Abstract
An effective measure to assess zinc status of humans has remained elusive, in contrast to iron, where a number of indicators of metabolism/function are available. Using monocytes, T lymphocytes, and granulocytes isolated by magnetic sorting and dried blood spots (DBS) derived from 50 μl of peripheral blood, we evaluated the response of metallothionein (MT), zinc transporter, and cytokine genes to a modest (15 mg of Zn per day) dietary zinc supplement in human subjects. Transcript abundance was measured by quantitative real-time RT-PCR (QRT-PCR). Zinc supplementation increased MT mRNA abundance by up to 2-fold in RNA from leukocyte subsets, and 4-fold in RNA from DBS. Transcript levels for the zinc transporter genes ZnT1 and Zip3 were increased and decreased, respectively, by zinc supplementation. Expression of the ZnT and Zip genes among leukocyte subsets differ by up to 270-fold. Monocytes and granulocytes from supplemented subjects were activated by LPS, whereas T lymphocytes were activated by mimicking antigen presentation. With zinc consumption, TNF-α and IL-1β expression was greater in activated monocytes and granulocytes, and IFN-γ mRNA levels were higher in activated T lymphocytes. These studies show that QRT-PCR is a tool to reliably measure transcript abundance for nutritionally responsive genes in human subjects, and that a small sample of whole dried blood, when appropriately collected, can be used as the source of total RNA for QRT-PCR analysis. The results obtained also show that zinc supplementation of human subjects programs specific leukocytic subsets to show enhanced cytokine expression upon activation by stimulators of immunity.
Keywords: granulocytes, monocytes, nutrition, quantitative RT-PCR, T lymphocytes
Methodological advances to assess gene expression have provided a new spectrum of research tools to identify individual genes and groups of genes that produce normal and altered physiology (1, 2). Quantitative real-time RT-PCR (QRT-PCR) provides a highly sensitive and reproducible method for measuring changes in expression of specific genes through transcript sequence detection. Analytical sensitivity of QRT-PCR rests with the fluorometric basis of this technology, which markedly reduces sample size requirements. The latter makes QRT-PCR an attractive method for clinical, survey, or field studies where the amount of sample is limited.
Small amounts of blood, spotted onto filter paper and dried, have been used to examine the blood cell levels of two vitamins (folic acid and retinol) for the purpose of nutritional status assessment of human subjects (3, 4). Furthermore, dried blood spots (DBS) extracted from collection cards have routinely been used for DNA amplification for population screening, e.g., genotyping and for DNA archiving (5, 6), but have not been widely used for RT-PCR (7–9). Such an approach of sample acquisition could similarly be used to measure transcript abundance for nutritionally regulated genes, and, therefore, be of value for nutrient assessment purposes. In this regard, we were successful in measuring metallothionein (MT) mRNA abundance by competitive PCR using total RNA extracted from DBS obtained from men taking a dietary zinc supplement, and showed the zinc responsiveness of transcript abundance for metallothionein (10). The combination of QRT-PCR analysis and DBS sample collection methods has the potential to be a powerful aid for evaluating the influence of environmental factors on gene expression. The experiments reported here use DBS sampling, as well as nonactivated and activated blood leukocyte populations separated by gradients and magnetic cell sorting, to examine by QRT-PCR transcript levels for MT, cytokines, and zinc transporter genes in response to elevated zinc consumption by human subjects.
Results
Plots of fluorescence produced by amplicon synthesis versus cycle number for MT mRNA detection by QRT-PCR are shown in Fig. 6A, which is published as supporting information on the PNAS web site These data were derived from total RNA extracted from DBS, and used the MT primers and probes described in Table 2, which is published as supporting information on the PNAS web site. There is a two-cycle number difference obtained with comparable amounts of total RNA from control subjects compared to RNA from zinc-supplemented subjects. This indicates that the MT mRNA abundance in these two samples differs by a factor of four. Also presented is a representative standard curve produced with RNA from human THP-1 cells (Fig. 6B). The intraassay coefficient of variation (CV) was 4.4% and the interassay CV was 9.3%. These values are comparable with other fluorogenic probe-based QRT-PCR results (11).
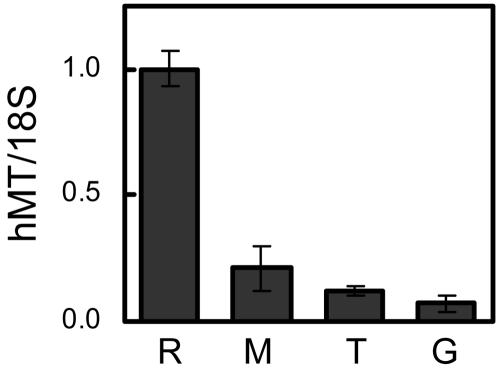
To evaluate the spectrum of zinc responsiveness of the MT genes in leukocyte populations, we first compared MT transcript levels in isolated granulocytes, monocytes, and T lymphocytes from control subjects. The cell purification methods used provided a purity of 98–100%. Transcript abundance values reported in Fig. 1, and elsewhere in this report, are relative to those levels obtained from the human reference RNA standard. Of note is that, when equal amounts of total RNA are compared, MT mRNA levels in leukocyte populations are relatively low. The most likely reason is the level of MT mRNA produced by the different cell lines used to prepare the reference RNA. Nevertheless, MT transcript abundance in monocytes is three times that found in granulocytes, and twice that found in T lymphocytes. This finding supports previous observations obtained with less quantitative methodology (10, 12).
