Abstract
The cDNA obtained by selective capture of transcribed sequences is a complex mixture that can be used in conjunction with microarrays to determine global gene expression by a pathogen during infection. We used this method to study genes expressed by Salmonella enterica serovar Typhi, the etiological agent of typhoid fever, within human macrophages. Global expression profiles of Typhi grown in vitro and within macrophages at different time points were obtained and compared. Known virulence factors, such as the SPI-1- and SPI-2-encoded type III secretion systems, were found to be expressed as predicted during infection by Salmonella, which validated our data. Typhi inside macrophages showed increased expression of genes encoding resistance to antimicrobial peptides, used the glyoxylate bypass for fatty acid utilization, and did not induce the SOS response or the oxidative stress response. Genes coding for the flagellar apparatus, chemotaxis, and iron transport systems were down-regulated in vivo. Many cDNAs corresponding to genes with unknown functions were up-regulated inside human macrophages and will be important to consider for future studies to elucidate the intracellular lifestyle of this human-specific pathogen. Real-time quantitative PCR was consistent with the microarray results. The combined use of selective capture of transcribed sequences and microarrays is an effective way to determine the bacterial transcriptome in vivo and could be used to investigate transcriptional profiles of other bacterial pathogens without the need to recover many nanograms of bacterial mRNA from host and without increasing the multiplicity of infection beyond what is seen in nature.
Keywords: microarrays, in vivo bacterial gene expression, host-pathogen interaction
Salmonella infections are a significant cause of morbidity in humans and animals. Salmonella enterica serovar Typhi (hereafter referred to as Typhi) is the etiological agent of typhoid fever, a major health problem in developing countries, and causes an estimated 16 million cases and 600, 000 deaths annually worldwide (1). Publications on human typhoid that use modern immunological and molecular techniques are scarce. We have a very limited knowledge of the pathogenesis of Typhi because Typhi infects only humans, resulting in a lack of virulence assays. Many studies have relied on a murine model of human typhoid that uses S. enterica serovar Typhimurium (hereafter referred to as Typhimurium), which causes a typhoid-like disease in mice. Consequently, what is known about Typhi pathogenicity has been largely extrapolated from studies of Typhimurium infections in mice.
The Salmonella chromosome possesses insertions of large regions of DNA, containing virulence genes. These Salmonella pathogenicity islands (SPIs) play a key role in Salmonella pathogenesis (2). Thus far, 10 SPIs have been identified in Typhi (3). SPI-1 and SPI-2, which are present in all S. enterica serovars, represent two major pathogenesis determinants that encode type III secretion systems (TTSS). SPI-1 and SPI-2 TTSS have distinct roles in Salmonella pathogenesis. SPI-1 effectors are injected into host cells via the TTSS and are required for invasion of epithelial cells (4), whereas SPI-2 contributes to Salmonella survival inside macrophages (5, 6). The ability to survive and replicate within macrophages is thought to be one of the major pathogenesis determinants for Salmonella (7, 8). After entry into macrophages, Salmonella adapts by modifying its gene expression to respond to the host cell environment. It was recently shown that Typhimurium alters the transcription of 20% of its genome (919 genes) once inside murine macrophages (9). It was previously shown that Typhimurium survives better in murine macrophages than in human macrophages, whereas Typhi survives better in human macrophages than in murine macrophages (10). It is possible that Typhi expresses specific factors for survival in human macrophages. Genomic comparison between Typhi strain CT18 and Typhimurium strain LT2 revealed that there are 601 genes unique to Typhi and 479 genes unique to Typhimurium (3, 11).
Microarray analysis is a powerful tool to expand our current knowledge of global bacterial gene expression, but there are very few studies on bacterial transcriptional response during infection. There are many factors that prevent using DNA microarrays effectively for studying bacterial gene expression in infected host cells or tissues, such as the low amount of bacterial RNA in vivo, the short half-life of bacterial mRNA, and the contamination of bacterial mRNA with rRNA and host RNA (12). To overcome these problems, researchers have used different approaches that can create additional problems, such as models of infection that do not necessarily represent conditions found within host cells or tissues during the infectious process, and which may cause artifactual bacterial gene expression.
Selective capture of transcribed sequences (SCOTS) has been used to identify genes expressed in vivo by bacterial pathogens such as Mycobacterium (13, 14), Salmonella (15–17), Actinobacillus (18), Helicobacter pylori (19), and avian pathogenic Escherichia coli (20). In this report, we demonstrate that SCOTS can be used to obtain high-quality transcript profiles from intracellular bacteria such as Typhi within human macrophages, and that the use of microarrays and SCOTS-cDNA hybridization provides an effective means to elucidate the global bacterial expression profile from infected host cells.
Results
SCOTS-cDNA.
