Abstract
Most analyses of single-molecule experiments consist of binning experimental outcomes into a histogram and finding the parameters that optimize the fit of this histogram to a given data model. Here we show that such an approach can introduce biases in the estimation of the parameters, thus great care must be taken in the estimation of model parameters from the experimental data. The bias can be particularly large when the observations themselves are not statistically independent and are subjected to global constraints, as, for example, when the iterated steps of a motor protein acting on a single molecule must not exceed the total molecule length. We have developed a maximum-likelihood analysis, respecting the experimental constraints, which allows for a robust and unbiased estimation of the parameters, even when the bias well exceeds 100%. We demonstrate the potential of the method for a number of single-molecule experiments, focusing on the removal of DNA supercoils by topoisomerase IB, and validate the method by numerical simulation of the experiment.
Keywords: constrained distribution, maximum-likelihood method, parameter estimation, single-molecule techniques
Over the past few years, single-molecule techniques have started to deliver on their promise as high-resolution tools for the study of biological systems. The activity of single proteins such as kinesin, myosin, and topoisomerases (1–3), among others, has been monitored in real time. A hallmark of such single-molecule experiments, in contrast to bulk experiments, is their unparalleled ability to yield the functional form of the distribution of experimental outcomes and not merely their averages (4) or other statistics (5). Estimating the parameter values that characterize these distributions often yields the information required to construct detailed mechanical models of the system under investigation.
To obtain these parameter values from an experiment, observables are typically binned into a histogram, and the histogram is fitted to the predictions of a model. An alternative method to obtain a distribution parameter is to use the maximum-likelihood method, in which one calculates the value of an unknown parameter in a distribution that maximizes the likelihood of the experimentally observed data (6, 7). The maximum-likelihood method has the advantage that one does not discard information, or introduce one's own biases, in the data through binning. Moreover, the histogram-fitting approach, at least when squared loss is used, ignores the fact that the errors induced in the construction of the histogram are themselves a function of the model and the number of counts represented in each bin of the histogram. Another important advantage of using the maximum-likelihood method, which we demonstrate below, is the possibility to build a model that is more faithful to experimental reality. Particularly in biophysical experiments where a multitude of factors, such as finite size or other experimental or biological constraints, unavoidably thwart the assumption that each individual observation is independent and identically distributed (referred to as the “i.i.d.” assumption below), the maximum-likelihood approach facilitates building a model that is both more experimentally sound and more statistically robust.
Frequently, constraints emerge because of experimental limitations in detecting all values of experimental outcomes in a distribution: one receives from the measurement a limited range of values instead of the entire domain. In some experiments, for example, the DNA translocation by the enzyme FtsK described in ref. 8, the experimental outcomes are uncoupled from one another and are i.i.d. However, in the scenario that experimental outcomes are coupled to each other by a global constraint, the range of values that can be detected varies with every new measurement taken. As we demonstrate below, the presence of global constraints is a factor that absolutely requires maximum-likelihood analysis if the biological parameters of a system are to be measured accurately.
In principle, the analysis of bulk experiments can be hampered as much as the analysis of single-molecule experiments. However, in single-molecule measurements, one can evaluate each experimental outcome with respect to the constraints. Armed with this knowledge, one can apply the mathematical treatment outlined here and counter the bias in the data accurately.
In this article, we illustrate the problem of global constraints by showing how the measurement process in a single-molecule study of DNA supercoil relaxation by the enzyme topoisomerase IB imposes global constraints on the probability distribution from which the experimental outcomes are drawn. Subsequently, we generalize the maximum-likelihood method for parameter estimation, enabling one to faithfully recover the unbiased estimate of the distribution parameter from data subject to global constraints. We also derive an expression for the standard deviation of the recovered parameter as a function of the available statistics. Numerical methods confirm our ability to recover the unbiased distribution parameter within the error estimation derived. Finally, we show that the method introduced here can play an important role in the extraction of biological parameters from several other single-molecule experiments.
Topoisomerase IB Steps Are Subjected to a Global Constraint
We first illustrate the concept of global constraints by using data obtained from the single-molecule analysis of topoisomerase IB (9). Topoisomerase IB is an enzyme that removes supercoils from a dsDNA molecule by transiently introducing a nick (10, 11). As long as the dsDNA molecule is nicked, torque present in the molecule will swivel the DNA about its intact strand. After a random number of supercoils are released, the enzyme religates the DNA, which terminates the removal of supercoils (9)
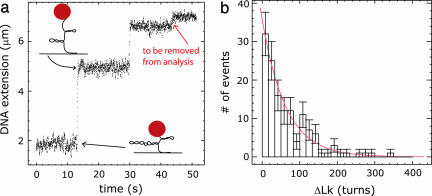
We can follow the action of the topoisomerase in real time by using magnetic tweezers (1). The experimental strategy is described elsewhere (12) and summarized in Fig. 1a. Each time the topoisomerase removes supercoils from the DNA molecule, one observes a discrete step in the height of a μm-sized bead attached to the molecule. The height of the bead is equal to the extension of the DNA molecule and is directly related to its linking number (Lk) and the number of supercoils present in the DNA. A small extension of the DNA corresponds to a large number of supercoils present, whereas a large extension corresponds to a few supercoils present in the DNA. Thus, each time the topoisomerase removes supercoils from the DNA, we observe a discrete step in the DNA extension, which is proportional to ΔLk. If the probability of religation (per turn) is constant, the distribution of ΔLk should be an exponential (Fig. 1b); the average of ΔLk, which is denoted 〈ΔLk〉, is the parametric description of topoisomerase activity we want to deduce from the experiment.
