Abstract
DNA ligase D (LigD) is a large polyfunctional protein that participates in a recently discovered pathway of nonhomologous end-joining in bacteria. LigD consists of an ATP-dependent ligase domain fused to a polymerase domain (Pol) and a phosphoesterase module. The Pol activity is remarkable for its dependence on manganese, its ability to perform templated and nontemplated primer extension reactions, and its preference for adding ribonucleotides to blunt DNA ends. Here we report the 1.5-Å crystal structure of the Pol domain of Pseudomonas LigD and its complexes with manganese and ATP/dATP substrates, which reveal a minimized polymerase with a two-metal mechanism and a fold similar to that of archaeal DNA primase. Mutational analysis highlights the functionally relevant atomic contacts in the active site. Although distinct nucleoside conformations and contacts for ATP versus dATP are observed in the cocrystals, the functional analysis suggests that the ATP-binding mode is the productive conformation for dNMP and rNMP incorporation. We find that a mutation of Mycobacterium LigD that uniquely ablates the polymerase activity results in increased fidelity of blunt-end double-strand break repair in vivo by virtue of eliminating nucleotide insertions at the recombination junctions. Thus, LigD Pol is a direct catalyst of mutagenic nonhomologous end-joining in vivo. Our studies underscore a previously uncharacterized role for the primase-like polymerase family in DNA repair.
Keywords: DNA repair, double-strand breaks, mycobacteria, Pseudomonas
The presumption that bacteria rely solely on homologous recombination to repair DNA double-strand breaks (DSB) has been overturned by evidence that Mycobacteria have a nonhomologous end-joining (NHEJ) system driven by a Ku homolog and a polyfunctional ATP-dependent DNA ligase (LigD) (1–4). LigD and Ku are jointly encoded by many diverse bacterial genera (1–6), suggesting that NHEJ is broadly relevant to bacterial physiology. LigD proteins differ with respect to the linear order of the three catalytic modules. Whereas Mycobacterium LigD consists of an N-terminal polymerase domain (Pol), a central phosphoesterase domain, and a C-terminal ligase domain (1–4), Pseudomonas LigD is composed of an N-terminal phosphoesterase domain, a central ligase domain, and a C-terminal polymerase domain (see Fig. 7A, which is published as supporting information on the PNAS web site) (7–9). The polymerase activity of LigD in vitro is manifest either as a nontemplated single-nucleotide addition to a blunt-end duplex DNA primer or as a templated extension of a 5′-tailed duplex DNA primer–template (2, 4, 7, 8). Because fill-in synthesis and the addition of a single nontemplated nucleotide are the molecular signatures of mutagenic mycobacterial NHEJ in vivo at 5′-overhang DSB and blunt-end DSB, respectively (2), it is plausible to invoke a direct role for the LigD polymerase as an agent of unfaithful repair. To address this hypothesis, we sought to define the structure and mechanism of the Pol domain and translate the findings to a genetic analysis of LigD Pol function.
Results and Discussion
Mutational Mapping of the Polymerase Active Site.
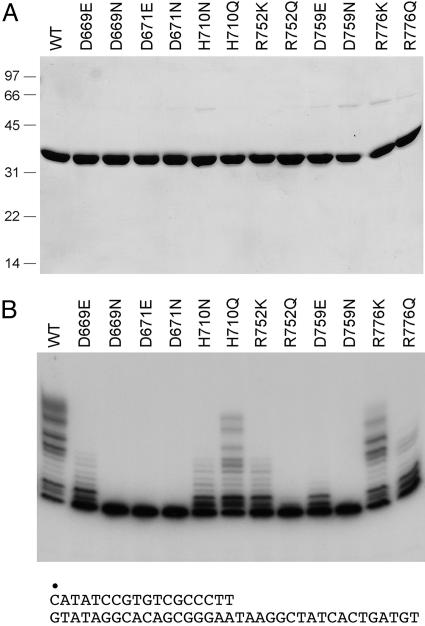
We performed an initial Ala scan of 18 aa of the autonomous Pol domain of Pseudomonas aeruginosa LigD (amino acids 533–840) that are conserved in LigD proteins from diverse bacterial species (Fig. 7B). We focused on acidic, basic, and polar side chains as candidates for functions in substrate and cofactor binding and/or catalysis of nucleotide addition. The wild-type and mutant proteins were produced as His10-tagged derivatives in Escherichia coli and purified from soluble extracts by Ni-agarose chromatography (Fig. 8A, which is published as supporting information on the PNAS web site). Template-directed DNA synthesis by purified Pol was gauged by a dNTP-dependent extension of a 5′ 32P-labeled 18-mer DNA strand primer annealed to a complementary 36-mer template strand (Fig. 8B). We found that 11 of the mutants retained polymerase activity comparable with wild-type: Q664A, S768A, K566A, E649A, K755A, S774A, R587A, H651A, K707A, R593A, and N763A. One mutant, S704A, had modestly reduced activity. We surmise that these 12 amino acids, although conserved, are not essential constituents of the DNA polymerase active site. Six of the Ala mutations either abolished or severely reduced the DNA polymerase activity: D669A, D671A, H710A, R752A, D759A, and R776A (Fig. S2B). The mutational studies highlighted Asp-669, Asp-671, His-710, Asp-759, Arg-752, and Arg-776 as candidate active site residues.