To verify whether SCOTS-derived cDNAs could be successfully used for microarray analysis, we hybridized each of the three rounds of SCOTS and the cDNA obtained from infected macrophages 2 h after infection (T2) to Salmonella arrays. The initial cDNA obtained from infected macrophages very weakly hybridized to the Salmonella microarrays, and only a few genes were detected (Fig. 1). This result demonstrates the inadequate sensitivity of labeled cDNA and illustrates limitations of direct identification of bacterial gene transcripts during interaction with host cells. The low number of genes detected in the initial cDNA and the low intensities of spots also confirmed the absence of contaminating bacterial genomic DNA, which was confirmed by PCR (data not shown), and the lack of host cDNA cross-hybridization. The number of detected genes increased in successive SCOTS rounds (Fig. 1, see numbers at the bottom of each column). This increasing complexity of transcripts was previously visualized by Southern blot (13). All cDNAs used for hybridization to microarrays were obtained by three rounds of SCOTS. Nearly 4, 000 genes were detected by using SCOTS-cDNA in each of the conditions, without regard to whether they are up-regulated or down-regulated relative to other growth conditions (see Fig. 1 legend for detection threshold). These results illustrate that the diversity of capture efficiencies were similar for the different conditions tested and that detection was largely independent of the quantity of bacteria recovered from samples (Fig. 4, which is published as supporting information on the PNAS web site). Similarly, detection of Mycobacterium tuberculosis transcripts by using microarrays identified 75% of genes that had measurable signal intensities (21).
Fig. 1.
Scan of the first subarray of the microarray slides hybridized with, from left to right, cDNA, 1× SCOTS, 2× SCOTS, and 3× SCOTS from 2 h. (A and B) Cy5 (cDNA) signal only (A) and both Cy5 (cDNA) and Cy3 (genomic DNA) signal (B). The number of genes detected after each round of SCOTS are at the bottom of each column. A set of 100 Typhimurium-specific genes were used to calculate the detection threshold. Genes with signals greater than the median of these 100 genes plus 3 standard deviations were considered to be detected.
The genomic content of Typhi strain ISP1820 was compared to strain CT18, the template used for the DNA array design. Seventy-six CT18 genes were missing or altered in the ISP1820 genome (Fig. 5, which is published as supporting information on the PNAS web site) and four regions (II, V, IX, and XIII) encoding prophage-like elements were not detected in strain ISP1820, as was observed in other Typhi strains (22).
In Vivo Global Transcription Profiles.
cDNA of Typhi present in the supernatants of infected macrophages was obtained by SCOTS and used as an in vitro control. Typhi cDNA was also obtained by SCOTS from infected macrophages after phagocytosis (T0), and intracellularly at 2 h (T2), 8 h (T8), and 24 h (T24) after infection. The gene expression profiles for intracellular Typhi were compared with the transcriptome of bacteria from the cell supernatants (Table 1, which is published as supporting information on the PNAS web site). During the time course of infection, 36% of the Typhi genome showed significant expression differences within macrophages compared to extracellular Typhi obtained from the cell supernatant. The number of Typhi genes down-regulated (2-fold) during infection, compared to bacteria from the cell supernatant, was 490, 474, 705, and 478 genes for T0, T2, T8, and T24, respectively. There were 1, 129 genes down-regulated by intracellular Typhi from at least one time point when compared to the supernatant (Table 2, which is published as supporting information on the PNAS web site), and 138 genes were repressed at all intracellular time points (Fig. 6A, which is published as supporting information on the PNAS web site).
Typhi gene transcripts corresponding to ≈300 different ORFs were more abundant (2-fold) at each time point (273, 309, 301, and 309 genes at T0, T2, T8, and T24, respectively). A total of 628 genes were up-regulated by intracellular Typhi at one or more time points (Table 3, which is published as supporting information on the PNAS web site). Some genes were more highly expressed only at one time point, others were up-regulated at two or more time points, and 117 Typhi genes were up-regulated at all intracellular time points (Fig. 6B). These intracellular expressed genes include SPI-2 genes (see below) and other genes involved in virulence, such as pagC, pagD, and mgtBC, and genes encoding osmotically induced proteins (osmC and osmE) and phage shock protein (psp).
SPI Expression.
SPI-1 encodes a TTSS required for invasion of nonphagocytic cells (4). We found that some SPI-1 genes were up-regulated during bacterial uptake by macrophages at T0, such as prg, sip, spa, and inv genes, and then down-regulated after internalization and for the rest of the infection (Fig. 2A). SPI-2 encodes a second TTSS that is required for Salmonella survival inside macrophages (5, 6). In our model, some SPI-2 genes were up-regulated after bacterial uptake by macrophages (T0), and most SPI-2 genes were up-regulated after 2 h of infection (Fig. 2B). Our results are consistent with the biological roles of both TTSS. Moreover, it is notable that genes encoding for SPI-1 effectors, such as sopE, sopE2, sopB(sigD), and sopD, or SPI-2 effectors such as pipB and sifB, demonstrated an expression profile similar to their respective TTSS even if encoded outside the SPIs. SPI-3-localized mgtBC genes, which encode a Mg2+ transporter required for full virulence of Typhimurium in mice (23), were found to be induced in macrophages. No significant difference in expression was observed for SPI-4 and SPI-9 components. The SPI-5 encoded gene pipD was overexpressed within macrophages at all time points. SPI-6 possesses a mixture of genes that were either up-regulated or repressed in vivo. A distinct feature of Typhi compared to Typhimurium is SPI-7, a 134-kb region (24) that encodes the type-IV pili involved in entry into intestinal cells (25) and the viaB locus responsible for the synthesis of the Vi-capsule, which is involved in the suppression of early inflammatory responses of intestinal cells (26). For SPI-7, 19 genes were up-regulated within macrophages, including sopE, a SPI-1 effector, and 47 genes were down-regulated within macrophages for at least one time point, including genes involved in Type-IV pili synthesis and genes involved in Vi capsule biosynthesis. Their role may be more important at the intestinal level than within macrophages. Most of the SPI-8- and SPI-10-encoded genes displayed signals below the background threshold.