Fig. 1.
Comparison of MT mRNA levels in monocytes (M), T lymphocytes (T), and granulocytes (G) of control human subjects with levels in a human reference RNA (R). Values are means ± SD (n = 3).
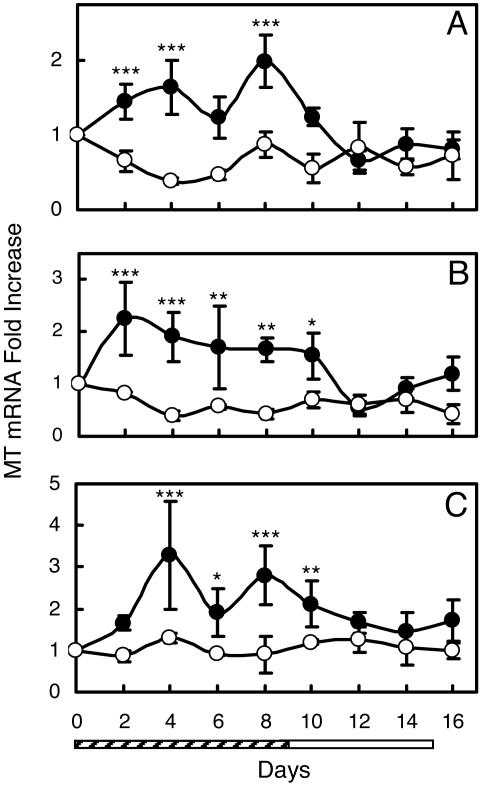
Supplementation of volunteer subjects with zinc at 15 mg/day produced an increase in MT transcript levels for each of the cell populations (Fig. 2). The kinetics of change in MT transcript abundance in these leukocyte populations during zinc supplementation and upon withdrawal are shown in Fig. 2. The kinetics differ by leukocyte subset but, in each case, there is a significant (P < 0.05 to <0.001) difference between supplemented and control subjects. No significant difference was shown among the control subjects. When the 10-day zinc supplementation period was over, the MT mRNA levels returned to the basal level within 6 days in each cell type. The biphasic response of MT mRNA in monocytes and granulocytes suggests that these cells receive zinc from similar pools (Fig. 2 A and C). This finding is in contrast to T lymphocyte MT mRNA, which was constant throughout zinc supplementation (Fig. 2B).
Fig. 2.
Relative MT mRNA levels in leukocyte subsets isolated from human subjects supplemented with dietary zinc. Values are means ± SD (n = 3–6 per group). Statistical difference from control is indicated (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001). Control subjects (open circles) were given placebo for 16 days. Supplemented subjects (filled) were given Zn (15 mg/day) for 10 days (hatched bar) and placebo for 6 days (open bar). (A) Monocytes. (B) T lymphocytes. (C) Granulocytes.
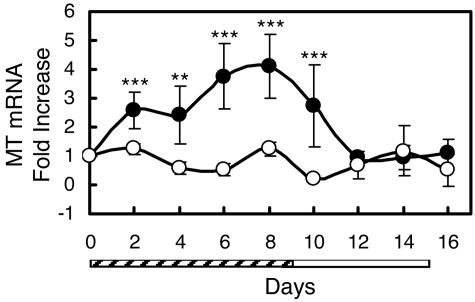
Total RNA from DBS can be used to collectively and rapidly assess the effect of the zinc supplement on MT transcript abundance in all blood cells. As shown in Fig. 3, the kinetics of transcript induction during zinc treatment and reduction when supplemental zinc is withdrawn is similar to those for isolated leukocyte populations. Although monocytes do not comprise a large proportion of total leukocytes, the induction of MT mRNA represented in RNA from DBS is similar to that found for monocytes (Fig. 2A). Of interest from an applied perspective is that MT transcripts in DBS total RNA increase 4-fold by day 8 of zinc supplementation. This response to zinc is greater than that observed with the three leukocyte subsets. This could result from the rapid stabilization of RNA with the DBS technique. Using RNA from DBS, the intra- and interassay coefficient of variation for determination of relative MT mRNA abundance was 8.7% (n = 3) and 13.8% (n = 3), respectively. We observed that storage of the DBS for up to 3 months at −75°C did not result in a loss of MT transcript abundance (data not shown).
Fig. 3.
Relative MT transcript levels in RNA from dried spots of whole blood (DBS) from human subjects supplemented with dietary zinc. Values are means ± SD (n = 3–6 per group). Statistically different from control (∗∗, P < 0.01; ∗∗∗, P < 0.001). Control subjects (open circles) were given placebo for 16 days. Supplemented subjects (filled circles) were given Zn (15 mg/day) for 10 days (hatched bar) and placebo for 6 days (open bar).