Fig. 1.
An example of a system that includes global constraints. (a) Topoisomerase IB removes DNA supercoils in steps. Each time the topoisomerase removes a number of supercoils, the DNA extension rises in a stepwise fashion. The final step that leads to the removal of the remaining supercoils in the DNA is artificially constrained and should be removed from the analysis. (b) The size of the steps (in units of change in ΔLk) is distributed exponentially. In the text, this is referred to as the measured distribution. This measured distribution may differ from an underlying true distribution because of the presence of global constraints.
The setup of the experiment, in which the DNA molecule only contains a limited number of supercoils, necessarily introduces global constraints on the distribution. Consequently, at some point the topoisomerase will inevitably remove the last few supercoils that remain in the DNA (red arrow, Fig. 1a). This final step toward the level of zero supercoils contains only limited information in comparison to previous steps, because the final step is artificially constrained by the fact that no more supercoils remain in the DNA for the topoisomerase to remove. Therefore, when drawing conclusions about the working of the enzyme, one should discard this final step. For convenience, we will define substeps as those steps that do not extend to the level of zero supercoils. Effectively, steps so large that they become the final step are discarded, whereas steps so small that they become substeps are not discarded, which leads to an overrepresentation of small steps. When one simply analyzes the surviving substeps, one obtains a skewed distribution with an incorrect parameter, which can hamper a proper interpretation of the system under investigation. In an actual experiment, one cannot distinguish between the “true” distribution and the distribution that is skewed as a result of the measurement. After all, all one has is the measured distribution (Fig. 1b), which is skewed. Fortunately, in a single-molecule measurement this skewing can be corrected for by the method we describe below.
Maximum-Likelihood and Domain Constraints
We briefly review the concept of parameter estimation by using the maximum-likelihood method (7). Let P(s|k) be a properly normalized probability density function (pdf) for step size s, with parameter k. The goal of the maximum-likelihood method is to obtain an estimate for the parameter of the pdf, which in this general case is k. Because the experimental outcomes are assumed to be statistically independent, the combined probability to find, in n measurements, the data s1, s2, …, sn is given by:
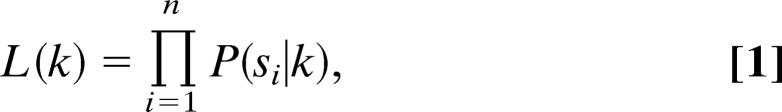 |
where L is the likelihood function. To avoid working with very large or small numbers that can cause computational inaccuracies, and to facilitate the analysis, one often works with the logarithmic likelihood
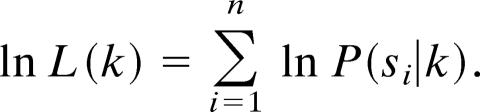 |
We now introduce k*, the value of k that maximizes L, otherwise known as the maximum-likelihood value. It is the best estimate for k and we want to calculate its value. We obtain k* by solving
(see Supporting Text, which is published as supporting information on the PNAS web site). Assuming that the shape of ln P(k|s) near k* is a Gaussian distribution (see Supporting Text), we can calculate the variance σ2 of k*, using
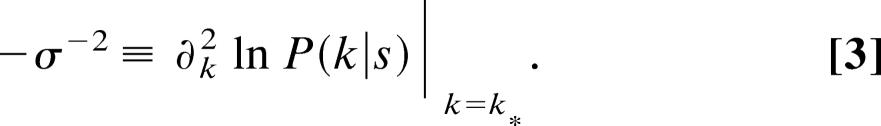 |
In many experimental scenarios, constraints apply to the measurable domain of P. In other words, it may not be experimentally possible to sample all possible values of s. A proper analysis of the data taken in this experimental scenario then requires P to be renormalized by a weighting function g(k):
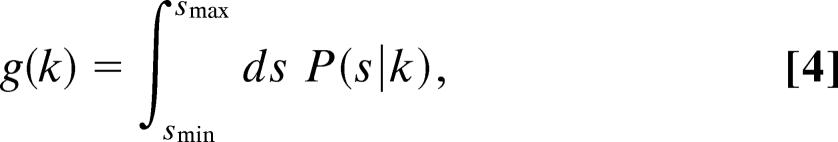 |
where smin is the minimum value for s that can be detected, and smax is the maximum value for s that can be detected. In this simplest case, smin and smax are constant for each measurement i of s. However, an alternative possibility is for a global constraint to couple all observations (indexed by i) of the variable s to each other. In this second case, one requires a weighting function that varies with each measurement i of s:
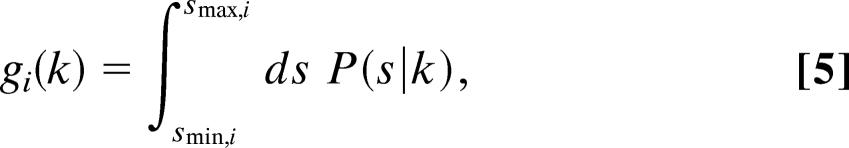 |
where smin,i and smax,i are again the minimum and maximum detectable values for s, respectively, but their values are not fixed for all measurements of s. Instead, smin,i represents the minimum detectable value for s that is valid only for the ith measurement of s. Similarly, smax,i represents the maximum detectable value for s that is valid only for the ith measurement of s. Analogously to the i.i.d. case, one can calculate the likelihood function for all of the constrained data, maximize this function, and obtain k*. The value for k* we obtain in this manner is then the unbiased estimate of the parameter of the distribution.