Fig. 2.
Number of genes (%) from SPI-1 (A), SPI-2 (B), iron transport (C), motility (D), antimicrobial peptide resistance (E), and peroxide-induced (F) that were significantly induced (■) or repressed (○). See text for details.
Functional Classes.
Typhi genes that were differentially expressed within macrophages were grouped by their function or genetic locus according to the Typhi functional classification scheme used by the Sanger Centre (www.sanger.ac.uk) and the percentage of genes that were up-regulated or down-regulated within macrophages in each of these groups was calculated (Fig. 7, which is published as supporting information on the PNAS web site). Genes involved in iron acquisition and transport (such as fes, fhu, feo, and ent) were down-regulated intracellularly (Fig. 2C). Genes involved in chemotaxis and motility, like flagella, were up-regulated during bacterial uptake by macrophages (T0) and then were reduced once bacteria were within macrophages (Fig. 2D). Moreover, the majority of genes involved in response to antimicrobial peptides, including phoP, pmrF, ugtL, pqaB, pgtE, mig-14, and somA (ybjX) were significantly up-regulated in macrophages (Fig. 2E). In addition, the pagP, virK, and pmrD genes were also more expressed within macrophages than within the supernatant. From a list of 92 genes induced after treatment of Typhi with peroxide (27), only 18 were significantly up-regulated in vivo, including phoH and mgtC (Fig. 2F). The sodB gene was up-regulated at 2 h and 24 h after infection, however, expression of the genes encoding OxyR, the oxidative stress regulator and katEG and sodA genes, were not up-regulated by Typhi in macrophages. Response of Typhi to acid was investigated by looking at a list of genes known to be acid-inducible (28, 29) or associated with the acid tolerance response (ATR) (30). No clear differential expression can be seen. However, regulators involved in ATR, such as rpoS and phoP, were induced but are also involved in other functions.
Salmonella has many transcriptional regulators that control virulence gene expression (31), but significant changes in expression levels were observed only for a subset of regulatory genes in Typhi. phoP and the ssrBA genes, which encode a two-component regulator of SPI-2 gene expression, were strongly up-regulated from T0 to T24. A 2-fold increase was observed for regulatory genes such as rpoS, rpoH, fur, soxS, slyA, and phoB during infection. The expression of other regulators, such as rpoE, rpoD, hilA, barA/sirA, oxyR, cyaA, and crp was not significantly changed.
Quantitative Real-Time PCR (qPCR) Analysis.
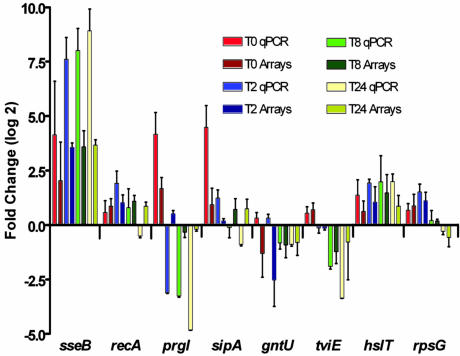
The results obtained with microarray experiments were confirmed by using qPCR. qPCR results were consistent with the microarray results (Fig. 3). As an example, both methods showed an increased expression of the SPI-2 gene sseB and a decreased expression of the SPI-1 gene prgI during infection. Normalization against 16s rRNA gave similar results (data not shown).
Fig. 3.
qPCR (light color) and microarray (dark color) results for a set of eight genes compared to supernatant. See text for details.
Discussion
This report investigated an approach to circumvent many of the technical problems encountered when studying bacterial gene transcription in vivo. We have demonstrated that SCOTS-cDNA represents a complex mixture that can be used for microarray analysis and allowed the study at different times after infection, not only those with relatively large numbers of bacteria, as required by other techniques (12). The transcriptomes of Typhi from macrophage cell culture supernatant (in vitro) or after macrophage uptake and survival at different times after infection were obtained and compared. Identification of Typhi gene transcription profiles within macrophages may elucidate the role of some Typhi genes, as adaptation and survival within macrophages was shown to be critical (8).