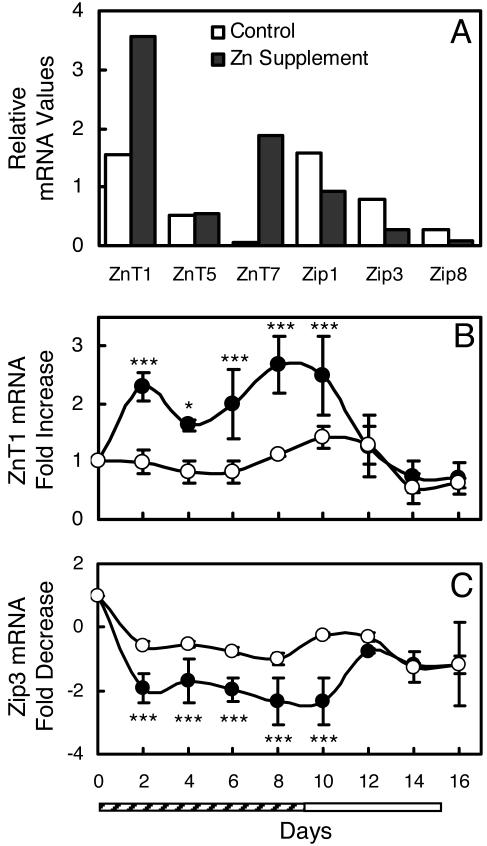
In rodent models, a number of zinc transporter genes have been shown to be responsive to dietary zinc intake (13–16). To investigate the zinc responsiveness of these genes in humans, the relative expression of 12 zinc transporter genes in DBS from whole blood was investigated (Fig. 4A). Of those, six mRNAs were of low abundance and did not amplify well with the limited amount of total RNA from the DBS. Because ZnT1 and Zip3 were quite responsive to zinc, but in opposite direction, we examined the abundance of these transcripts in total RNA from DBS during the entire 16-day supplementation protocol. Upon receiving zinc, the ZnT1 mRNA levels increased to 2.5-fold that found in control subjects (Fig. 4B). Decreased Zip3 transcripts were found in DBS RNA from zinc-supplemented subjects (Fig. 4C).
Fig. 4.
Relative ZnT and Zip transcript levels in RNA from dried spots of whole blood (DBS) from human subjects supplemented with dietary zinc or the placebo. (A) Pooled DBS from subjects that were supplemented for 8 days. (B and C) Control subjects (open circles) were given placebo for 16 days. Supplemented subjects (filled circles) were given Zn (15 mg/day) for 10 days (hatched bar) and placebo for 6 days (open bar). Values are means ± SD (n = 3–4 per group). Statistically different from control (∗, P < 0.05; ∗∗∗, P < 0.001). (B) ZnT1 mRNA. (C) Zip3 mRNA.
Abundance of ZnT and Zip mRNAs among the leukocyte subsets varied markedly. These differences are illustrated in Table 1, where relative transcript levels for nine of these genes in leukocyte subsets from a representative blood sample are shown. The values shown are quantities for each transporter mRNA relative to that of the human reference RNA. Efficiency of QRT-PCR for each primer/probe set may vary, so the absolute abundance may vary among the compared transcripts. Nevertheless, the data point out major differences in zinc transporter gene expression among human leukocytic subsets. Monocytes have the greatest abundance of ZnT1, Zip1, and Zip3 mRNAs. Expression of ZnT5, ZnT6, ZnT7, and Zip4 is relatively uniform in each cell type. Abundance of Zip2, Zip5, and ZnT2 mRNAs were below the level of detection. Most striking in these comparisons is much greater expression of Zip8 and Zip14 in T lymphocytes. Between granulocytes and T lymphocytes, Zip8 and Zip14 mRNA levels differ by 269- and 98-fold, respectively. These major differences must have some teleologic basis, and will be a new avenue of inquiry.
Table 1.
Comparison of relative expression of genes from the ZnT and Zip families in human monocytes, T lymphocytes, and granulocytes
| Transcript | Monocytes | T lymphocytes | Granulocytes |
|---|---|---|---|
| ZnT1 | 14.2 ± 0.6 | 1.3 ± 0.1 | 2.8 ± 0.4 |
| ZnT5 | 1.9 ± 0.1 | 1.4 ± 0.1 | 1.2 ± 0.2 |
| ZnT6 | 0.5 ± 0.03 | 1.6 ± 0.3 | 0.4 ± 0.07 |
| ZnT7 | 4.1 ± 0.4 | 3.7 ± 0.5 | 1.7 ± 0.2 |
| Zip1 | 7.5 ± 1.5 | 1.2 ± 0.1 | 2.7 ± 0.5 |
| Zip3 | 7.1 ± 0.3 | 2.1 ± 0.4 | 1.4 ± 0.2 |
| Zip4 | 0.05 ± 0.01 | 0.02 ± 0.006 | 0.06 ± 0.008 |
| Zip8 | 1.0 ± 0.3 | 10.2 ± 1.6 | 0.04 ± 0.007 |
| Zip14 | 0.05 ± 0.02 | 0.39 ± 0.03 | 0.004 ± 0.0007 |
Values are mean ± SD, n = 3. Transcript abundance has been normalized to 18S rRNA and calibrated to a human reference total RNA, where quantities for each transcript were placed at 1.0.