To illustrate the method explicitly, we use an exponential function as a pdf, the appropriate model for a topoisomerase that removes supercoils from DNA with constant probability per turn of religation. The normalized pdf for 0 < s < ∞ is then given by
where k = 1∕〈s〉. Here and in the following, brackets indicate averages over the experimental observations. The corresponding likelihood function is given by
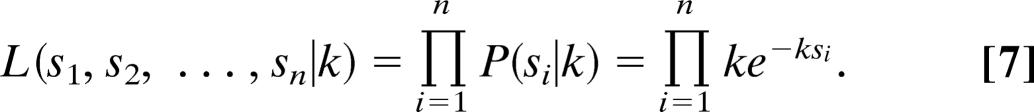 |
In the case of i.i.d. observations, we obtain
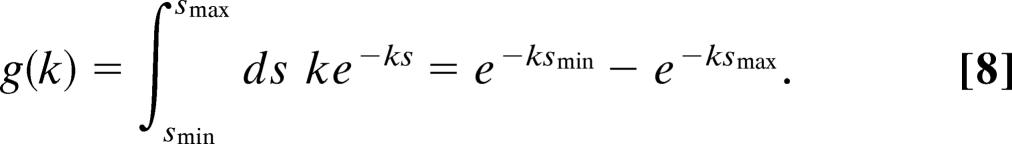 |
However, in the case of global constraints, we obtain:
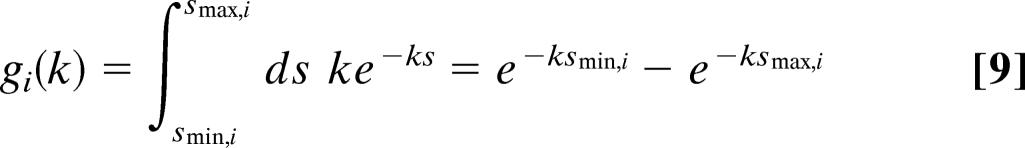 |
and the values for s are drawn from
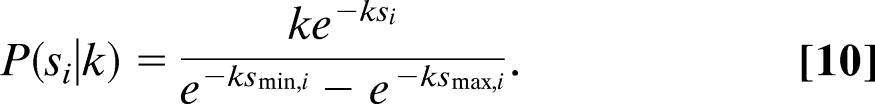 |
Having obtained a relation for P, we can calculate the corresponding likelihood. The probability of the data in the presence of global constraints is
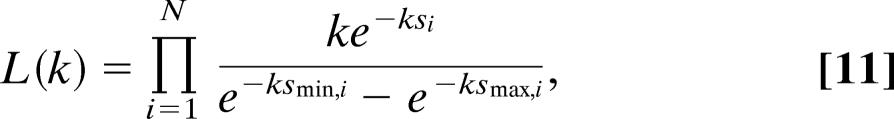 |
where N is the number of experimental outcomes of s. The logarithm of L is given by
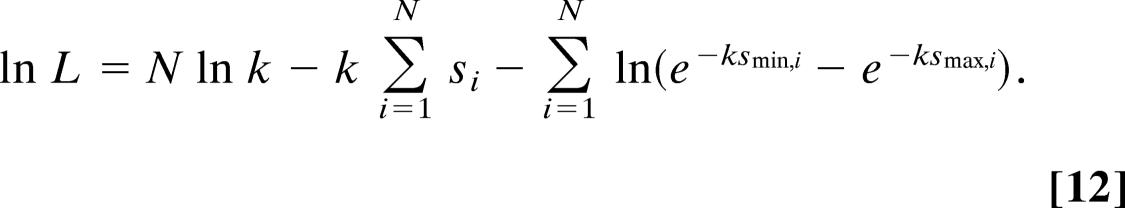 |
The parameter measured in the topoisomerase IB experiment is the average change in 〈ΔLk〉, which is equal to 〈s〉 in the terminology used above. Because 〈ΔLk〉 = 1∕k, we take the derivative of Eq. 12 with respect to 1∕k:
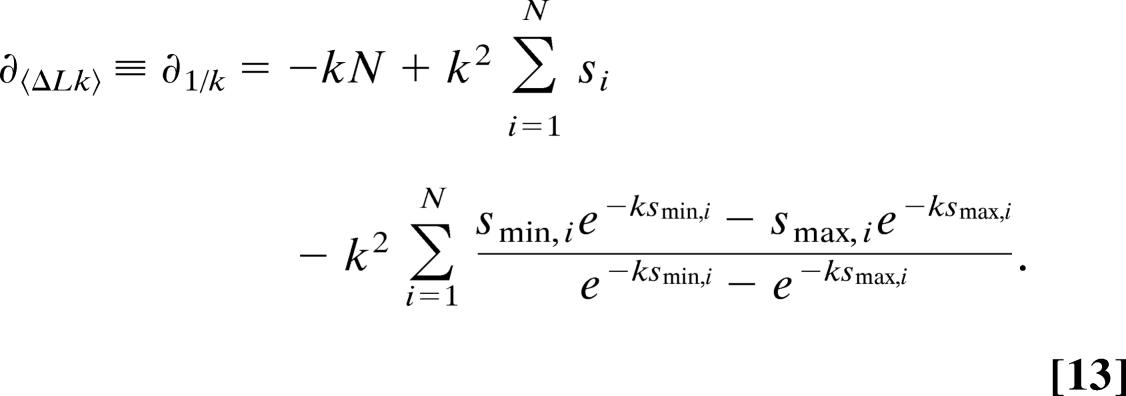 |
We find the maximum in the likelihood by setting Eq. 13 equal to zero,