We determined that ≈300 Typhi genes were up-regulated at each time point compared to supernatant; however, only 117 genes were invariably up-regulated at the three intracellular time points. These results indicate that genes overexpressed at a defined time point may be required at different stages of internalization or survival within macrophages or may represent a nonsynchronization of the infection process even though all Typhi examined were internalized in the first 30 min. As an example, SPI-1 and SPI-2 TTSS have very different expression profiles, consistent with their respective function. The results obtained from microarray experiments were corroborated by qPCR. Two different technologies applied on different biological samples were consistent, supporting the validity of SCOTS-microarray analysis as an effective way to study host–pathogen interaction.
The molecular mechanism of typhoid fever has predominantly been indirectly derived from the Typhimurium murine typhoid fever model because these serovars share many virulence genes. Our data suggest that many virulence factors and global regulators common to both Typhi and Typhimurium are similarly up-regulated inside macrophages. The genes coding for the flagellar apparatus, chemotaxis, and iron transport systems were down-regulated by Typhi within macrophages, as described for Typhimurium in ref. 9, indicating that iron is available from the vacuole, which could explain why Typhi can afford to harbor many mutations in iron acquisition genes (3).
SPI-1 encodes a TTSS involved mainly in cell invasion and is an essential system for infection of epithelial cells of the gut (4). SPI-1 genes were down-regulated after bacterial internalization in macrophages as expected (Fig. 2A), and the expression profile of prgI, a SPI-1 encoded gene, was confirmed by qPCR (Fig. 3). The up-regulation of SPI-1 at the earliest times after invasion was not previously observed by Eriksson et al. (9), because the earliest time point investigated in that study was 4 h after infection. It is interesting to note that SPI-1 and genes involved in motility have similar expression patterns (Fig. 2 A and D), and transcription of flagella and SPI-1 genes are coregulated by fliZ (32), which is consistent with our results. SPI-1 genes up-regulated during uptake of Typhi by macrophages (T0) may contribute to Typhi invasion of macrophages, as shown for Typhimurium in ref. 33. SPI-1 effector SopE, which is present in some Typhimurium strains, recruits Rab5 to the Salmonella-containing phagosome, promoting continuous fusion with early endosomes and prevents fusion with lysosomes (34). In addition, macrophage apoptosis is thought to be mediated, in part, by SPI-1 effectors such as SipB (35). Thus, induction of SPI-1 during or immediately after invasion is not surprising. Another possible role of SPI-1 during macrophage infection was recently demonstrated by the loss of transposon mutants within SPI-1, such as inv, sip, and prg, after passage into macrophages (36). We may also hypothesize that some other genes that are not currently known to be effectors may be translocated by the TTSS system of SPI-1 and may contribute to the initial interaction of Typhi with macrophages. Two genes encoding for unknown protein, STY1482 and STY1353, that were up-regulated by Typhi within macrophages were recently identified as new TTSS effectors in Typhimurium (37) and named steA and steC, respectively. The detection of these genes confirms that our method can identify new virulence genes. Many cDNAs corresponding to genes with unknown functions or unique to Typhi were up-regulated inside human macrophages, including STY1361-STY1367, STY2000-STY2002, and some SPI-6 genes, and will be important to consider for future studies to elucidate the intracellular lifestyle of this human-specific pathogen.
Genes encoding the SPI-2 TTSS and the Mg2+ transport system (mgtBC located on SPI-3) were more highly expressed in vivo compared to supernatant, as was observed with Typhimurium inside murine macrophages in ref. 9. SPI-2 is well known to facilitate Salmonella survival inside macrophages (5, 6). Many genes encoded on SPI-2 were already up-regulated after bacterial uptake (T0), and other SPI-2 encoded genes were up-regulated only after 2 h of infection. Most of the SPI-2-encoding genes were overexpressed at all time points, consistent with their implication in creating and maintaining the Salmonella-containing vacuole. Some SPI-2 genes were not differentially expressed, such as the ttr genes, which encode a tetrathionate reduction system (38). In addition, transposon mutants of ttr genes of Typhimurium are not lost during infection of macrophages (36).
Most of the genes responsible for the SOS response were not differentially expressed by Typhi inside human macrophages compared to the supernatant. The SOS genes (recA, umuCD, uvrABY, sulA, and mutH) were induced by Typhimurium inside murine macrophages (9). However, strong inductions of dinI, the repressor of the SOS response, and no induction of other DNA repair systems were observed. It is unclear whether Typhi encounters oxidative stress inside human macrophages as systems involved to protect against oxidative stress, such as oxyR and katG, were not differentially expressed (Fig. 2F). No clear induction of genes involved in acid response was observed in Typhi. It is possible that Typhi already experiences some acid or oxidative stress and that the SOS response was already activated by Typhi cells in contact with macrophages or their metabolic products, present in the supernatant, our in vitro control.