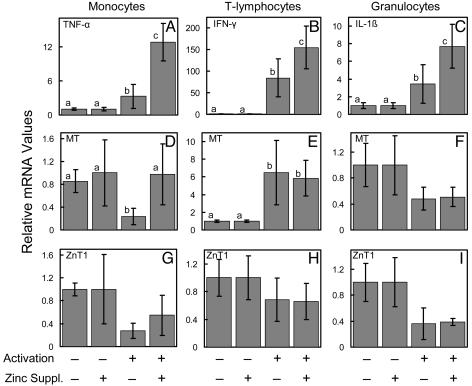
Activation of the monocytes and granulocytes with LPS and T lymphocytes by antigen presentation produces elevation of TNF-α, IL-1β, and IFN-γ transcripts, respectively. These are marker cytokines for these leukocytic subsets. As shown in Fig. 5, at day 4, cells from zinc-supplemented subjects had a greater (P < 0.05 to P < 0.001) abundance of these cytokine transcripts than those cells from controls (Fig. 5 A–C). This in vivo effect of dietary zinc was not evident in nonactivated cells. It is of interest that the effects of zinc supplementation of the subjects in this study on MT and ZnT1 expression observed with freshly isolated cells (Figs. 2 and 4) does not remain after the cells are placed in culture for activation studies (Fig. 5 D–I). Activation decreased (P < 0.05) MT mRNA in monocytes of the control subjects (Fig. 5D), whereas activation increased (P < 0.05) MT transcripts in T lymphocytes from control and zinc-supplemented subjects (Fig. 5E). No other effects (P > 0.05) of activation were found.
Fig. 5.
Relative selected cytokine, MT, and ZnT1 transcript levels in nonactivated and activated monocytes, T lymphocytes, and granulocytes from human subjects supplemented with dietary zinc. Values are means ± SD (n = 3–6 per group). Within each panel, statistical differences at P < 0.05 are indicated by a different superscript. In vitro activation was assessed for monocytes, T lymphocytes, and granulocytes by TNF-α (A), IFN-γ (B), and IL-1β (C) expression, respectively. MT transcripts for these leukocyte subsets (D–F, respectively) are also shown. ZnT1 transcripts are in G–I, respectively. The cells were obtained from human subjects who were given Zn (15 mg/day) or a placebo for 4 days. The cells were then activated in vitro.
Discussion
Zinc has been widely used as an effective agent for decreasing the morbidity of infectious disease worldwide (17). Unfortunately, the only laboratory measure of the effectiveness of these interventions is the plasma/serum zinc concentration. Over a period of nearly 50 years, plasma (serum) zinc concentrations have repeatedly been shown to be a poor index of zinc nutritional status (10, 12, 18, 19). In some contexts, the measure may be of some value (20–23). The exceptions are during acute illness or acute starvation, where they transiently decrease and increase (19). A transient increase in plasma zinc levels also occurs during zinc supplementation (10). This increase may be more evident if status is initially low (22). We have demonstrated, through the data presented here, that a modest supplement of zinc produces a variety of changes in transcript abundance of MT and zinc transporter genes in leukocyte subsets and whole blood (DBS) that are measurable by QRT-PCR. These provide targets that could be exploited as alternative zinc-responsive measures.
Whole blood and specific blood cell subsets have received recent attention as sources of RNA for microarray experiments to obtain information about disease-specific global changes in gene expression. Both whole blood and leukocyte subsets have been used (1 2). Among the information obtained is that genes involved in protein synthesis were more highly expressed in mononuclear cells; individual gene expression patterns depended on time of day, but variation within subjects was relatively low; and leukocyte subsets differ considerably in expression patterns. These basic characteristics should transfer to transcript abundance measurements of specific individual genes, such as those using QRT-PCR. As with any new technology, QRT-PCR is undergoing continued refinements (24, 25), but this methodology provides a dynamic range and sensitivity needed for measuring subtle differences in expression of nutritionally responsive genes with very small samples of blood (26, 27).
We have shown previously that MT mRNA is responsive to dietary zinc supplementation in humans (10, 12). Studies with rodents have shown that MT mRNA levels in many tissues are responsive to the amount of zinc provided in the diet. In mice, pancreatic MT mRNA is responsive to both deficient and excessive intakes in a linear fashion (28). It is apparent from the experiments here that zinc consumption by human subjects increases MT mRNA in the three major leukocyte subsets in peripheral blood. The differential MT mRNA expression shown within leukocyte subsets could reflect their divergent origins. For example, monocytes and granulocytes show similar peaks of maximum expression. This could originate with the common progenitor stem cells in the bone marrow, which differentiate into specific cell types through stimulation with different colony-stimulating factors. The mechanistic origin of these biphasic responses is not clear. The difference in half-life (6 h for granulocytes and 72 h for monocytes) could be a factor. That the highest-fold levels were found in granulocytes could also be related to a more rapid cell turnover. The plateau in MT mRNA levels in T lymphocytes during zinc supplementation could reflect the source of zinc providing the signal for transcription, which may originate from one pool that provides a constant elevated supply of zinc.