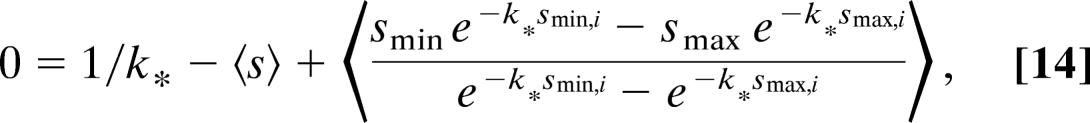 |
where k* is again the maximum-likelihood value for k, the value that solves Eq. 14 (the summation signs in Eq. 13 have been replaced by brackets in Eq. 14 to denote averages). Eq. 14 can be evaluated numerically to yield 〈ΔLk〉*, the maximum-likelihood value of 〈ΔLk〉. We deduce that the variance of 〈ΔLk〉∗ is given by
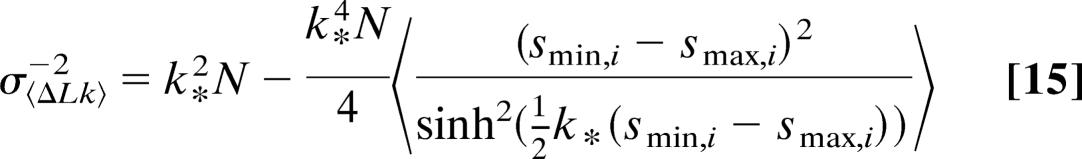 |
(see also Supporting Text). Comparing Eqs.14 and 15 to the case in which no constraints apply, or smax = ∞ and smin = 0, we recover
and
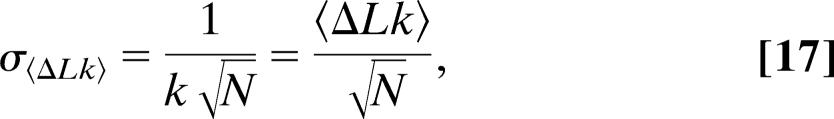 |
as expected.
Eqs. 14 and 15 can be directly applied to the single-molecule data of DNA supercoil relaxation by topoisomerase IB (Fig. 1b) to determine the true parameter of the underlying true distribution and its associated standard deviation as a function of the number of experimental outcomes N
Numerical Simulation and the Consequences of Ignoring Global Constraints
We simulate the measurement process in a single-molecule experiment to quantify the biasing effect on a true distribution as a result of the global constraints and the “sampling error” inherent in the finite number of observations performed. The applicability of Eqs.14 and 15 to the determination of the value and the standard deviation of the distribution parameter can therefore be assessed.
