Abstract
Protein dynamics, including conformational switching, are recognized to be crucial for the function of many systems. These motions are more challenging to study than simple static structures. Here, we present evidence suggesting that in the enzyme adenylate kinase large “hinge bending” motions closely related to catalysis are regulated by intrinsic properties of the moving domains and not by their hinges, by anchoring domains, or by remote allosteric-like regions. From a pair of highly homologous mesophilic and thermophilic adenylate kinases, we generated a series of chimeric enzymes using a previously undescribed method with synthetic genes. Subsequent analysis of the chimeras has revealed unexpected spatial separation of stability and activity control. Our results highlight specific contributions of dynamics to catalysis in adenylate kinase. Furthermore, the overall strategy and the specific mutagenesis method used in this study can be generally applied.
Keywords: chimeric protein, protein flexibility, thermostability
Structural biology is moving beyond simple analysis of the average structures of proteins to include dynamic components. Mounting evidence suggests that dynamic motions of proteins play specific and essential roles in function (1–6), but the mechanism is rarely clear. Adenylate kinase (AK) is an excellent target for the study of connections between dynamics and function of protein. It is a small enzyme catalyzing reversible conversions between ATP/AMP and two ADP molecules (7). Structures have been solved of various states of the enzyme from various organisms and revealed a large conformational rearrangement of the enzyme during its catalytic cycle (8, 9). Among the three defined characteristic AK domains CORE, AMPbind, and LID (10), the AMPbind and LID domains are directly involved in the dynamic event and close over the enzyme’s AMP- and ATP-binding sites, respectively (Fig. 1). A recent NMR experiment suggested that the opening of the AMPbind and/or LID domains upon product release is the rate-limiting step, with opening times commensurate with the turnover rate (11). It also was proposed that the dynamic motion of AK may involve catastrophic events such as cracking and subsequent reassembly (12, 13). Other than the two mobile domains, two loops in the CORE domain (Fig. 1) displayed substantially increased flexibility upon substrate binding as in allostery and have been suggested to serve as a counterweight balancing the substrate-binding energy (9, 14).
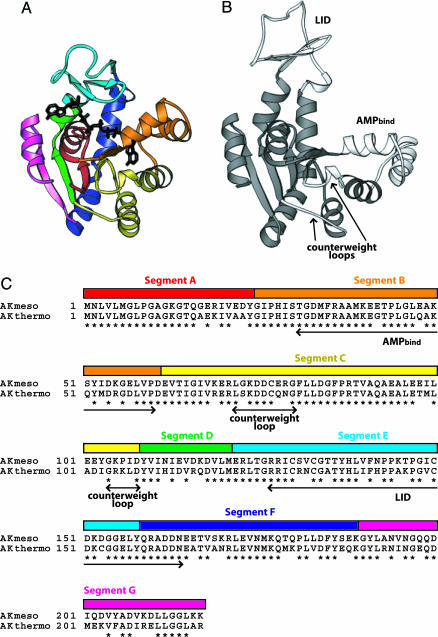
Fig. 1.
Description of the divide and swap method with structures and sequences of AKs. (A) Seven segments divided by eight restriction sites are illustrated in different colors on the closed conformation of AKmeso (18). The bound inhibitor P1,P5-di(adenosine-5′) pentaphosphate is also shown (black). (B) Two mobile domains of AK, the AMPbind and the LID, and the counterweight loops are highlighted (white) on the open conformation of E. coli AK (9). (The E. coli AK structure is used to depict the open form because structures of AKmeso and AKthermo in the open state are not available.) (C) The seven segments are represented as colored bars along with the sequence alignment of AKmeso and AKthermo. The colors used for the seven segments are the same as in A. Conserved residues are indicated by asterisks. The AMPbind and LID domains and the counterweight loops are also indicated.
Clues to connections between amino acid sequence, structure, dynamics, and catalysis can be obtained by comparing and contrasting highly similar proteins from psychrophiles, mesophiles, and thermophiles (15–17). We have previously reported crystal structures, thermal stabilities, and temperature activity profiles for three such proteins from the genus Bacillus (18). As might be expected, the catalytic activities and thermal transitions scale with the operating temperatures of the source organisms. There is, however, no a priori reason to expect a direct connection between the temperature dependence of enzymatic catalysis and that of a major unfolding transition. As pointed out in a previous study with chimeric archaeal trimers of AK, changes in activity are not necessarily coupled to overall thermal stability (19).