Structure–activity relationships were illuminated by introducing conservative substitutions (Fig. 1). Isosteric Asn changes abolished activity at all three essential Asp positions. Whereas Glu partially restored activity in lieu of Asp-669 and, to a lesser extent, at Asp-759, there was no salutary effect of Glu at Asp-671. These findings are consistent with a role for Asp-669, Asp-671, and Asp-759 in binding the manganese cofactor. Replacing His-710 with Asn elicited a loss of function comparable to that of H710A (Figs. 1B and 8B), but Gln resulted in a gain of activity, suggesting that His-710 engages in hydrogen-binding interactions (via Nε) rather than serving as a general acid–base catalyst. Positive charge appears to be the relevant feature at Arg-752 and Arg-776, insofar as the Gln mutants were as defective as the respective Ala mutants, whereas Lys resulted in partial restoration of function (Fig. 1B).
Fig. 1.
Structure–activity relationships at essential residues of LigD Pol. (A) Aliquots (5 μg) of the Ni-agarose preparations of wild-type Pol and the indicated mutants were analyzed by SDS/PAGE. Polypeptides were visualized by staining the gel with Coomassie blue dye. The positions and sizes (in kDa) of marker polypeptides are indicated on the left. (B) Reaction mixtures (20 μl) containing 50 mM Tris·HCl (pH 7.5), 5 mM DTT, 5 mM MnCl2, 1 pmol of 5′ 32P-labeled 18/36-mer primer–template (shown below the gel in B), 50 μM each of dATP, dGTP, dCTP, and dTTP, and 200 ng of wild-type or mutant LigD Pol were incubated for 20 min at 37°C. The radiolabeled products were resolved by denaturing PAGE and visualized by autoradiography.
Crystal Structure of LigD Pol.
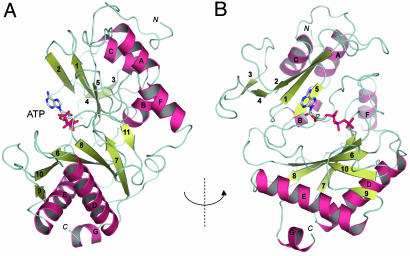
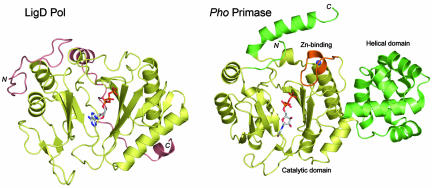
To extend the structure–function analysis of the LigD Pol domain to atomic resolution, we crystallized selenomethionine (SeMet)-substituted Pol and solved its structure at 1.5 Å (see Materials and Methods; see also Table 1, which is published as supporting information on the PNAS web site). Additional 1.9-Å cocrystal structures were obtained with manganese and nucleotides bound at the active site (Table 1). LigD Pol is a globular protein composed of 11 β-strands organized mainly as two central sheets flanked by seven α-helices on the periphery. A six-strand antiparallel sheet (comprising β-strands 9, 10, 6, 8, 7, and 11), a three-strand antiparallel sheet (consisting of strands 2, 1, and 5), and the respective interstrand loops form a central cleft to which a nucleoside triphosphate substrate binds in the absence of a DNA primer or primer–template (Fig. 2A). In the ATP-bound Pol, the adenine base is solvent-exposed (Fig. 2A), whereas the ribose hydroxyls are oriented toward the enzyme surface (Fig. 2B). The fold of the LigD Pol domain most closely resembles that of the catalytic domain of the DNA primases of the archea Pyrococcus horikishii (Fig. 3) and Pyrococcus furiosus (10, 11). The LigD Pol and Pho primase structures align in dali (12) with a Z score of 11.8 and an rms deviation of 3.7 Å at 203 Cα positions. A UTP nucleotide soaked into the crystal of Pho primase in the absence of divalent cation occupies a position in the cleft roughly analogous to that seen for ATP in LigD Pol. However, the nucleoside conformation in the primase–UTP complex (11) is quite different from that of ATP in LigD, insofar as the UTP ribose hydroxyls are solvent-exposed and the base faces inward toward the enzyme (Fig. 3). LigD Pol lacks the distinctive helical domain appended to archaeal primase as an insert to the sequence of the catalytic domain; it also lacks the zinc-binding motif located within the primase catalytic domain (Fig. 3). We surmise that LigD Pol is a minimized version of an archaeal primase-type polymerase. LigD Pol is also structurally similar to a DNA primase-polymerase domain encoded by the pRN1 plasmid from Sulfolobus islandicus (dali Z score of 6.4 with an rms deviation of. 3.7 Å at 145 Cα positions) (13). The LigD Pol fold is unrelated to that of the Pol X polymerase family (14), members of which have been implicated in eukaryal NHEJ (15–18).