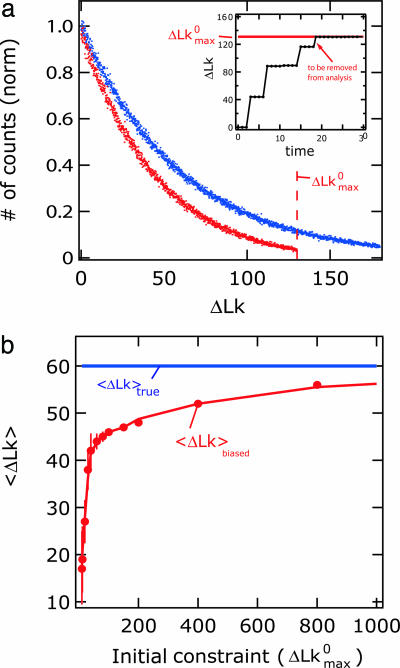
We start by generating an exponential distribution characterized by a parameter that we define as the “true parameter” and is denoted 〈ΔLk〉true. We arbitrarily set it to 〈ΔLk〉true = 60. 〈ΔLk〉true represents the parameter of the distribution that would be measured in the absence of constraints. We call this unbiased distribution the true distribution. Because it is unbiased, we can think of this distribution as representing the physics governing the workings of the enzyme. We then simulate the process of removing supercoils from a DNA molecule that has a maximum of 130 supercoils present (the global constraint ΔLkmax0, Fig. 2a Inset). We use these values for all simulations. The number of supercoils that the topoisomerase removes each time from the DNA is randomly drawn from our true distribution. As described above, all final steps are subsequently discarded, and the substeps that remain are displayed in a histogram. This histogram reflects what we would measure experimentally and we call it the “measured distribution,” characterized by a “measured parameter,” which is biased and therefore denoted 〈ΔLk〉biased. The true distribution is shown in blue in Fig. 2a, and the measured distribution is shown in red. As can be clearly seen from Fig. 2a, the two distributions are not identical. The true distribution obviously yields an average of 60 (in units of ΔLk). We obtain 〈ΔLk〉biased by fitting the measured distribution to an exponential in the range between zero and ΔLkmax0. In fact, the functional form is altered slightly because of the global constraints, as discussed formally in Supporting Text and Fig. 5, which is published as supporting information on the PNAS web site. Note that the value for 〈ΔLk〉biased is thus biased because of a combination of factors: (i) the presence of global constraints, (ii) the number of experimental outcomes N, and (iii) the analysis by histogram fitting rather than maximum likelihood. For the particular values for 〈ΔLk〉true and ΔLkmax0 we used, 〈ΔLk〉biased was 46, which is an underestimate of ≈23% in comparison to 〈ΔLk〉true. Indeed, the measurement process has biased small steps over large steps, skewing the measured distribution toward lower values. We now focus more closely on the relationship between the magnitude of the constraint and the resulting degree of bias. Fig. 2b plots 〈ΔLk〉biased as a function of the severity of the global constraint ΔLkmax0. We plot 〈ΔLk〉true as a blue horizontal line in Fig. 2b. The discrepancy between 〈ΔLk〉biased and 〈ΔLk〉true caused by the global constraints is thus reflected graphically as the distance between the red curve and the blue line in Fig. 2b; in the absence of any biasing effect, all values for 〈ΔLk〉biased would fall on top of the blue line. We describe three salient features of Fig. 2b. First, as the constraint becomes less severe (ΔLkmax0 increases), the magnitude of the bias decreases. Conversely, as the constraint becomes more severe (ΔLkmax0 decreases), the magnitude of the bias increases. Second, the discrepancy between 〈ΔLk〉biased and 〈ΔLk〉true is very large (>100%) for small values of ΔLkmax0. For example, for ΔLkmax0 = 20, 〈ΔLk〉biased = 27, which constitutes an underestimation of 〈ΔLk〉true by ≈120%. Although this is an example that might not generally be observed experimentally, we include it to emphasize that our method can recover 〈ΔLk〉true robustly even in the case of extreme bias, as we show below. The third feature of Fig. 2b highlights that in a regime where one naively would expect virtually no biasing effect because of the constraints, the bias is significant nevertheless. Indeed, for ΔLkmax0 = 800, which is well over an order of magnitude larger than 〈ΔLk〉true, one still observes that 〈ΔLk〉true is underestimated by ≈7%. This surprising behavior stems from the fact that the constraints on the distribution vary from step to step and are on average smaller than ΔLkmax0. We now describe how we can nonetheless obtain an accurate value for 〈ΔLk〉true, even in cases where 〈ΔLk〉true is severely underestimated.
Fig. 2.