An important host defense mechanism involves production of antimicrobial peptides. Responses to antimicrobial peptides involved the phoPQ two-component system regulator, the pmr operon (39), ugtL (40), pagP (41), pqaB (42), pgtE (43), virK and its homologue somA (ybjX) (44), and mig-14 (45, 46). These genes were up-regulated by Typhi in human macrophages (Fig. 2E). These data imply that Typhi strongly responds to antimicrobial peptides inside the vacuole of human macrophages by modifying its lipid A, as described for Typhimurium in ref. 47. However, among all of these genes, only pgtE up-regulation was identified by microarray analysis of gene expression of Typhimurium from murine macrophages (9). By contrast, most of the drug resistance systems, such as marRAB, emrRAB, aac, and aadA, were repressed or not differentially expressed by Typhi, whereas these systems were up-regulated by Typhimurium (9).
Other differences in gene expression between Typhi and Typhimurium were also identified from the microarray data (Table 4, which is published as supporting information on the PNAS web site). It is important to mention that the model from Erickson et al. (9) contains many technical differences from our model, including opsonization of bacterial cells, a higher multiplicity of infection and number of host cells, a murine macrophage cell line (J774-A.1), a different in vitro control (cell culture medium), a different way to obtain cDNA (differential lysis), and the time course only overlapped at 8 h after infection. Thus differences observed may be caused by one or more of these factors instead of the different serovar used.
There are two major regulators of Salmonella virulence genes required for survival in macrophages: slyA and phoP (8, 48–51). We detected significant induction of phoP and slyA in Typhi inside the macrophage, which was not seen in Typhimurium (9). A recent study identified 22 genes that were coregulated by SlyA and PhoP in Typhimurium (52). Among these genes, 62% of them were significantly up-regulated by Typhi within macrophages, including well known virulence determinants such as the SPI-2 genes, sseAB and their regulator ssrA, and many genes involved in resistance to antimicrobial peptides. In contrast, Typhimurium expressed only 25% of these genes.
It was also proposed that once inside macrophages, Typhimurium uses gluconate and related carbohydrates as carbon sources (9). However, Typhi lacks the dgo operon involved in the utilization of this sugar, and genes gntT and gntU, encoding transport proteins, were not up-regulated in vivo. Repression of gntU was confirmed by using qPCR (Fig. 3). Other sugar transport systems (hexose and fructose) were also repressed. Therefore, Typhi likely uses different carbon sources inside macrophages than Typhimurium. Recent studies showed that isocitrate lyase, which allow the utilization of fatty acid as a carbon source via the glyoxylate bypass, is needed for persistence of Mycobacterium tuberculosis in human macrophages and Typhimurium in the mouse model (53, 54). The isocitrate lyase gene, aceA or icl, was found by SCOTS to be expressed at 48 h after infection of macrophages by two species of Mycobacterium in refs. 13 and 14. In Typhi, the aceA gene was significantly induced starting at 8 h after infection. In Typhimurium, aceA induction was not detected 12 h after infection (9).
Our study demonstrates that by using SCOTS and microarray analysis, the transcriptome of intracellular bacteria can be obtained without altering the existing infection model. The method is an effective way to determine global bacterial gene expression profiling in the context of host infection. Its application on tissue biopsies from infected patients, as demonstrated with H. pylori in human biopsy samples (19), will be a potential means of further elucidating the bacterial genes expressed during infection of the human host. The SCOTS-cDNA mixture displayed an expected expression profile of Typhi virulence genes from infected macrophages. Our data suggest that inside THP-1 human macrophages Typhi responds to antimicrobial peptides but does not encounter an acidic or oxidative environment and that Typhi may use fatty acids as a carbon source as soon as 8 h after infection. Our global expression analysis identified many hypothetical and characterized Typhi genes that may contribute to adaptation and survival within macrophages, and such data are of importance for future experimental studies to elucidate the intracellular lifestyle of this human-specific pathogen.
Materials and Methods
Cell Culture and Bacterial Infection Model.
The human monocyte cell line THP-1 (ATCC TIB-202) was maintained and infected as described in ref. 55. For RNA extraction, cells were seeded at 2 × 107 in 100-mm-wide tissue culture dishes. The wild-type virulent Typhi strain ISP1820 (obtained from D. M. Hone, Aeras Global Tuberculosis Vaccine Institute, Rockville, MD) was used because it is a well characterized strain that is sensitive to most antibiotics that are used for bacterial genetics, whereas CT18 is multidrug-resistant. The strain was grown at 37°C overnight as a static culture in Luria–Bertani (LB) broth to an OD600 of 0.6 (≈3 × 108 colony-forming units/ml). Typhi cells were added to the THP-1 cell monolayer at a multiplicity of infection of 10 per cell, a low density that does not affect cell viability (data not shown). After incubation for 30 min at 37°C, the supernatant was centrifuged and the pellet was lysed with TRIzol reagent (Invitrogen) and frozen at −70°C. The infected cells were washed three times and incubated with complete RPMI medium 1640 (Invitrogen) initially containing 100 μg/ml gentamicin, which was reduced to 12 μg/ml after 2 h, to kill extracellular bacteria. At each time point, 0, 2, 8, and 24 h after infection, macrophage monolayers were washed, lysed in TRIzol, and stored at −70°C.