Fig. 2.
Structure of the LigD Pol domain. The overall fold of the Pol domain of Pseudomonas LigD is depicted as a ribbon diagram with α-helices colored magenta and β-strands colored green. The N and C termini are indicated. ATP is bound within a cleft formed by two central β-sheets; the cleft is viewed from the side in A and from above in B.
Fig. 3.
Comparison of the LigD Pol and archaeal DNA primase folds. The structure of the ATP cocrystal of LigD Pol was aligned to that of the UTP-soaked crystal of Pyroccocus horikoshii DNA primase (Protein Data Bank ID code IV34). The aligned structures are offset horizontally and depicted as ribbon traces, with the N and C termini as indicated. ATP and UTP are drawn as stick models in Corey–Pauling–Koltun atomic coloring. The structurally conserved elements are colored gold. The unique helical domain emanation from the primase catalytic domain and the C-terminal helix of primase are colored green. The Zn-binding module within the primase catalytic domain is colored brown; the zinc ion is a blue sphere. Structural elements of LigD Pol not found in primase are also colored brown.
Nucleotide and Manganese Binding in the Polymerase Active Site.
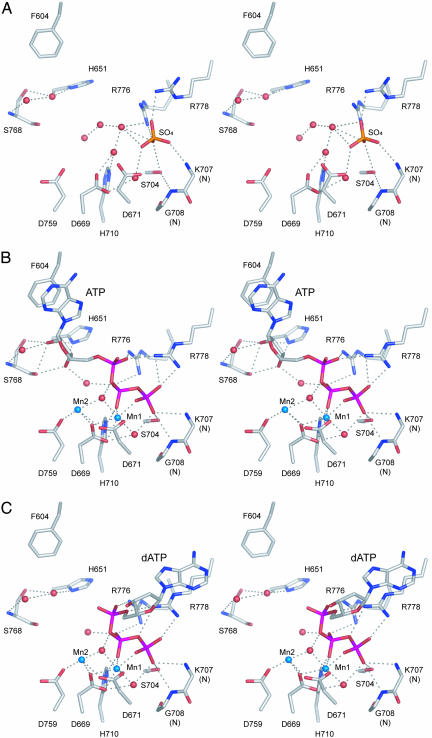
Fig. 4 highlights the architecture and atomic contacts in the active site of LigD Pol, which was defined by the location of the nucleotide and two manganese ions in the Mn-ATP and Mn-dATP cocrystals (Fig. 4 B and C; see also Fig 9, which is published as supporting information on the PNAS web site). A sulfate ion in the apoenzyme (Fig. 4A) occupies a position corresponding to the γ-phosphate of ATP/dATP. (The sulfate ion was acquired during the ammonium sulfate precipitation step that immediately preceded crystallization.) Five of the side chains defined by Ala scanning as essential or important for Pol activity (Asp-669, Asp-671, His-710, Asp-759, and Arg-776) are bona fide constituents of the Mn-NTP binding site (Fig. 4B). The sixth essential residue, Arg-752, is on the surface of the active site cleft and a candidate for binding the primer–template. The Mn-NTP site can be divided into three components that interact with (i) the two manganese ions, (ii) the triphosphate moiety of the nucleotide, and (iii) the nucleoside moiety. The conformation of the triphosphate, the positions of the metals, and the contacts to manganese and the triphosphate group are nearly identical in the ATP and dATP cocrystals (Fig. 4 B and C). Mn1 is sequestered in an octahedral coordination complex to nonbridging oxygens of the β- and γ-phosphates, the essential Asp-669 and Asp-671 side chains, and two waters that coordinate the bridging α–β-oxygen and the γ-phosphate, respectively (Fig. 4B). Asp-671 undergoes a conformational switch during the transition from apoenzyme, where it is pointing away from the Mn1-binding site (Fig. 4A), to the Mn-NTP complex. The coordination complex of Mn2 is partially filled by Asp-669, the essential Asp-759 side chain, and a water shared with Mn1 (Fig. 4B). We infer from the LigD Pol structure that Mn1 stabilizes a productive conformation of the PPi leaving group, whereas Mn2 is likely to activate the 3′-OH primer terminus for attack on the α-phosphorus of the nucleotide substrate. The two-metal mechanism is a conserved theme among DNA polymerases (19, 20).