The DBS produced in these studies were collected on Whatman FTA cards. These are paper based, and are prepared with a proprietary formula that lyses cell membranes (including those of pathogens) denatures proteins, and stabilizes nucleic acids. They have been used as a source for RNA in virology and the plant sciences (9), but have not been widely used for studies with human cellular RNA. The experiments reported here show mRNA integrity is sufficient to allow for amplification by QRT-PCR. However, the latter does not have the stringent criteria for integrity as Northern analysis, because partially fragmented RNA can be analyzed by QRT-PCR. Furthermore, the PCR primers used were designed to generate amplicons of <100 nt in length. Nevertheless, the RNA must be free of salts and other contaminants. The DBS extraction procedure outlined in Supporting Text, which is published as supporting information on the PNAS web site, produces the RNA quality needed for QRT-PCR. Of note in this regard is the difference in magnitude observed in MT mRNA abundance between DBS-derived RNA vs. that from the leukocyte subsets. The latter require up to 3 h to prepare, whereas cells placed on FTA cards are lysed rapidly and RNase activity appears to be minimal. That difference could account for the difference in magnitude of transcript abundance. It is also remotely possible that some other cell type (e.g., beta cells, eosinophils, or platelets) not represented in the granulocyte, monocyte, and T lymphocyte populations examined here produce large amounts of MT mRNA. Differences in gene expression profiles were also found by others when whole blood vs. isolated leukocytes were compared (1, 2).
Variation encountered with QRT-PCR has been the subject of considerable discussion (9, 24, 25). The variation encountered in these experiments was relatively low, despite a modest number of subjects in the two treatment groups. The longitudinal data were analyzed by repeated-measures ANOVA. However, the variation encountered was such that large numbers of samples were not required. A number of factors may have contributed to this result. Molecular biology-grade solvents were used for RNA extraction steps, optimization steps led to low salt content of the final RNA sample, linear acrylamide was used as a precipitation aid, and one analyst performed all of the assays. In addition, we devised sequences for an 18S rRNA primer/pair set. This improved our assays where some residual salt remained.
A unique feature of the data reported here is that, whereas leukocyte MT and ZnT1 respond to dietary zinc in a positive mode, Zip1 and Zip3 respond in a negative mode. This finding suggests that, when dietary zinc intake is below an adequate amount, MT and ZnT1 transcript levels would decrease, and Zip1 and Zip3 transcripts would increase. This finding agrees with the roles of ZnT1 in regulating zinc efflux from cells, of MT in providing a pool of intracellular zinc, and of the Zip transporters in influencing zinc uptake. The mechanism for the positive mode of zinc regulation of MT and ZnT1 genes is believed to be via zinc occupancy of a binding site of the metal-responsive transcription factor MTF-1 (29). In contrast, regulation of the Zip genes is not clear. An induction of Zip4 expression in mice occurs during dietary zinc depletion (14, 16). Mutations of the human Zip4 gene leads to zinc malabsorption in a phenotype called Acrodermatitis Enteropathica (30). The down-regulation of Zip1 and Zip3 described in these zinc-supplemented human subjects suggests that a common zinc-sensing mode of regulation may be common to many genes of the Zip family. Previously, we observed an up-regulation of Zip2 expression in zinc-depleted THP-1 cells (31). A series of zinc-responsive genes, showing positive and negative modes of regulation, could provide flexibility for zinc status assessment.
A considerable literature base exists for the influence of zinc on host defense. A particular focus has been on cells involved with immune function (32–35). Much of the data have been acquired from peripheral blood mononuclear cells (PBMC) of human subjects or cultured human leukocytic cell lines. For example, in vitro addition of zinc at 50–450 μM to human PBMC cultures increases IFN-γ, IL-1β, and TNF-α production and TNF-α mRNA expression (35). Not all studies have shown an in vitro effect of zinc, as the responses are concentration dependent (32, 33). In vivo experiments suggest that zinc deficiency decreases chemotaxis and oxidative burst by neutrophils and phagocytosis by macrophages (33). Our experiments in this report show that consumption of a modest amount of supplemental zinc has a measurable influence on immune cell activation as demonstrated through changes in cytokine gene transcript abundance. For our studies, normalization was to 18S rRNA, which proved to be satisfactory. In some studies, cytokine transcript analyses by QRT-PCR have used normalization to housekeeping genes (11, 36). In addition to possible roles in immunity, it has been proposed that cytoplasmic Zn2+ acts in the signal transduction pathway for T lymphocyte activation. Specifically, upon antigen presentation, the cysteine-rich motif of the T-cell receptor (CD4) coordinates Zn2+ binding with a comparable motif of cytoplasmic protein-tyrosine kinase (p56lck) (37). The zinc-responsive enhanced T lymphocyte activation we found with the zinc-supplemented subjects (Fig. 5B) supports such a mechanism. Because immune activation produced some changes in the transcript levels for MT shown in these studies, future studies will need to evaluate the effect of infection on the zinc-responsive measures (MT, ZnT1, and Zip3 mRNAs).
The increase in cytokine expression in the activated leukocyte subsets produced by zinc supplementation is of interest from a clinical perspective. It has been observed, for example, that zinc administered in supplemental amounts via parenteral nutrition increases the febrile response of the acute phase response of patients (38). Because TNF-α and IL-1β are among the cytokines characterized as having endogenous pyrogen activity, their increased expression in the zinc-supplemented subjects in this study supports the immunostimulatory role for zinc. In addition, the antiapoptotic effects of zinc have been used as a therapy during sepsis. However, zinc administration during the proinflammatory stage of septic shock produces increases in TNF-α that influences sepsis-related organ failure (39). Consequently, the increases in expression of the marker proinflammatory cytokines during zinc supplementation, as observed in the present studies, may not always be of benefit to the host. These effects of zinc need to be further explored.