SCOTS.
RNA from each condition was extracted according to the TRIzol reagent manufacturer’s instructions (Invitrogen). RNA samples were then treated with RNase-free DNase (Ambion). Each RNA sample was converted to cDNA as described in refs. 13 and 55 (Fig. 8, which is published as supporting information on the PNAS web site). Briefly, 5 μg of total RNA was converted to first-strand cDNA by random priming by using a conserved primer containing a defined 5′ end and random nonamer at the 3′ end (16), with Superscript II (Invitrogen), according to the manufacturer’s instructions. Second-strand cDNA was synthesized by using Klenow fragment (New England Biolabs) according to the manufacturer’s instructions. Bacterial transcripts were separated from host cDNA by SCOTS, a selective hybridization to bacterial genomic DNA as described in refs. 13 and 55. Captured cDNA was eluted, precipitated, and amplified by PCR with the conserved primer added during reverse transcription. Noncompetitive PCR amplification of short random-primed DNA fragments with a single primer has been shown to yield a generally unbiased population of amplicons, even when applied to complex nucleic acid pools (56, 57). Three rounds of this capture hybridization were used to generate cDNA mixtures used as templates for microarray analysis.
Salmonella ORF Array.
Typhi strain CT18 array description, labeling and hybridization conditions, data acquisition, and normalization are described in ref. 27. Bacterial genomic DNA was used as the reference channel on each slide to allow comparison of each time point and different samples (58). The log ratio (base 2) was used for statistical analysis by using the ANOVA function with a standard Bonferonni correction by using tmev software (59). Data have been deposited in the NCBI Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession nos. GSE3094, GSE3095, and GSE3096.
qPCR Analysis.
The results obtained from microarray experiments were corroborated by qPCR experiments on a previously undescribed series of infection experiments (biological replicates). Infection and RNA extraction was performed as described above. cDNA was synthesized in triplicate by using SuperScript II (Invitrogen) with random hexamers (Sigma), according to the manufacturer’s instructions. For each sample, a no-reverse-transcriptase reaction served as a no-template control. qPCR was performed by using QuantiTect SybrGreen PCR Kit (Qiagen) according to the manufacturer’s instructions. Primers are described in Table 5, which is published as supporting information on the PNAS web site. For each qPCR run, the calculated threshold cycle (Ct) was normalized to the Ct of the internal control rpoD gene amplified from the corresponding sample, and the fold change was calculated as described in ref. 60. The alternative sigma factor rpoD was chosen as an internal control because no significant variation of expression for this gene in either Typhi (this study) or Typhimurium (9) was observed inside macrophages. In addition, expression of rpoD in E. coli was shown to be independent of growth phase (61).
Supplementary Material
Acknowledgments
This work was supported by the Banting Research Foundation and Canadian Natural Sciences and Engineering Research Council (NSERC) Grants (to F.D.). M.M. and S.P. were supported by National Institutes of Health Grants AI034829, AI057733, and AI052237. C.M.D. was supported by a Canada Research Chair and by the Canada Foundation for Innovation. S.P.F. was supported by an award from Fonds de la Recherche en Santé du Québec and NSERC.
Abbreviations
- qPCR
quantitative real-time PCR
- SCOTS
selective capture of transcribed sequences
- SPI
Salmonella pathogenicity island
- TTSS
type three secretion system
- Typhi
S. enterica serovar Typhi
- Typhimurium
S. enterica serovar Typhimurium
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The microarray data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE3094–GSE3096).