In this study, we produced a series of chimeras from AKs of the mesophile Bacillus subtilis (AKmeso) (20) and the thermophile Bacillus stearothermophilus (AKthermo) (21). AKmeso and AKthermo share a high sequence identity (74%; Fig. 1C), and their structures are very similar (18). This similarity allowed specific regions of the AKs to be exchanged, producing fully functional chimeric AKs. We subsequently analyzed them for temperature dependence of stability and activity to determine the roles of different regions in stability and catalysis of the enzyme and to ultimately study the relationship between structure, function, and dynamics of AK.
Results
Construction of Chimeric AKs.
To efficiently produce a large number of different chimeras with desired linkage points from the wild-type (WT) AKmeso and AKthermo, we used a “divide and swap” method with synthetic genes (Fig. 1). First, AKmeso and AKthermo genes were commercially synthesized to have eight unique restriction sites dividing the AKs into seven segments (see Fig. 4, which is published as supporting information on the PNAS web site). Names of the seven segments have been assigned alphabetically from N terminus as segments A to G. Segments B and E correspond closely to the previously defined AMPbind (residues 31–60) and LID (residues 127–164) domains, respectively. The other five segments (A, C, D, F, and G) nearly match the CORE (residues 1–30, 61–126, and 165–217) domain. Segment C includes the counterweight loops (residues 72–80 and 104–108) (9), and segments D and F contain putative hinge residues for motion of the LID domain (22). We did not define exchangeable segments for the hinges of the AMPbind domain because their sequences are precisely conserved between AKmeso and AKthermo.
Eight AK chimeras (AKc1–AKc8) were produced by exchanging one or more segments of the two WT AKs using the introduced restriction sites (Fig. 2). AKc1 and AKc2 were generated by swapping segments B and E. Although the two segments do not exactly match the defined domains, the resulting chimeras (AKc1 and AKc2) essentially have the swapped AMPbind and LID domains because of the exact sequence identity at the ends of the domains. In AKc3 and AKc4, segment C including the counterweight loops was exchanged. AKc5 and AKc6 were constructed by substituting the two mobile domains and the counterweight loop region together. AKc7 and AKc8 are chimeras with the swapped hinge regions for their LID domains. The constructed chimeric genes were confirmed by DNA sequencing, and the expressed and purified chimeric proteins were verified by mass spectrometry.
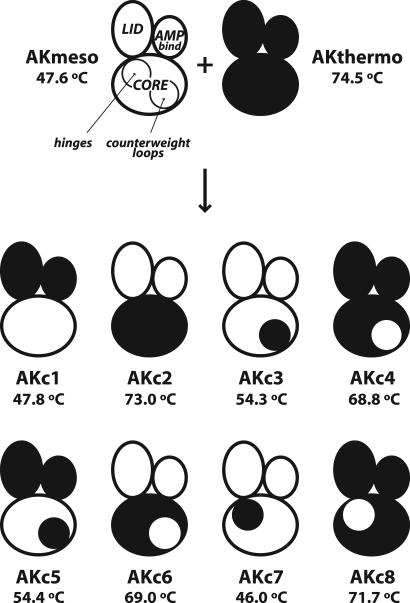
Fig. 2.
Schematic description of WT and chimeric AKs with their Tm values. “Mickey Mouse”-like figures represent AK structures. Right and left “ears” and “faces” stand for the AMPbind, LID, and CORE domains, respectively. The hinge regions for the LID domain and the counterweight loop regions are represented by the circles inside the CORE domains at the upper left and lower right, respectively. Mesophilic and thermophilic parts of the proteins are shown in white and black, respectively. Tm values for AKmeso and AKthermo were obtained from previous studies (18,21).
Thermal Stabilities of Chimeric AKs.
Differential scanning calorimetry (DSC) was used to measure the thermal stabilities of the chimeras (see Fig. 5, which is published as supporting information on the PNAS web site). Thermal denaturation midpoints (Tm values) of the chimeric and two WT AKs are presented in Fig. 2 with their schematic descriptions. The most striking finding was that thermal stabilities of the chimeric AKs were not significantly affected by the AMPbind or LID domain and were defined almost exclusively by the CORE domain. A chimera composed of the mesophilic CORE and the thermophilic AMPbind and LID (AKc1) has essentially the same Tm as AKmeso, and a chimera having thermophilic CORE and mesophilic AMPbind and LID (AKc2) displayed a very similar Tm to that of AKthermo. Two chimeras whose counterweight regions were swapped (AKc3 and AKc4) showed considerable changes in Tm in the expected directions, i.e., an increase for AKc3 from AKmeso and a decrease for AKc4 from AKthermo. These results are consistent with the previous finding because the counterweight region is a part of the CORE domain.