Materials and Methods
Subject Supplementation and Blood Collection and Processing.
Young, healthy men between 19 and 31 years of age gave informed consent for this study, which was approved by the University of Florida Institutional Review Board. Exclusion criteria and subject guidelines were as reported (10). For the first 10 days of the 16-day study, each subject consumed a zinc supplement (15 mg of Zn as ZnSO4) or a placebo between 7:00 and 8:00 a.m. after an overnight fast. Compliance was assessed by observation. All subjects received the placebo capsule for the final 6 days of the study period. Blood was sampled before the first supplement and every other day thereafter. Venous blood (10 ml) was collected in Vacutainer tubes with K3 EDTA. Heparin inhibits Taq DNA polymerase (40), and was not used as an anticoagulant.
DBS were prepared by using aliquots (50 μl) of whole blood applied to FTA Mini Cards (Whatman) and allowed to air dry for 3–4 h. The cards were stored at −75°C. To isolate RNA, DBS of 12 mm diameter were placed in buffer (100 mM Tris·Cl/0.1 mM EDTA/2 mM DTT) and incubated on ice. After 15 min, TRIzol Reagent (Invitrogen) was added (2 ml), and DBS were incubated at 37°C for 30 min. After subsequent extraction with chloroform and additions of linear acrylamide (Ambion) and isopropyl alcohol, the RNA was precipitated overnight at −20°C. The final total RNA sample was stored in water at −75°C. Details of the isolation procedure and purity assessment can be found in Supporting Text.
Remaining whole blood (>9 ml) was mixed (1:1, vol/vol) with PBS, and the diluted blood was placed on top of 10 ml of Histopaque 1.077 (Sigma) and centrifuged at 600 × g at room temperature for 30 min. The peripheral blood mononuclear cells at the interface were removed and washed two times with PBS using 250 × g for 10 min, then suspended in 70 μl of MACS buffer (PBS, pH 7.2, with 0.5% BSA and 2 mM EDTA). An aliquot (30 μl) was mixed with a cocktail of antibodies against nonmonocytic cells bound to magnetically labeled microbeads (Miltenyi Biotec) for isolation of untouched monocytes. The remaining cells (40 μl) were similarly labeled with other antibodies/microbeads (Miltenyi Biotec) to isolate untouched T lymphocytes. After purification by negative magnetic selection as recommended by the manufacturer, the respective cell populations were harvested at 310 × g (10 min) and placed in TRIzol (1 ml). Immediately after removal of the peripheral blood mononuclear cell-rich interface, the leukocytes in the layer above the red blood cell pellet were collected in a lysis buffer (155 mM NH4Cl/10 mM KHCO3/0.1 mM EDTA), washed in PBS as above, and suspended in MACS buffer. Antibodies to human CD15, conjugated to microbeads (Miltenyi Biotech), were added. The CD15-expressing cells (granulocytes) were recovered by positive magnetic selection, harvested at 310 × g (10 min), and placed in TRIzol (1 ml). Purity of monocytes, T lymphocytes, and granulocytes isolated by these methods was evaluated by flow cytometry using FITC-labeled antibodies against CD14, CD3, and CD15, respectively.
To evaluate the effects of activation, monocytes and granulocytes were placed in RPMI medium 1640 containing 10% FBS or 15% human AB serum, respectively. After 3 h at 37°C and 5% CO2, fresh medium containing LPS (10 μg/ml; Sigma; E. coli serotype 0127:B8) was added for up to 2 h. Similarly, T lymphocytes were placed in X-VIVO15 medium (Cambrex) supplemented with 5% human AB serum. For activation, the T lymphocytes were incubated for 2 days with microbeads conjugated to antibodies against human CD2, CD3, and CD28 (Miltenyi) to simulate antigen presentation. After activation of each cell type, the media were removed and TRIzol (1 ml) was added to each culture dish. Total RNA was isolated as described above.
Quantitative RT-PCR.
To protect against residual DNA contamination, all RNA samples were treated with Turbo DNA-free reagents (Ambion) as described by the manufacturer. Primers and TaqMan probes were designed by using primer express, version 2.0 (Applied Biosciences). The protein coding region of hMT-1 and -2 (GenBank accession nos. NM005950 and X97260) was originally targeted to generate QRT-PCR primers and probes. Although these were satisfactory for samples with relatively high amounts of MT mRNA (31), there was a marked loss of amplification efficiency in small samples with low abundance. Furthermore, MT transcript abundance data from human mononuclear cells obtained by microarray analysis (Affymetrix U-133A) revealed the most highly expressed MT mRNAs were for MT1X, -1H-like, -1F, -1H, -1L, -1G, and MT2 (31). No common amplicon region for all of these isoforms was identified. A combination of two forward primers, two reverse primers, and two TaqMan probes provided matches for MT1H, -1H-like, -1G, -1L, -1E, -1A, and MT2 (Table 2). For the assay, the four primers were at 450 nM each, and the two probes were each at 125 nM. Compared to the original MT mRNA assay (31), these reagents allowed for better detection from DBS and shifted the threshold cycle (Ct) down two cycles (i.e., fourfold increase in sensitivity. Primer/probes for hZnT2, hZnT4, hZip5, hZip8, hZip14, hTNF-α, hIFN-γ, and hIL-1β are shown in Table 2, and those for other ZnT and Zip transporter genes were described in ref. 31. These were used at 900 and 250 nM, respectively. All assays were conducted as one-step reverse-transcriptase reactions (Applied Biosystems), with fluorescence measured with Bio-Rad or Applied Biosystems instruments. Relative quantitation for all assays used 18S rRNA as the normalizer, and a human reference total RNA (Stratagene) was used as the interassay calibrator.