References
- 1.Parry C. M., Hien T. T., Dougan G., White N. J., Farrar J. J. N. Engl. J. Med. 2002;347:1770–1782. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- 2.Marcus S. L., Brumell J. H., Pfeifer C. G., Finlay B. B. Microbes. Infect. 2000;2:145–156. doi: 10.1016/s1286-4579(00)00273-2. [DOI] [PubMed] [Google Scholar]
- 3.Parkhill J., Dougan G., James K. D., Thomson N. R., Pickard D., Wain J., Churcher C., Mungall K. L., Bentley S. D., Holden M. T., et al. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 4.Galan J. E. Curr. Opin. Microbiol. 1999;2:46–50. doi: 10.1016/s1369-5274(99)80008-3. [DOI] [PubMed] [Google Scholar]
- 5.Cirillo D. M., Valdivia R. H., Monack D. M., Falkow S. Mol. Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 6.Hensel M., Shea J. E., Waterman S. R., Mundy R., Nikolaus T., Banks G., Vazquez-Torres A., Gleeson C., Fang F. C., Holden D. W. Mol. Microbiol. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 7.Schwan W. R., Huang X. Z., Hu L., Kopecko D. J. Infect. Immun. 2000;68:1005–1013. doi: 10.1128/iai.68.3.1005-1013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fields P. I., Swanson R. V., Haidaris C. G., Heffron F. Proc. Natl. Acad. Sci. USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eriksson S., Lucchini S., Thompson A., Rhen M., Hinton J. C. Mol. Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 10.Schwan W. R., Kopecko D. J. Adv. Exp. Med. Biol. 1997;412:277–278. doi: 10.1007/978-1-4899-1828-4_46. [DOI] [PubMed] [Google Scholar]
- 11.McClelland M., Sanderson K. E., Spieth J., Clifton S. W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., et al. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 12.Hinton J. C., Hautefort I., Eriksson S., Thompson A., Rhen M. Curr. Opin. Microbiol. 2004;7:277–282. doi: 10.1016/j.mib.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Graham J. E., Clark-Curtiss J. E. Proc. Natl. Acad. Sci. USA. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou J. Y., Graham J. E., Clark-Curtiss J. E. Infect. Immun. 2002;70:3714–3726. doi: 10.1128/IAI.70.7.3714-3726.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrow B. J., Graham J. E., Curtiss R., III Infect. Immun. 1999;67:5106–5116. doi: 10.1128/iai.67.10.5106-5116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daigle F., Graham J. E., Curtiss R., III Mol. Microbiol. 2001;41:1211–1222. doi: 10.1046/j.1365-2958.2001.02593.x. [DOI] [PubMed] [Google Scholar]
- 17.Faucher S. P., Curtiss R., III, Daigle F. Infect. Immun. 2005;73:5217–5221. doi: 10.1128/IAI.73.8.5217-5221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baltes N., Gerlach G. F. Infect. Immun. 2004;72:6711–6716. doi: 10.1128/IAI.72.11.6711-6716.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham J. E., Peek R. M., Jr., Krishna U., Cover T. L. Gastroenterology. 2002;123:1637–1648. doi: 10.1053/gast.2002.36589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dozois C. M., Daigle F., Curtiss R., III Proc. Natl. Acad. Sci. USA. 2003;100:247–252. doi: 10.1073/pnas.232686799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talaat A. M., Lyons R., Howard S. T., Johnston S. A. Proc. Natl. Acad. Sci. USA. 2004;101:4602–4607. doi: 10.1073/pnas.0306023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyd E. F., Porwollik S., Blackmer F., McClelland M. J. Clin. Microbiol. 2003;41:3823–3828. doi: 10.1128/JCM.41.8.3823-3828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanc-Potard A. B., Groisman E. A. EMBO J. 1997;16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bueno S. M., Santiviago C. A., Murillo A. A., Fuentes J. A., Trombert A. N., Rodas P. I., Youderian P., Mora G. C. J. Bacteriol. 2004;186:3202–3213. doi: 10.1128/JB.186.10.3202-3213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X. L., Tsui I. S., Yip C. M., Fung A. W., Wong D. K., Dai X., Yang Y., Hackett J., Morris C. Infect. Immun. 2000;68:3067–3073. doi: 10.1128/iai.68.6.3067-3073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma A., Qadri A. Proc. Natl. Acad. Sci. USA. 2004;101:17492–17497. doi: 10.1073/pnas.0407536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porwollik S., Frye J., Florea L. D., Blackmer F., McClelland M. Nucleic Acids Res. 2003;31:1869–1876. doi: 10.1093/nar/gkg298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park Y. K., Bearson B., Bang S. H., Bang I. S., Foster J. W. Mol. Microbiol. 1996;20:605–611. doi: 10.1046/j.1365-2958.1996.5441070.x. [DOI] [PubMed] [Google Scholar]
- 29.Lin J., Smith M. P., Chapin K. C., Baik H. S., Bennett G. N., Foster J. W. Appl. Environ. Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Audia J. P., Webb C. C., Foster J. W. Int. J. Med. Microbiol. 2001;291:97–106. doi: 10.1078/1438-4221-00106. [DOI] [PubMed] [Google Scholar]
