti-Flag. (C) MSCs were infected with vector control or LEF1ΔC adenoviruses. Ectopic expression of LEF1ΔC was detected with an anti-HA antibody. (D) Cytosolic and nuclear fraction of protein lysates were isolated from LEF1ΔC or vector control adenovirus-infected MSCs untreated or treated with TGF-β for 2 h. Western blots were performed with anti-β-catenin antibody or anti-HA antibody. (E) Proliferation of human MSCs infected with the LEF1ΔC or the vector control adenoviruses was examined by 3H-thymidine incorporation after the cells were untreated or treated with TGF-β1 for 24 h. Relative proliferation activities are shown and error bars were calculated for standard deviation from three duplicate experiments. (F) ALP activity was probed for osteogenic differentiation after MSCs were infected with LEF1ΔC or the vector control adenoviruses and cultured in OS medium for 5 d. (G) BLK mRNA levels in vector control and LEF1ΔC adenovirus-infected MSCs treated or untreated with TGF-β1 for 2 h were quantified by real-time PCR. Fold induction is shown and error bars were calculated for standard deviation from three duplicate experiments. (H) BAX mRNA levels in vector control and LEF1ΔC adenovirus-infected MSCs treated or untreated with TGF-β1 for 2 h were quantified by real-time PCR. Fold induction is shown and error bars were calculated for standard deviation from three duplicate experiments. (I) Protein levels of PAI-1 in vector control and LEF1ΔC adenovirus-infected MSCs untreated or treated with TGF-β1 for indicated time were determined by Western blot with anti-PAI-1 antibody.
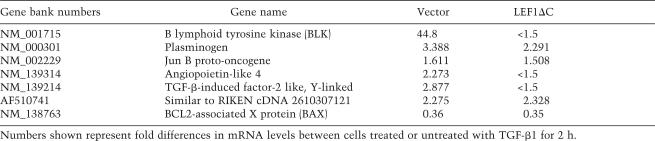
Both β-catenin and Smad3 are transcription factors that participate in the activation or repression of target gene expression when they translocate into the nucleus. To investigate whether expression of specific target genes is regulated by this β-catenin-mediated TGF-β signaling pathway in MSCs, we carried out microarray analysis on RNA samples isolated from vector control or LEF1ΔC adenovirus-infected MSCs with or without the treatment of TGF-β1 for 2 h. Under such conditions, we expected to identify TGF-β1-regulated target genes that depended on the activity of nuclear β-catenin. Indeed, we discovered several TGF-β1-regulated genes whose expression profiles were altered by the presence of LEF1ΔC. As shown in Table 1, which contains a partial list of TGF-β1-regulated genes, one notable target gene was B lymphoid tyrosine kinase (BLK). BLK was induced >40-fold in response to TGF-β1 treatment, but this induction was completely blocked in cells expressing the dominant-negative form of LEF1, even though LEF1ΔC expression also slightly increased the basal expression level of BLK (Fig. 4G). This result suggests that nuclear β-catenin is required for the TGF-β1-mediated induction of BLK. On the other hand, expression of plasminogen and Bcl-associated X protein (BAX) remains equally responsive to TGF-β1 in both the control and LEF1ΔC-expressing cells, suggesting that TGF-β1-induced modulation on the expression of these genes does not require nuclear β-catenin. We further verified the expression profile of BLK and BAX in response to TGF-β1 in the vector control and LEF1ΔC-expressing cells by real-time PCR. As shown in Figure 4, G and H, BLK expression was unable to be induced by TGF-β1 in LEF1ΔC-expressing cells, whereas the expression of BAX was similarly inhibited by TGF-β1 in both types of cells, results that are consistent with the microarray data. In addition, the inducibility of PAI-1 expression, a TGF-β1 target gene known to be regulated by the Smad pathway (Dong et al. 2001), was unaffected by LEF1ΔC expression (Fig. 4I), supporting the notion that LEF1ΔC expression only affects selected TGF-β-responsive genes in MSCs. Thus, although the expression of LEF1ΔC as a dominant-negative inhibitor to block β-catenin nuclear translocation could have unknown effects, taken together, these results suggest that this β-catenin-mediated TGF-β1 signaling pathway regulates the expression of specific target genes without affecting the expression pattern of other genes that are likely under the sole control of Smad proteins.
Table 1.