Fig. 4.
Active site of LigD Pol. Stereo views are shown of the active site constituents of the apoenzyme (A), the Mn-ATP cocrystal (B), and the Mn-dATP cocrystal (C). Potential hydrogen-bonding interactions are denoted by dashed lines. Manganese ions and waters are rendered as blue and red spheres, respectively. Amino acids and sulfate (A), ATP (B), or dATP (C) ligands are labeled and shown in stick representation.
The triphosphate moiety is coordinated by side chains Arg-776 (which contacts nonbridging oxygens of the α- and β-phosphates), His-710 (β-phosphate via Nε), Arg-778 (γ-phosphate), and Ser-704 (γ-phosphate). Whereas Ala substitutions for His-710 and Arg-776 inhibit Pol activity, the more modest contributions of the hydrogen bond of Ser-704 to the γ-phosphate might reflect functional redundancy with the hydrogen bonds between the backbone amides of Lys-707 and Gly-708 to the same oxygen atom contacted by Ser-704 (Fig. 4B).
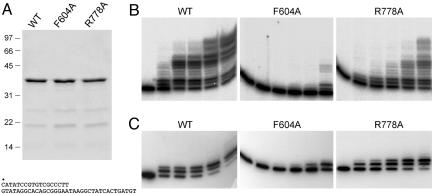
The nucleosides adopt different conformations in the ATP versus dATP cocrystals (Fig. 4 and 9). The adenosine of ATP extends away from the triphosphate group and is engaged to the enzyme via stacking of the adenine base on Phe-604 and a network of direct and water-mediated contacts to the ribose sugar, including (i) hydrogen bonds between the ribose O2′ and the Ser-768 Oγ, His-651 Nδ, and Ser-768 backbone carbonyl atoms, as well as a water coordinated by the Ser-768 backbone amide and (ii) hydrogen bonds between the ribose O3′ and the His-651 Nδ and Ser-768 backbone carbonyl atoms, plus a water shared with the α–β bridging oxygen of the triphosphate (Fig. 4B). In contrast, the deoxyadenosine of dATP, although disordered over the ribose moiety in the electron density map (Fig. 9) appears to have rotated about the bridging Pα–O bond to Pβ so that the adenine is nearly folded back over the triphosphate. In the dATP complex, the adenine base is modeled into density (Fig. 9) such that it stacks on the guanidinium moiety of the Arg-778 side chain (a π-cation interaction) (Fig. 4C). In this conformation, there are no apparent contacts available to the sugar 3′O (Fig. 4C). In the dATP-bound active site (and in the apoenzyme), a water occupies the position that was filled by the ribose O2′ in the ATP complex (Fig. 9) and engages in a similar network of polar interactions (Fig. 4 A and C). We surmise that there might be two nucleotide binding modes involving alternative base stacking partners, the choice of which during crystallization is likely influenced by contacts to the ribose O2′ that favor the extended conformation with stacking on Phe-604. Consistent with this hypothesis, we observed that, in cocrystals of LigD Pol with Mn-CTP and Mn-UTP, the nucleosides adopt a predominant ATP-like conformation with the pyrimidine base stacked on Phe-604 (data not shown).
Which nucleotide binding mode is productive for polymerase activity, or are both modes productive, with one favored for deoxynucleotide incorporation and the other for ribonucleotide addition? To gauge the relative contributions of the two binding modes, we introduced Ala in lieu of the Phe-604 and Arg-778 side chains that stack on the adenine base in the ATP and dATP complexes, respectively. The recombinant F604A and R778A Pol proteins (Fig. 5A) were tested for templated DNA synthesis. Whereas the R778A change had only a mild impact on activity, the F604A mutation strongly suppressed dNMP incorporation (Fig. 5B). The templated RNA polymerase activity of wild-type LigD Pol is limited to only a few cycles of ribonucleotide addition to a DNA primer (8) (Fig. 5C). The F604A protein was defective for rNMP addition to the primer–template, whereas R778A remained active with rNTP substrates (Fig. 5C). These results suggest that, although ribonucleosides and deoxynucleosides can assume different conformations in the substrate-binding cleft, the ATP mode is likely to be the productive one for templated nucleotide addition. Conceivably, the two binding modes are in equilibrium, and dNTPs switch from the dATP conformation to the ATP conformation when the primer–template is present.