Statistical Analysis.
Data are expressed as means ± SD. Comparisons among subject groups used ANOVA with Student–Newman–Keuls post test. Longitudinal data were analyzed by repeated-measures ANOVA. Significance was set at P < 0.05.
Supplementary Material
Acknowledgments
We thank Mitchell D. Knutson, Juan P. Liuzzi, Bobbi Langkamp-Henken, and Calvert L. Green for helpful discussions. We thank Bhavna Bhardwaj of the University of Florida Flow Cytometry Core Facility for evaluation of leukocyte subset purity and Virginia Mauldin for help with the granulocyte preparation. This research was supported in part by National Institutes of Health Grant DK 31127 and Boston Family Endowment Funds (to R.J.C.).
Abbreviations
- QRT-PCR
quantitative RT-PCR
- DBS
dried blood spots
- MT
metallothionein.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Whitney A. R., Diehn M., Popper S. J., Alizadeh A. A., Boldrick J. C., Relman D. A., Brown P. O. Proc. Natl. Acad. Sci. USA. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cobb J. P., Mindrinos M. N., Miller-Graziano C., Calvano S. E., Baker H. V., Xiao W., Laudanski K., Brownstein B. H., Elson C. M., Hayden D. L., et al. Proc. Natl. Acad. Sci. USA. 2005;102:4801–4806. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Broin S. D., Gunter E. W. Am. J. Clin. Nutr. 1999;70:359–367. doi: 10.1093/ajcn/70.3.359. [DOI] [PubMed] [Google Scholar]
- 4.Craft N. E., Bulux J., Valdez C., Li Y., Solomons N. W. Am. J. Clin. Nutr. 2000;72:450–454. doi: 10.1093/ajcn/72.2.450. [DOI] [PubMed] [Google Scholar]
- 5.Kline M. C., Duewer D. L., Redman J. W., Butler J. M., Boyer D. A. Anal. Chem. 2002;74:1863–1869. doi: 10.1021/ac015715e. [DOI] [PubMed] [Google Scholar]
- 6.Steinberg K., Beck J., Nickerson D., Garcia-Closas M., Gallagher M., Caggana M., Reid Y., Cosentino M., Ji J., Johnson D., et al. Epidemiology. 2002;13:246–254. doi: 10.1097/00001648-200205000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Matsubara Y., Ikeda H., Endo H., Narisawa K. Nucleic Acids Res. 1992;20:1998. doi: 10.1093/nar/20.8.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers C. D. G., Burgoyne L. A. Biotechnol. Appl. Biochem. 2000;31:219–224. doi: 10.1042/ba19990113. [DOI] [PubMed] [Google Scholar]
- 9.Natarajan P., Trinh T., Mertz L., Goldsborough M., Fox D. K. BioTechniques. 2000;29:1328–1333. doi: 10.2144/00296pf01. [DOI] [PubMed] [Google Scholar]
- 10.Cao J., Cousins R. J. J. Nutr. 2000;130:2180–2187. doi: 10.1093/jn/130.9.2180. [DOI] [PubMed] [Google Scholar]
- 11.Overbergh L., Valckx D., Waer M., Mathieu C. Cytokine. 1999;11:305–312. doi: 10.1006/cyto.1998.0426. [DOI] [PubMed] [Google Scholar]
- 12.Sullivan V. K., Burnett F. R., Cousins R. J. J. Nutr. 1998;128:707–713. doi: 10.1093/jn/128.4.707. [DOI] [PubMed] [Google Scholar]
- 13.McMahon R. J., Cousins R. J. Proc. Natl. Acad. Sci. USA. 1998;95:4841–4846. doi: 10.1073/pnas.95.9.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dufner-Beattie J., Kuo Y. M., Gitschier J., Andrews G. K. J. Biol. Chem. 2004;279:49082–49090. doi: 10.1074/jbc.M409962200. [DOI] [PubMed] [Google Scholar]
- 15.Liuzzi J. P., Blanchard R. K., Cousins R. J. J. Nutr. 2001;131:46–52. doi: 10.1093/jn/131.1.46. [DOI] [PubMed] [Google Scholar]
- 16.Liuzzi J. P., Bobo J. A., Lichten L. A., Samuelson D. A., Cousins R. J. Proc. Natl. Acad. Sci. USA. 2004;101:14355–14360. doi: 10.1073/pnas.0406216101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker C. F., Black R. E. Annu. Rev. Nutr. 2004;24:255–275. doi: 10.1146/annurev.nutr.23.011702.073054. [DOI] [PubMed] [Google Scholar]