- 31.Rhen M., Dorman C. J. Int. J. Med. Microbiol. 2005;294:487–502. doi: 10.1016/j.ijmm.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Iyoda S., Kamidoi T., Hirose K., Kutsukake K., Watanabe H. Microb. Pathog. 2001;30:81–90. doi: 10.1006/mpat.2000.0409. [DOI] [PubMed] [Google Scholar]
- 33.Cherayil B. J., McCormick B. A., Bosley J. Infect. Immun. 2000;68:5567–5574. doi: 10.1128/iai.68.10.5567-5574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee K., Parashuraman S., Raje M., Mukhopadhyay A. J. Biol. Chem. 2001;276:23607–23615. doi: 10.1074/jbc.M101034200. [DOI] [PubMed] [Google Scholar]
- 35.Hersh D., Monack D. M., Smith M. R., Ghori N., Falkow S., Zychlinsky A. Proc. Natl. Acad. Sci. USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan K., Kim C. C., Falkow S. Infect. Immun. 2005;73:5438–5449. doi: 10.1128/IAI.73.9.5438-5449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geddes K., Worley M., Niemann G., Heffron F. Infect. Immun. 2005;73:6260–6271. doi: 10.1128/IAI.73.10.6260-6271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hensel M., Hinsley A. P., Nikolaus T., Sawers G., Berks B. C. Mol. Microbiol. 1999;32:275–287. doi: 10.1046/j.1365-2958.1999.01345.x. [DOI] [PubMed] [Google Scholar]
- 39.Gunn J. S., Ryan S. S., Van Velkinburgh J. C., Ernst R. K., Miller S. I. Infect. Immun. 2000;68:6139–6146. doi: 10.1128/iai.68.11.6139-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Y., Cromie M. J., Hsu F. F., Turk J., Groisman E. A. Mol. Microbiol. 2004;53:229–241. doi: 10.1111/j.1365-2958.2004.04107.x. [DOI] [PubMed] [Google Scholar]
- 41.Guo L., Lim K. B., Poduje C. M., Daniel M., Gunn J. S., Hackett M., Miller S. I. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 42.Baker S. J., Gunn J. S., Morona R. Microbiology. 1999;145:367–378. doi: 10.1099/13500872-145-2-367. [DOI] [PubMed] [Google Scholar]
- 43.Guina T., Yi E. C., Wang H., Hackett M., Miller S. I. J. Bacteriol. 2000;182:4077–4086. doi: 10.1128/jb.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Detweiler C. S., Monack D. M., Brodsky I. E., Mathew H., Falkow S. Mol. Microbiol. 2003;48:385–400. doi: 10.1046/j.1365-2958.2003.03455.x. [DOI] [PubMed] [Google Scholar]
- 45.Brodsky I. E., Ernst R. K., Miller S. I., Falkow S. J. Bacteriol. 2002;184:3203–3213. doi: 10.1128/JB.184.12.3203-3213.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brodsky I. E., Ghori N., Falkow S., Monack D. Mol. Microbiol. 2005;55:954–972. doi: 10.1111/j.1365-2958.2004.04444.x. [DOI] [PubMed] [Google Scholar]
- 47.Gibbons H. S., Kalb S. R., Cotter R. J., Raetz C. R. Mol. Microbiol. 2005;55:425–440. doi: 10.1111/j.1365-2958.2004.04409.x. [DOI] [PubMed] [Google Scholar]
- 48.Libby S. J., Goebel W., Ludwig A., Buchmeier N., Bowe F., Fang F. C., Guiney D. G., Songer J. G., Heffron F. Proc. Natl. Acad. Sci. USA. 1994;91:489–493. doi: 10.1073/pnas.91.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watson P. R., Paulin S. M., Bland A. P., Libby S. J., Jones P. W., Wallis T. S. Infect. Immun. 1999;67:4950–4954. doi: 10.1128/iai.67.9.4950-4954.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller S. I., Kukral A. M., Mekalanos J. J. Proc. Natl. Acad. Sci. USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bijlsma J. J., Groisman E. A. Mol. Microbiol. 2005;57:85–96. doi: 10.1111/j.1365-2958.2005.04668.x. [DOI] [PubMed] [Google Scholar]
- 52.Navarre W. W., Halsey T. A., Walthers D., Frye J., McClelland M., Potter J. L., Kenney L. J., Gunn J. S., Fang F. C., Libby S. J. Mol. Microbiol. 2005;56:492–508. doi: 10.1111/j.1365-2958.2005.04553.x. [DOI] [PubMed] [Google Scholar]
- 53.McKinney J. D., Honer zu Bentrup K., Munoz-Elias E. J., Miczak A., Chen B., Chan W. T., Swenson D., Sacchettini J. C., Jacobs W. R., Jr., Russell D. G. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 54.Fang F. C., Libby S. J., Castor M. E., Fung A. M. Infect. Immun. 2005;73:2547–2549. doi: 10.1128/IAI.73.4.2547-2549.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daigle F., Hou J. Y., Clark-Curtiss J. E. Methods Enzymol. 2002;358:108–122. doi: 10.1016/s0076-6879(02)58083-6. [DOI] [PubMed] [Google Scholar]
- 56.Teng D. H., Hsu F., Peterson I., Cardon K. E., Caponigro G., Kamb A. BioTechniques. 2001;30:868–877. doi: 10.2144/01304rr04. [DOI] [PubMed] [Google Scholar]
- 57.Ko M. S., Ko S. B., Takahashi N., Nishiguchi K., Abe K. Nucleic Acids Res. 1990;18:4293–4294. doi: 10.1093/nar/18.14.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Talaat A. M., Howard S. T., Hale W., IV, Lyons R., Garner H., Johnston S. A. Nucleic Acids Res. 2002;30:e104. doi: 10.1093/nar/gnf103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 60.Livak K. J., Schmittgen T. D. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 61.Jishage M., Ishihama A. J. Bacteriol. 1995;177:6832–6835. doi: 10.1128/jb.177.23.6832-6835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.