Fig. 5.
Phe-604 is essential for ribonucleotide and deoxyribonucleotide addition. (A) Aliquots (7 μg) of wild-type Pol domain and the F604A and R778A mutants were analyzed by SDS/PAGE. Polypeptides were visualized by staining the gel with Coomassie blue dye. Polymerase reaction mixtures (20 μl) containing 50 mM Tris·HCl (pH 7.5), 5 mM DTT, 5 mM MnCl2, 1 pmol 5′ 32P-labeled 18/36-mer primer–template (shown below the gel in A), 50 μM each of dATP, dGTP, dCTP, and dTTP (B) or ATP, GTP, CTP, and UTP (C), and (from left to right in each titration series) 0, 12, 25, 50, 100, or 200 ng of wild-type or mutant LigD Pol were incubated for 20 min at 37°C. The radiolabeled products were resolved by denaturing PAGE.
LigD Pol Is a Catalyst of Mutagenic NHEJ in Vivo.
To query the role of the LigD polymerase during NHEJ in vivo, we turned to Mycobacterium smegmatis, a genetically tractable bacterium in which the Ku and LigD requirements for DSB repair by NHEJ in vivo were first established and in which the mutagenic quality of the pathway was demonstrated (2). The MsmLigD and PaeLigD Pol domains align with 64 positions of side chain identity plus 43 positions of side chain conservation in the 228-aa segment shown in Fig. 7B. Moreover, the biochemical properties of the PaeLigD and MsmLigD polymerases are the same with respect to manganese-dependent catalysis of templated fill-in synthesis at a 5′-overhang and single-nucleotide addition at a blunt DSB end (2, 7).
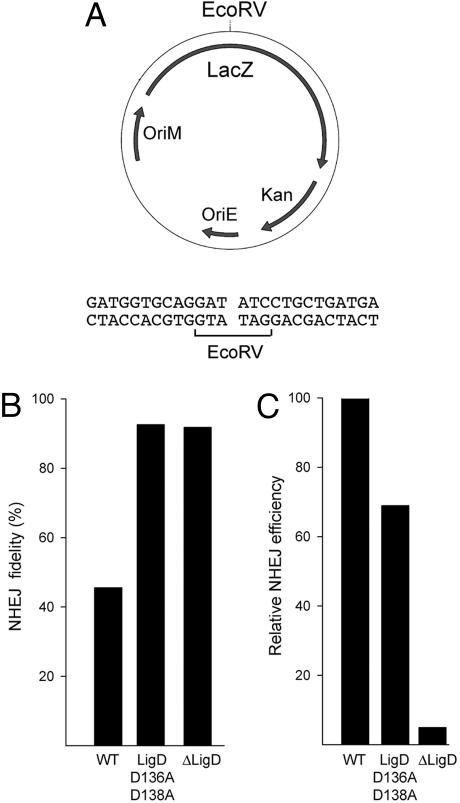
We performed an allelic replacement in M. smegmatis, whereby a mutant gene encoding a polymerase-defective, ligase-active MsmLigD protein, D136A–D138A (2), lacking two of the essential manganese ligands of the Pol active site was reintroduced at the ligD locus of a ΔligD strain. As a control, we constructed an isogenic strain in which the wild-type ligD gene was reintegrated at the ΔligD locus. NHEJ fidelity was assayed by transformation with a kanamycin-resistance plasmid that was linearized within a lacZ reporter gene by digestion with EcoRV, which generates a blunt-ended DSB (Fig. 6A). Successful transformation of M. smegmatis to kanamycin-resistance by the linear plasmid depends on ligation of the ends to produce a circular DNA molecule. Faithful resealing of the original ends with no gain or loss of nucleotides will restore the lacZ reading frame and result in blue colony color on agar medium containing X-gal. However, unfaithful sealing after insertion or deletion of nucleotides will usually result in a frame-shift mutation that inactivates the lacZ gene and elicits a white colony phenotype (2). The fidelity of NHEJ is defined as the percent of transformants that have blue colony color.
Fig. 6.