- 18.Fung E. B., Kawchak D. A., Zemel B. S., Ohene-Frempong K., Stallings V. A. Nutr. Clin. Pract. 2002;17:365–372. doi: 10.1177/0115426502017006365. [DOI] [PubMed] [Google Scholar]
- 19.King J., Cousins R. J. In: Modern Nutrition in Health and Disease. Shils M. E., Shike M., Ross A. C., Caballero B., Cousins R. J., editors. Baltimore: Lippincott Williams and Wilkins; 2005. pp. 271–285. [Google Scholar]
- 20.Andree K. B., Kim J., Kirschke C. P., Gregg J. P., Paik H., Joung H., Woodhouse L., King J. C., Huang L. J. Nutr. 2004;134:1716–1723. doi: 10.1093/jn/134.7.1716. [DOI] [PubMed] [Google Scholar]
- 21.Sazawal S., Bentley M., Black R. E., Dhingra P., George S., Bhan M. K. Pediatrics. 1996;98:1132–1137. [PubMed] [Google Scholar]
- 22.Baqui A. H., Walker C. L., Zaman K., Arifeen S. E., Chowdhury H. R., Wahed M. A., Black R. E., Caulfield L. E. J. Nutr. 2005;135:2187–2191. doi: 10.1093/jn/135.9.2187. [DOI] [PubMed] [Google Scholar]
- 23.Tamura T., Munger R. G., Corcoran C., Bacalao J. Y., Nepomuceno B., Solon F. Birth Defects Res. A Clin. Mol. Teratol. 2005;73:612–616. doi: 10.1002/bdra.20179. [DOI] [PubMed] [Google Scholar]
- 24.Wong M. L., Medrano J. F. BioTechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 25.Bar T., Muszta A. BioTechniques. 2005;39:333–334, 336, 338. doi: 10.2144/05393ST01. [DOI] [PubMed] [Google Scholar]
- 26.Cousins R. J. In: Modern Nutrition in Health and Disease. Shils M. E., Shike M., Ross A. C., Caballero B., Cousins R. J., editors. Baltimore: Lippincott Williams and Wilkins; 2005. pp. 615–626. [Google Scholar]
- 27.Rimbach G., Fuchs J., Packer L., editors. Nutrigenomics. Boca Raton, FL: CRC Press; 2005. [Google Scholar]
- 28.Moore J. B., Blanchard R. K., McCormack W. T., Cousins R. J. J. Nutr. 2001;131:3189–3196. doi: 10.1093/jn/131.12.3189. [DOI] [PubMed] [Google Scholar]
- 29.Wimmer U., Wang Y., Georgiev O., Schaffner W. Nucleic Acids Res. 2005;33:5715–5727. doi: 10.1093/nar/gki881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K., Zhou B., Kuo Y. M., Zemansky J., Gitschier J. Am. J. Hum. Genet. 2002;71:66–73. doi: 10.1086/341125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cousins R. J., Blanchard R. K., Popp M. P., Liu L., Cao J., Moore J. B., Green C. L. Proc. Natl. Acad. Sci. USA. 2003;100:6952–6957. doi: 10.1073/pnas.0732111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wellinghausen N., Rink L. J. Leukoc. Biol. 1998;64:571–577. doi: 10.1002/jlb.64.5.571. [DOI] [PubMed] [Google Scholar]
- 33.Rink L., Gabriel P. Proc. Nutr. Soc. 2000;59:541–552. doi: 10.1017/s0029665100000781. [DOI] [PubMed] [Google Scholar]
- 34.Fraker P. J., King L. E. Annu. Rev. Nutr. 2004;24:277–298. doi: 10.1146/annurev.nutr.24.012003.132454. [DOI] [PubMed] [Google Scholar]
- 35.Wellinghausen N., Driessen C., Rink L. Cytokine. 1996;8:767–771. doi: 10.1006/cyto.1996.0102. [DOI] [PubMed] [Google Scholar]
- 36.Häartel C., Bein G., Muller-Steinhardt M., Klüuter H. J. Immunol. Methods. 2001;249:63–71. doi: 10.1016/s0022-1759(00)00334-3. [DOI] [PubMed] [Google Scholar]
- 37.Huse M., Eck M. J., Harrison S. C. J. Biol. Chem. 1998;273:18729–18733. doi: 10.1074/jbc.273.30.18729. [DOI] [PubMed] [Google Scholar]
- 38.Braunschweig C. L., Sowers M., Kovacevich D. S., Hill G. M., August D. A. J. Nutr. 1997;127:70–74. doi: 10.1093/jn/127.1.70. [DOI] [PubMed] [Google Scholar]
- 39.Krones C., Klosterhalfen B., Fackeldey V., Junge K., Rosch R., Schwab R., Stumpf M., Klinge U., Schumpelick V. J. Invest. Surg. 2004;17:249–256. doi: 10.1080/08941930490502817. [DOI] [PubMed] [Google Scholar]
- 40.Beutler E., Gelbart T., Kuhl W. BioTechniques. 1990;9:166. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.