A partial list of TGF-β1 target genes in MSCs
Discussion
In this study, we demonstrated the existence of a novel form of cross-talk between the TGF-β and Wnt signaling pathways. We have shown that TGF-β1-induced β-catenin nuclear translocation is mediated by a novel mechanism that is independent of changes in β-catenin stability and phosphorylation status. We have provided evidence that the effector of TGF-β signaling, Smad3, plays an essential role in shuttling β-catenin into the nucleus, likely through a TGF-β1-induced change in the dynamics and composition of protein complexes. In this model, the signaling process is initiated by the TGF-β receptor-mediated phosphorylation of Smad3, leading to the disruption of the protein complex as indicated by the reduced interactions between Smad3 and GSK-3β. Dissociation of this protein complex allows cotranslocation of β-catenin and Smad3 into the nucleus, with Smad3 acting as a chaperone, a scenario consistent with the constitutive interactions between the two proteins and the established rapid time course of Smad3 nuclear translocation.
It is intriguing that this novel signaling pathway is found in the MSCs but not other cell types examined, even though the interactions between Smad3, β-catenin, and other components of the Axin/CKI/GSK-3 complex have been documented in those cell types, suggesting that the cellular context of MSCs may be a prerequisite for the existence of the unique cross-talk between the two prominent signaling pathways. Furthermore, our studies show that TGF-β elicits unique biological effects in this specific cellular context of human MSCs, as it stimulates the proliferation of those cells (Fig. 3A), which is in contrast to the potent antiproliferative effect of TGF-β on many other cell types, suggesting that TGF-β1 may induce specific biological responses through the combined actions of β-catenin and Smad proteins in the nucleus to regulate the expression of a specific set of TGF-β1 target genes. It remains to be determined whether TCF/LEF transcription factors participate in the mediation of TGF-β-induced proliferative response in MSCs once β-catenin is cotranslocated with Smad3 into the nucleus. It is possible that the two sets of partners, β-catenin/Smad3 and β-catenin/TCF/LEF, act to regulate the expression of different target genes, and together they mediate the biological effects of TGF-β in this cellular context. In this regard, we found that activation of this unique pathway by TGF-β1 leads to a substantial increase in the expression of BLK, a member of the Src tyrosine kinase family that potently promotes pre-B-cell leukemia development when its kinase activity is constitutively activated (Malek et al. 1998). Although further experiments are necessary to demonstrate a causative relationship, this result suggests that the induction of BLK by the β-catenin/Smad3-mediated signaling pathway may be directly linked to the stimulatory effect of this pathway on the proliferation of MSCs. In the meantime, the β-catenin-independent repression of BAX expression may also contribute to the execution of the biological program of TGF-β in MSCs by suppressing apoptosis (von Haefen et al. 2004). MSCs have been shown to secrete proteins of the TGF-β superfamily, and expression of at least four different Wnt proteins was detected in those cells (data not shown), suggesting that these two signaling pathways are likely involved in regulating the biological activities of MSCs. With the recent discovery that the Wnt signaling pathway plays an important role in the regulation of self-renewal of hematopoitic stem cells (Reya et al. 2003), subsequent research on the molecular composition of MSCs that permits the existence of this distinct TGF-β signaling pathway and on the functional characterization of downstream targets of this unique pathway should provide critical insights into the mechanisms underlying self-renewal and differentiation programs of MSCs.
Materials and methods
Human MSCs
Three separate batches of MSCs and growth medium were purchased from Cambrex Bio Science (PT-2501, PT-2500). The cells were cultured according to the protocols provided by the manufacturer.
Nuclear and cytoplasmic fractionation
MSCs were collected by trypsin digestion and washed with PBS one time before addition of the cytoplasmic lysis buffer (10 mM Hepes at pH 8.0, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 300 mM sucrose, 0.1% NP-40, 10 mM NaF, 20 mM β-glycerophosphate, 10 mM Na3VO4, 1× protease inhibitors, 0.5 mM PMSF). Cells were lysed for 10 min on ice and then quick-spun for 15 sec to collect cytosolic lysate. Pellets were washed two times with cytoplasmic lysis buffer and then lysed with nuclear lysis buffer (50 mM Hepes at pH 7.9, 250 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.1% NP-40, 0.1% glycerol, 10 mM NaF, 10 mM Na3VO4, 1 mM DTT, 1× protease inhibitors, 0.5 mM PMSF) for 30 min on ice. The lysates were spun for 20 min at 14,000 rpm at 4°C to collect nuclear lysates. Lysates were run in SDS-PAGE for Western blot analysis.
Immunofluorescence
Immunofluorescence staining was carried out as described previously (Eckner et al. 1996).