Role of the Pol activity in blunt-end NHEJ in vivo. (A) The NHEJ reporter plasmid contains a mycobacterial replication origin (OriM), a kanamycin-resistance gene (Kan), and a lacZ gene with a unique restriction site for endonuclease EcoRV, which generates the blunt DSB end structure shown. (B) NHEJ in M. smegmatis was assayed by parallel transformations with linear and circular plasmid DNA as described in ref. 2. NHEJ fidelity (percent of blue transformants) is shown for wild-type M. smegmatis, the Pol-defective ligD mutant D136A–D138A, and the ΔligD-null strain. The fidelity values are derived from pooled counts of blue and white colonies from three separate experiments entailing nine independent transformations. The number of colonies scored was 9,194 for wild-type, 4,355 for D136A–D138A, and 820 for ΔligD. (C) Relative NHEJ efficiency is the ratio of the transformation efficiencies (colonies per μg) for linear and circular DNAs normalized to the wild-type value (100%). Relative efficiency values are the mean of three independent transformations.
Blunt-end NHEJ in M. smegmatis reconstituted with wild-type LigD was characteristically error prone (45% fidelity). However, fidelity increased to 92% in the strain containing the polymerase-defective LigD protein, thereby phenocopying the increased fidelity seen in the ΔligD strain (Fig. 6B). Whereas the elimination of the LigD protein resulted in a 20-fold decrement in the efficiency of blunt-end DSB repair, the active-site mutation of the Pol domain reduced NHEJ efficiency by only one-third (Fig. 6C). We determined the nucleotide sequences of rejoined EcoRV-cut plasmids from 20 independent lacZ− transformants of the wild-type LigD strain (Fig. 10, which is published as supporting information on the PNAS web site). Half of the junctions contained nontemplated single nucleotide insertions at the otherwise unperturbed blunt ends, and 10% (2/20) contained a single nontemplated nucleotide at one intact EcoRV end and a deletion at the other terminus. In contrast, all 22 of the imprecise NHEJ junctions recovered from the Pol-defective D136A–D138A strain entailed deletions at one or both ends, and none of the junctions contained a nontemplated nucleotide insert. These results indicate that the polymerase component of LigD is a major determinant of mutagenic blunt-end DSB repair in vivo by virtue of its direct role in adding nontemplated nucleotide(s) at the junctions. The Pol activity is apparently not required for the joining step of LigD-dependent blunt-end NHEJ.
Conclusions
By integrating biochemical, structural, and genetic approaches, we illuminate the Pol component of LigD as a bona fide case of an archaeal primase-type polymerase within the bacterial domain of life, define its active site with respect to nucleotide and metal binding, and provide clear evidence of a biological role for a member of this polymerase family in a specific DNA repair pathway (one that is unrelated to the priming of lagging-strand DNA replication).
Materials and Methods
Purification of SeMet-Substituted LigD Pol.
A gene fragment encoding LigD (533–840) (Pol domain) was amplified by PCR with primers that introduced BamHI restriction sites over the start codon and 3′ of the stop codon, thereby facilitating in-frame insertion into vector pET28-His10-Smt3. SeMet-substituted His10-Smt3-LigDPol protein was purified from a soluble bacterial extract of a 4-liter culture of E. coli B834/pET28-His10-Smt3-Pol that had been grown in complete LeMaster medium (21) containing 0.06 mg/ml kanamycin and 0.05 mg/ml SeMet until the A600 reached 0.5, then adjusted to 0.3 mM isopropyl β-d-thiogalactoside/2% ethanol and incubated for 16 h at 22°C. Cells were harvested by centrifugation, and the pellet was stored at −80°C. All subsequent procedures were performed at 4°C. Preparation of the soluble extract (150 ml; 1.8 g of protein) and purification of the fusion protein by Ni-agarose affinity chromatography were performed as described in ref. 22 with minor modifications. His10-Smt3-Pol was recovered in the 300 mM imidazole eluate, which was dialyzed against buffer containing 50 mM Tris·HCl (pH 7.5), 0.2 M NaCl, and 5 mM DTT. The dialysate was clarified by centrifugation and then digested with the Smt3-specific protease Ulp1 (23) for 3 h at 4°C (at a Pol:Ulp1 ratio of 1,000:1). The tag-free Pol domain was separated from the His10-Smt3 tag by passage of the digest over a Ni-agarose column. The Pol protein recovered in the flow-through fraction was precipitated by adding ammonium sulfate to 70% saturation. The precipitate (32 mg protein) was recovered by centrifugation and dissolved in 20 mM Tris·HCl, pH 7.5/100 mM NaCl/5 mM DTT/10% glycerol to attain a protein concentration of 17 mg/ml. Aliquots were stored at −80°C.
Crystallization and Structure Determination.