Simulated step-size distributions for the enzymatic removal of supercoils from the DNA molecule. (a) The number of supercoils that the enzyme removes each time from the DNA molecule is randomly drawn from a generated exponential distribution, called the true distribution (blue dots). The true distribution is characterized by an average of 60 (units of ΔLk). After discarding the final steps leading to the level of zero supercoils ΔLkmax0 = 130, see text), one obtains a measured distribution (red dots) whose parameter is underestimated (〈ΔLk〉 = 46). (Inset) Numerical simulation of the enzymatic removal of supercoils. The size of each step is drawn from the true distribution. As in reality, the DNA molecule simulated contains only a limited number of supercoils. The level at which no supercoils are present is depicted as a horizontal red line and acts as a constraint for the removal of supercoils by the enzyme. Because the final step toward the level of zero supercoils (red arrow) is artificially constrained, this final step is removed from the data analysis (see text). (b) The degree to which the measured parameter is underestimated is a function of the constraints (the initial maximum number of supercoils in the DNA, denoted ΔLkmax0). As the constraints become more pronounced, the underestimation grows. In some cases, the underestimation of 〈ΔLk〉 caused by global constraints is severe (>100%). The true value for 〈ΔLk〉 is depicted as a horizontal blue line, which the measured value for 〈ΔLk〉 (red dots) approaches asymptotically (red line is a spline through the data points).
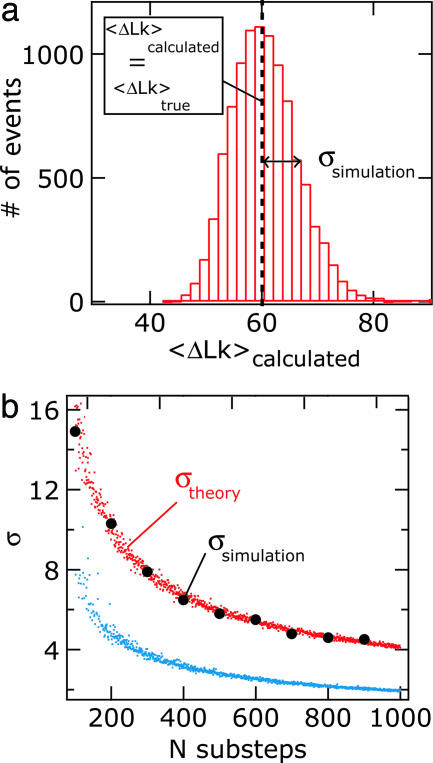
By monitoring the DNA extension, either in a real experiment or the simulation discussed here, we know the number of supercoils that remain in the DNA molecule before the topoisomerase removes a number of supercoils; that is to say, we know the constraints that apply to the measurement of each substep. The important point is that although the constraints vary for each step, they are known, and we can therefore substitute their values for smax,i in Eq. 10. We also know the value of smin,i, which is the minimum detectable number of supercoils removed and is determined by the noise in the height of the bead. This is beyond the scope of this work, and for all practical purposes, we give smin,i the fixed value of zero. We now solve Eq. 14 and call the solution 〈ΔLk〉calculated. In this calculation, we have used N = 105 substeps. To get an idea of the reproducibility in 〈ΔLk〉calculated, we repeat the calculation Q = 105 times and build a histogram of the solutions (Fig. 3a). Importantly, we calculate that the mean of the distribution of 〈ΔLk〉calculated is 60, which is identical to the value we have chosen as 〈ΔLk〉true, the true parameter of the true distribution. Therefore, we conclude that the analysis method accurately recovers the true parameter, despite the biasing effect of the measurement.
Fig. 3.
Recovery and error calculation of the true distribution parameter by using the maximum-likelihood method (see text). (a) The distribution of the calculated distribution parameter is generated by solving Eq. 14 for 〈ΔLk〉 106 times and binning the outcome of the calculation into bins. The distribution is peaked at the value that characterizes the unbiased step-size distribution (〈ΔLk〉true). Importantly, the method thus successfully recovers the unbiased parameter despite the biasing effect of global constraints. The standard deviation of the distribution, σ, is numerically calculated. (b) The theoretical standard deviation σ, obtained by solving Eq. 15, as a function of the number of substeps per exponential distribution. The theoretical standard deviation is calculated for constrained (maximum initial ΔLkmax0 = 130, red points) and unconstrained (maximum initial ΔLkmax0 = ∞, blue points) distributions. The theoretical error in the case of the constrained distribution is compared with the error as calculated from simulations as in a and is shown as black solid circles. The theoretical error calculated by using Eq. 15 predicts the measured error very well.