Osteogenic differentiation and ALP staining
MSCs were cultured in OS medium for osteogenic differentiation. OS medium: 100 nM dexamethasone, 0.25 mM ascorbic acid, and 10 mM β-glycerophosphate. ALP staining kit was purchased from Sigma (86C-1KT) and staining was performed according to the manual provided with the kit.
Antibodies and immunoprecipitation
Immunoprecipitation and Western blot were carried out as described (Shen et al. 1998). Antibodies to β-catenin (Transduction Lab), GSK-3β (Santa Cruz), Smad3 I-20 (Santa Cruz), Smad3 (Zymed), phspho-Smad2 (Cell Signaling), Lamin (Santa Cruz), β-tubulin (Santa Cruz), LEF1 (Oncogene), HA Y-11 (Santa Cruz), and Myc 9E10 (Roche) were used to detect proteins and for immunoprecipitation. HRP-conjugated antibodies to mouse or rabbit IgG were purchased from Zymed.
Quantitive real-time RT–PCR
Real-time RT–PCR was performed as described (Bai et al. 2004). GAPDH gene was used as the internal control for normalization. The primer sequences for BLK were as follows: forward, 5′-GCAGATTGTCACTCCCAAGGC-3′, and reverse, 5′-CT GTCGATGATTCGAGCCAAG-3′. The primer sequences for BAX were as follows: 5′-GCTGACATGTTTTCTGACGGC-3′, and reverse, 5′-AAGTCCAATGTCCAGCCCATG-3′. Standard curves for BLK and BAX primers were constructed using a serial dilution of cDNA to verify equal amplification efficiency.
Microarray analysis
The RNA samples used for microarray analysis were collected from vector control or LEF1ΔC-expressing MSCs untreated or treated with TGF-β for 2 h. The RNA samples were purified by RNeasy mini kit (Qiagen). For further information about preparation of the slides for microarrays, synthesis of fluorescent-labeled cDNA, hybridization, scanning and data acquisition, and quality control steps, visit the Duke University Microarray Core Facility at http://microarray.genome.duke.edu. Data were analyzed using GeneSpring 6.1 (Silicon Genetics).
Acknowledgments
We thank T. Reya, R. Wechsler-Reya, A.W. Duncan, D. Zhang, P. Casey, and D. Kaplan for various reagents, and J. Nevins and C. Counter for critically reading the manuscript. This work was supported by NIH grant DK064113 to X.F.W. and a DOD grant DAMD 17-02-1-0369 to H.J.
Footnotes
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1388806
References
- Bai X., Alekseyenko A.A., Kuroda M.I. Sequence-specific targeting of MSL complex regulates transcription of the roX RNA genes. EMBO J. 2004;23:2853–2861. doi: 10.1038/sj.emboj.7600299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth A.I., Stewart D.B., Nelson W.J. T cell factor-activated transcription is not sufficient to induce anchorage-independent growth of epithelial cells expressing mutant β-catenin. Proc. Natl. Acad. Sci. 1999;96:4947–4952. [Google Scholar]
- Cadigan K.M., Nusse R. Wnt signaling: A common theme in animal development. Genes & Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Caplan A.I. Mesenchymal stem cells and gene therapy. Clin. Orthop. Relat. Res. 2000:S67–S70. doi: 10.1097/00003086-200010001-00010. [DOI] [PubMed] [Google Scholar]
- Derynck R., Zhang Y., Feng X.H. Smads: Transcriptional activators of TGF-β responses. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- Dong C., Zhu S., Yoon W., Wang T., Alvarez R.J., Goldschmidt-Clermont P.J. Upregulation of PAI-1 is mediated through TGF β/SMAD pathway in transplant arteriopathy. J. Heart Lung Transplant. 2001;20:219. doi: 10.1016/s1053-2498(00)00480-0. [DOI] [PubMed] [Google Scholar]
- Eckner R., Yao T.P., Oldread E., Livingston D.M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes & Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- Furuhashi M., Yagi K., Yamamoto H., Furukawa Y., Shimada S., Nakamura Y., Kikuchi A., Miyazono K., Kato M. Axin facilitates Smad3 activation in the transforming growth factor β signaling pathway. Mol. Cell. Biol. 2001;21:5132–5141. doi: 10.1128/MCB.21.15.5132-5141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T.C., Zhou S., da Costa L.T., Yu J., Kinzler K.W., Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Semenov M., Tamai K., Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/β-catenin signaling: Arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- Hino S., Michiue T., Asashima M., Kikuchi A. Casein kinase Iε enhances the binding of Dvl-1 to Frat-1 and is essential for Wnt-3a-induced accumulation of β-catenin. J. Biol. Chem. 2003;278:14066–14073. doi: 10.1074/jbc.M213265200. [DOI] [PubMed] [Google Scholar]