Crystals of SeMet-substituted LigD Pol were grown at 22°C via the hanging drop vapor diffusion method after mixing the Pol solution with an equal volume of the well solution containing 100 mM sodium cacodylate, pH 6.5, 0.2 M sodium acetate, 5 mM DTT, and 26–32% polyethylene glycol 8000. Crystals grew over 7–8 days to form 200- to 300-μm rods. Cocrystals were grown by adjusting the protein solution to 10 mM MnCl2 and 5 mM ATP or dATP before mixing with the well solution. The crystals were cryoprotected with well solution containing 20% ethylene glycol (with Mn plus NTP for the cocrystals) before flash-freezing in liquid nitrogen. x-ray diffraction data were collected at the National Synchrotron Light Source beamline X29 at a wavelength of 0.9792 Å. Data were indexed, integrated, and scaled by using denzo and scalepack (24) and reduced with ccp4 (25). LigD crystals belonged to orthorhombic space group P21212, with a = 71.75, b = 203.58, and c = 44.20 Å. The structure of LigD was determined by the single wavelength anomalous diffraction method employing the anomalous signal from Se atoms as implemented in solve (26). Ten selenium atoms were located, and phases derived from these anomalous centers were used for density modification as implemented in resolve. The asymmetric unit contained two molecules of LigD Pol. Noncrystallographic symmetry was used for further phase improvement. Manual model building was accomplished using o (27). Refinement of the model was accomplished with refmac (28) and cns (29). The protein structures of the Mn-ATP-LigD and Mn-dATP cocrystals were determined with refmac by refining the diffraction data sets against the apoenzyme structure. Models for ATP and dATP were built into the electron density maps obtained after initial refinement. Simulated annealing omit maps were calculated with cns. The LigD Pol structure comprised a continuous polypeptide from amino acid 543 to 837; no electron density was observed for the N-terminal 10-aa segments and the C-terminal 3-aa segments. The final model statistics and crystallographic data for the apoenzyme, Mn-ATP, and Mn-dATP cocrystals are reported in Table 1 and were deposited in the Protein Data Bank under ID codes 2FAO, 2FAQ, and 2FAR, respectively. The three models exhibited excellent geometry with no amino acids in the disallowed regions of the Ramachandran plot. The structure images were prepared with pymol (30).
Pol Mutants.
pET-PaeLigD- (533–840) encodes a His10-tagged Pol domain of LigD. Missense mutations were introduced by two-stage PCR overlap extension as described in ref. 7; the plasmid inserts were sequenced completely to exclude the acquisition of unwanted changes. Wild-type Pol and Pol mutants were purified from soluble extracts of isopropyl β-d-thiogalactoside-induced E. coli BL21(DE3) by Ni-agarose chromatography as described in ref. 7. The protein concentrations were determined with the Bio-Rad dye reagent with BSA as the standard.
Construction of a Pol-Defective LigD Strain of M. smegmatis.
Plasmid pMSG346 (marked with selectable and counterselectable hyg and sacB genes) was designed to facilitate allelic exchange at the ΔligD locus of the M. smegmatis null mutant described in ref. 2. pMSG346 contains 503 bp of genomic DNA 5′ of the ligD ORF and 490 bp of genomic DNA 3′ of the ORF, with an NdeI site introduced at the start codon and a BamHI site introduced 37 bp 3′ of the stop codon. The ligD-(D136A–D138A) gene encoding a Pol-defective, ligase-active M. smegmatis LigD protein (2) was excised from a pET vector and inserted between the NdeI and BamHI sites of pMSG346. The resulting plasmid was transformed into the ΔligD strain; allelic exchange was executed by the two-step selection/counterselection strategy (2). A control allelic exchange was performed by using pMSG346 containing a wild-type M. smegmatis ligD insert. Restoration of the ligD locus was confirmed by Southern hybridization. The D136A–D138A mutation was verified by PCR-amplifying and sequencing the Pol coding region from the Pol-defective ligD strain.
Supplementary Material
Acknowledgments
Beamline X29A at the National Synchrotron Light Source is supported by the Offices of Biological and Environmental Research and of Basic Energy Sciences of the U.S. Department of Energy and the National Center for Research Resources of the National Institutes of Health. This work was supported by National Institutes of Health Grants GM63611 and AI064693. S.S. is an American Cancer Society Research Professor. C.D.L. is a Rita Allen Foundation Scholar. M.S.G. is the recipient of an Investigator in Pathogenesis of Infectious Disease Award from the Burroughs-Wellcome Fund. J.A. is supported by National Institutes of Health Medical Scientist Training Program Grant GM07739.