In an experiment, it is not only important to recover the true parameter of the distribution but also to know its associated standard deviation as a function of the number of experimental outcomes N. We have therefore calculated the standard deviation of the 〈ΔLk〉calculated distribution (Fig. 3a) according to
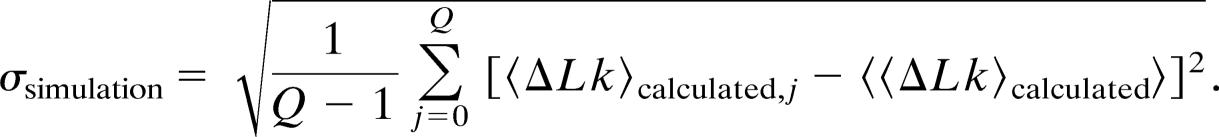 |
This procedure is repeated for nine different values of N and their values are plotted as solid black circles in Fig. 3b. They can be compared with the theoretically predicted values for σ, denoted as σtheory, calculated by using Eq. 15. Fig. 3b also plots σtheory as a function of N as red and blue dots. Red dots are calculations of σtheory with global constraints, and blue dots are calculations of σtheory in the absence of constraints (ΔLkmax0 = ∞). As is evident from Fig. 3b, the solid circles fall on top of the theoretical prediction σtheory given by Eq. 15. Thus, we have shown that Eq. 15 predicts the standard deviation associated with 〈ΔLk〉true accurately. From this result, we can draw an important conclusion, namely that in any given situation with global constraints, an experimenter can assess whether enough statistics have been obtained to determine the unbiased value of the true distribution to the desired accuracy.
Application of the Method
The method outlined above deals with global constraints in the domain of the distribution of experimental outcomes. Therefore, the method should in principle be used in all experiments that involve global constraints and whose experimental outcomes are not distributed like (a series of) delta functions. An example of outcomes distributed like a delta function is the fixed step size of 37 nm with which a myosin protein walks over an actin filament (3). Although experimentally it seems that one measures a Gaussian distribution of observables, the Gaussian shape in fact arises from stochastic fluctuations around a fixed true value. The function describing these processes is a delta function, peaked at the fixed true value of the observable. Mathematically, this process implies that the weight function [gi(k), Eq. 9] is always equal to one, and consequently the pdf is unaltered by constraints. Therefore, the likelihood function and the observable that maximizes it remain unaffected, and one is not required to use this method.
We expect that the analysis method outlined here could guide the proper design and analysis of experiments including assays of the processivity of helicases, polymerases, and other translocation enzymes, single-molecule Föorster resonance energy transfer (FRET) measurements, and real-time single-molecule tracking of DNA condensation. For clarity, we describe a few of these experiments in more detail below.
Processivity Measurements on Limited Substrate
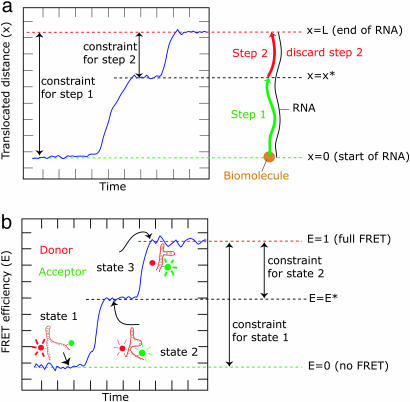
Some substrates, such as ssRNA or dsRNA molecules, are practically hard to prepare in lengths longer than a few kb if they are to be used in single-molecule techniques (13). If one wants to measure the distribution of the processivity of a biomolecule that tracks along the RNA, one may find that the processivity exceeds the length of the RNA. In such a case, one is required to discard the final processive action, because it is artificially constrained by the fact that there is no more dsRNA substrate for the biomolecule to move on. The constraint is global, because the length of the RNA molecule that is available for the biomolecule shrinks as it proceeds. This dilemma is summarized in Fig. 4a. An example of an enzyme translocating on RNA is the RNA-dependent RNA polymerase P2 from φ6 bacteriophage (14). This polymerase can perform an RNA synthesis reaction by using either dsRNA or ssRNA as a template. The processivity, which can be only roughly estimated from experiments, is on the order of 10 kb or more (D. Bamford, personal communication) and is comparable to the length of the RNA substrate. In single-molecule processivity measurements for P2 polymerase and other enzymes, we expect that our treatment would be instrumental in determining the mean processivity correctly.
Fig. 4.
Sketch of experiments in which global constraints can bias parameter estimation. (a) Processivity of a biomolecule (beige circle) along a short biopolymer such as a ssRNA or dsRNA molecule. When the biomolecule starts its procession, it has the total length of the RNA molecule (the global constraint) at its disposal (smax,0 = L). It then moves a distance x and stops. From there, it can start moving again, but the biomolecule can now only travel a length smax,0 = L − x*, before falling off the RNA. The constraint on the distance the biomolecule can travel along the RNA is different for the first and the second step. (b) Conformational changes in e.g., an RNA molecule studied by using FRET. The FRET efficiency is defined between 0 and 1, which is the global constraint for the experiment. E.g., at state 1, the FRET efficiency E = 0. From this state, the FRET efficiency can only change by 1 at maximum (smax,0 = L). However, from an arbitrary intermediate state 2 (at E = E*), it can increase its E only by smax,0 = ΔE = 1 − E*. The constraint on the change in FRET efficiency is thus different for the first and the second state, as described in the text.