- Hsieh J.C., Rattner A., Smallwood P.M., Nathans J. Biochemical characterization of Wnt–frizzled interactions using a soluble, biologically active vertebrate Wnt protein. Proc. Natl. Acad. Sci. 1999;96:3546–3551. doi: 10.1073/pnas.96.7.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe E., Letamendia A., Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-β and wnt pathways. Proc. Natl. Acad. Sci. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan C.Y., Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Malek S.N., Dordai D.I., Reim J., Dintzis H., Desiderio S. Malignant transformation of early lymphoid progenitors in mice expressing an activated Blk tyrosine kinase. Proc. Natl. Acad. Sci. 1998;95:7351–7356. doi: 10.1073/pnas.95.13.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGF-β signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Nishita M., Hashimoto M.K., Ogata S., Laurent M.N., Ueno N., Shibuya H., Cho K.W. Interaction between Wnt and TGF-β signalling pathways during formation of Spemann’s organizer. Nature. 2000;403:781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- Orford K., Crockett C., Jensen J.P., Weissman A.M., Byers S.W. Serine phosphorylation-regulated ubiquitination and degradation of β-catenin. J. Biol. Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., Moorman M.A., Simonetti D.W., Craig S., Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Prieve M.G., Guttridge K.L., Munguia J.E., Waterman M.L. The nuclear localization signal of lymphoid enhancer factor-1 is recognized by two differentially expressed Srp1-nuclear localization sequence receptor proteins. J. Biol. Chem. 1996;271:7654–7658. doi: 10.1074/jbc.271.13.7654. [DOI] [PubMed] [Google Scholar]
- Reya T., Duncan A.W., Ailles L., Domen J., Scherer D.C., Willert K., Hintz L., Nusse R., Weissman I.L. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- Shen X., Hu P.P., Liberati N.T., Datto M.B., Frederick J.P., Wang X.F. TGF-β-induced phosphorylation of Smad3 regulates its interaction with coactivator p300/CREB-binding protein. Mol. Biol. Cell. 1998;9:3309–3319. doi: 10.1091/mbc.9.12.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuli R., Tuli S., Nandi S., Huang X., Manner P.A., Hozack W.J., Danielson K.G., Hall D.J., Tuan R.S. Transforming growth factor-β-mediated chondrogenesis of human mesenchymal progenitor cells involves N-cadherin and mitogen-activated protein kinase and Wnt signaling cross-talk. J. Biol. Chem. 2003;278:41227–41236. doi: 10.1074/jbc.M305312200. [DOI] [PubMed] [Google Scholar]
- Uhl M., Aulwurm S., Wischhusen J., Weiler M., Ma J.Y., Almirez R., Mangadu R., Liu Y.W., Platten M., Herrlinger U., et al. SD-208, a novel transforming growth factor β receptor I kinase inhibitor, inhibits growth and invasiveness and enhances immunogenicity of murine and human glioma cells in vitro and in vivo. Cancer Res. 2004;64:7954–7961. doi: 10.1158/0008-5472.CAN-04-1013. [DOI] [PubMed] [Google Scholar]
- de van Wetering M., Oving I., Muncan V., Pon Fong M.T., Brantjes H., van Leenen D., Holstege F.C., Brummelkamp T.R., Agami R., Clevers H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Haefen C., Gillissen B., Hemmati P.G., Wendt J., Guner D., Mrozek A., Belka C., Dorken B., Daniel P.T. Multidomain Bcl-2 homolog Bax but not Bak mediates synergistic induction of apoptosis by TRAIL and 5-FU through the mitochondrial apoptosis pathway. Oncogene. 2004;23:8320–8332. doi: 10.1038/sj.onc.1207971. [DOI] [PubMed] [Google Scholar]
- Waddell D.S., Liberati N.T., Guo X., Frederick J.P., Wang X.F. Casein kinase Iε plays a functional role in the transforming growth factor-β signaling pathway. J. Biol. Chem. 2004;279:29236–29246. doi: 10.1074/jbc.M400880200. [DOI] [PubMed] [Google Scholar]
- Zhou S., Eid K., Glowacki J. Cooperation between TFG-β and Wnt pathways during chondrocyte and adipocyte differentiation of human marrow stromal cells. J. Bone Miner. Res. 2004;19:463–470. doi: 10.1359/JBMR.0301239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material is available at http://www.genesdev.org.