Abbreviations
- DSB
double-strand break
- NHEJ
nonhomologous end-joining
- LigD
DNA ligase D
- Pol
polymerase domain
- SeMet
selenomethionine.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2FAO, 2FAQ, and 2FAR).
References
- 1.Gong C., Martins A., Bongiorno P., Glickman M., Shuman S. J. Biol. Chem. 2004;279:20594–20606. doi: 10.1074/jbc.M401841200. [DOI] [PubMed] [Google Scholar]
- 2.Gong C., Bongiorno P., Martins A., Stephanou N. C., Zhu H., Shuman S., Glickman M. S. Nat. Struct. Mol. Biol. 2005;12:304–312. doi: 10.1038/nsmb915. [DOI] [PubMed] [Google Scholar]
- 3.Weller G. R., Kysela B., Roy R., Tonkin L. M., Scanlan E., Della M., Devine S. K., Day J. P., Wilkinson A., di Fagagna F., et al. Science. 2002;297:1686–1689. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- 4.Della M., Palmbos P. L., Tseng H. M., Tonkin L. M., Daley J. M., Topper L. M., Pitcher R. S., Tomkinson A. E., Wilson T. E., Doherty A. J. Science. 2004;306:683–685. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- 5.Aravind L., Koonin E. V. Genome Res. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doherty A. J., Jackson S. P., Weller G. R. FEBS Lett. 2001;500:186–188. doi: 10.1016/s0014-5793(01)02589-3. [DOI] [PubMed] [Google Scholar]
- 7.Zhu H., Shuman S. J. Biol. Chem. 2005;280:418–427. doi: 10.1074/jbc.M410110200. [DOI] [PubMed] [Google Scholar]
- 8.Zhu H., Shuman S. J. Biol. Chem. 2005;280:25973–25981. doi: 10.1074/jbc.M504002200. [DOI] [PubMed] [Google Scholar]
- 9.Zhu H., Shuman S. J. Biol. Chem. 2005;280:33707–33715. doi: 10.1074/jbc.M506838200. [DOI] [PubMed] [Google Scholar]
- 10.Augustin M. A., Huber R., Kaiser J. T. Nat. Struct. Biol. 2001;8:57–61. doi: 10.1038/83060. [DOI] [PubMed] [Google Scholar]
- 11.Ito N., Nureki O., Shirouzu M., Yokoyama S., Hanaola F. Genes Cells. 2003;8:913–923. doi: 10.1111/j.1365-2443.2003.00693.x. [DOI] [PubMed] [Google Scholar]
- 12.Holm L., Sander C. Nucleic Acids Res. 1999;27:244–247. doi: 10.1093/nar/27.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipps G., Weinzerl A. O., von Scheven G., Buchen C., Cramer P. Nat. Struct. Mol. Biol. 2004;11:157–162. doi: 10.1038/nsmb723. [DOI] [PubMed] [Google Scholar]
- 14.Ramadan K., Shevelev I., Hübscher U. Nat. Rev. Mol. Cell. Biol. 2004;5:1038–1043. doi: 10.1038/nrm1530. [DOI] [PubMed] [Google Scholar]
- 15.McElhinny S. A. N., Havener J. M., Garcia-Diaz M., Juárez R., Bebenek K., Kee B. L., Blanco L., Kunkel T. A., Ramsden D. A. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Daley J. M., Vander Lann R. L., Suresh A., Wilson T. E. J. Biol. Chem. 2005;280:29030–29037. doi: 10.1074/jbc.M505277200. [DOI] [PubMed] [Google Scholar]
- 17.Tseng H. M., Tomkinson A. E. J. Biol. Chem. 2002;277:45630–45637. doi: 10.1074/jbc.M206861200. [DOI] [PubMed] [Google Scholar]
- 18.Wilson T. E., Lieber M. R. J. Biol. Chem. 1999;274:23599–23609. doi: 10.1074/jbc.274.33.23599. [DOI] [PubMed] [Google Scholar]
- 19.Steitz T. A. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 20.Sawaya M. R., Prasad R., Wilson S. H., Kraut J., Pelletier H. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 21.Hendrickson W. A., Horton J. R., LeMaster D. M. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho C. K., Wang L. K., Lima C. D., Shuman S. Structure (London) 2004;12:327–339. doi: 10.1016/j.str.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Mossessova E., Lima C. D. Mol. Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Collaborative Computational Project. Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 26.Terwilliger T. C., Berendzen J. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Acta Crystallogr. A. 1991;47:110–118. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov G. N., Vagin A. A., Dodson E. J. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.DeLano W. L. The pymol User’s Manual. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.