Transitions in FRET Efficiency
FRET efficiency depends on the distance between a donor dye and acceptor dye and ranges between zero (no FRET) and one (maximum FRET) (4,15) (in practice, the range in which meaningful FRET measurements can be performed is even smaller because of the lack of sensitivity close to both the no-FRET and the maximum-FRET regimes). Changes in FRET efficiency can in theory be used to quantify conformational changes in biomolecules (e.g., in the folding of RNA molecules or in proteins). Future experiments measuring distributions of changes in FRET efficiency could be biased because of the global constraint imposed by the limited meaningful range in FRET efficiency. For example, one could measure a series of conformational changes in an RNA molecule in which each conformational change is associated with a transition in FRET efficiency between a donor and acceptor dye attached to two parts of the RNA molecule (e.g., refs. 16–18), as schematically depicted in Fig. 4b. In such an experiment, one would be required to discard those transitions that extend to or exceed the limits of the FRET efficiency range. To correct for the ensuing bias toward small FRET transitions and thus for a correct analysis of the distribution, one needs to apply the method described here.
Concluding Remarks
Experimental outcomes that are nonglobally constrained in that they can be assumed to be i.i.d. can be relatively easily analyzed in their measured range. However, when such analysis is performed on outcomes that are coupled by global constraints, severe bias in the parameter estimation can occur. We have therefore generalized the maximum-likelihood method for parameter estimation to include distributions that have global constraints. Using this method, we robustly recover the unbiased distribution parameter from biased data, independent of the severity of the bias. In addition, we have adapted the relation describing errors in the estimation for distribution parameters for the case of global constraints, which allows an experimenter to assess whether enough data points have been accumulated to predict the true parameter to the desired accuracy. Finally, we show that global constraints can occur in a variety of experiments, all of which would benefit from using this method.
Supplementary Material
Acknowledgments
We thank Ulrich Keyser for stimulating discussions and Cees Dekker and Thijn van der Heijden for a critical reading of the manuscript. This work was supported by the Technische Universiteit Delft, the Foundation for Fundamental Research on Matter, and the Netherlands Organization for Scientific Research.
Abbreviations
- i.i.d.
independent and identically distributed
probability density function
- FRET
Föorster resonance energy transfer.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Strick T. R.,, Croquette V., Bensimon D. Nature. 2000;404:901–904. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- 2.Visscher K., Schnitzer M. J., Block S. M. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 3.Yildiz A., Forkey J. N., McKinney S. A., Ha T., Goldman Y. E., Selvin P. R. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 4.Weiss S. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 5.Svoboda K., Mitra P. P., Block S. M. Proc. Natl. Acad. Sci. USA. 1994;91:11782–11786. doi: 10.1073/pnas.91.25.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hald A. Stat. Sci. 1999;14:214–222. [Google Scholar]
- 7.Rice J. A. Mathematical Statistics and Data Analysis. Pacific Grove, CA: Wadsworth & Brooks/Cole Advanced Books & Software; 1988. [Google Scholar]
- 8.Saleh O. A., Perals C., Barre F. X., Allemand J. F. EMBO J. 2004;23:2430–2439. doi: 10.1038/sj.emboj.7600242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koster D. A., Croquette V., Dekker C., Shuman S., Dekker N. H. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 10.Stivers J. T., Harris T. K., Mildvan A. S. Biochemistry. 1997;36:5212–5222. doi: 10.1021/bi962880t. [DOI] [PubMed] [Google Scholar]
- 11.Wang J. C. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 12.Strick T. R., Allemand J. F., Bensimon D., Bensimon A., Croquette V. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 13.Dekker N. H., Abels J. A., Veenhuizen P. T., Bruinink M. M., Dekker C. Nucleic Acids Res. 2004;32:e140. doi: 10.1093/nar/gnh138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bamford D. H. EMBO Rep. 2002;3:317–318. doi: 10.1093/embo-reports/kvf074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ha T., Ting A. Y., Liang J., Caldwell W. B., Deniz A. A., Chemla D. S., Schultz P. G., Weiss S. Proc. Natl. Acad. Sci. USA. 1999;96:893–898. doi: 10.1073/pnas.96.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanchard S. C., Kim H. D., Gonzalez R. L., Jr., Puglisi J. D., Chu S. Proc. Natl. Acad. Sci. USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhuang X. Annu. Rev. Biophys. Biomol. Struct. 2005;34:399–414. doi: 10.1146/annurev.biophys.34.040204.144641. [DOI] [PubMed] [Google Scholar]
- 18.Zhuang X., Kim H., Pereira M. J., Babcock H. P., Walter N. G., Chu S. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.