The exclusive control of the highly dominant thermal transition by the CORE domain also was found for the other chimeras. Despite the exchanged AMPbind and LID domains, AKc5 has nearly identical Tm values to that of AKc3, which has the same CORE domain. The same relationship is true for AKc6 and AKc4, which have the same CORE but different AMPbind and LID domains. Although the hinge regions are defined as parts of the CORE domain, the hinge-swapped chimeras (AKc7 and AKc8) displayed only small changes from the WT enzymes. This result indicates that the hinge regions may not be as important for the overall stability as the other parts of the CORE domain including the counterweight region. The hinge-swapped chimeras are also different from the others in that the swapping caused a decrease in Tm even in the case of AKc7, in which the two hinge regions were replaced with those of AKthermo. This finding suggests that interactions between the hinge regions and the other regions in the CORE domain are optimized differently in AKmeso and AKthermo, and exchanging only one partner of the cooperative interactions may result in destabilization.
Activity Profiles of WT and Chimeric AKs.
To examine the temperature dependence of the catalytic activity of AKs, we performed activity assays of the WT and chimeric AKs at various temperatures and compared their activity profiles (Fig. 3). AKmeso and AKthermo showed maximum activities at disparate temperatures (Fig. 3A), while the gap in the temperatures was smaller than that of their Tm values. This difference is because the activity of AKmeso increased beyond its Tm (47.6°C), whereas AKthermo started to be inactivated around its Tm (74.5°C), which can be explained by proposing different inactivation mechanisms between the two AKs (see Discussion). The descending portions of their profiles (inactivation profiles) after reaching maximum activity seem to result from thermal denaturation. However, the difference in the ascending portions of the profiles (activation profiles), especially the reduced activity of AKthermo at low temperatures, is not explained by the difference in stabilities of AKthermo and AKmeso. A comparison between the activity profiles of AKc1 and AKc2 and those of the WT AKs suggests a possible source of the difference in the activation profiles (Fig. 3B). AKc1 consists of the mesophilic CORE domain and the thermophilic AMPbind and LID domains, and its inactivation profile matches that of AKmeso, whereas the activation profile closely resembles that of AKthermo. Conversely, the profile of AKc2 composed of the thermophilic CORE and the mesophilic AMPbind and LID displayed the opposite pattern, combining the increased activity of AKmeso at low temperatures and the inactivation profile of AKthermo. These results suggest that the activity of AK is controlled by the AMPbind and LID domains until limited by unfolding of its overall structure.
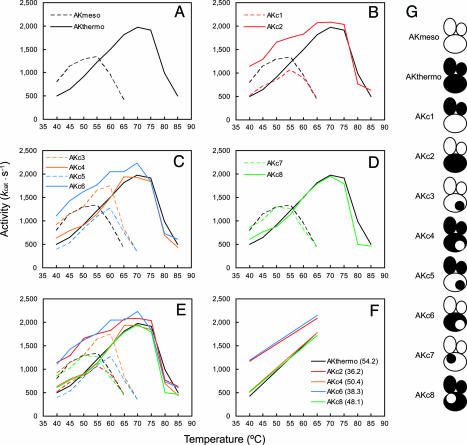
Fig. 3.
Temperature dependence of activities of WT and chimeric AKs. (A) Activity profiles of AKmeso and AKthermo. (B–D) Activity profiles of chimeric AKs are represented with those of the WT AKs. (E) Activity profiles of the WT and the chimeric AKs in A–D are shown all together. (F) Lines were fitted to ascending portions (<70°C) of the profiles of AKthermo and chimeric AKs with the AKthermo-like CORE using the least squares method. Values of their slopes are shown in parentheses. (G) Schematic descriptions of WT and chimeric AKs are displayed for convenience.
The results from the other chimeras support the importance of the two mobile domains in activity (Fig. 3 C and D). Regardless of the identity of the CORE domain, it is the AMPbind and LID domains that determine their activation profiles. The chimeras with the thermophilic AMPbind and LID domains have almost identical activation profiles to that of AKthermo, whereas the others containing the mesophilic AMPbind and LID domains showed increased activity at low temperatures like AKmeso. Conversely, their CORE domains seem important for their inactivation profiles. This finding makes sense because the inactivation is most likely the result of the denaturation of the overall structure, which can be measured by DSC. However, the differences in the Tm values of the chimeras do not completely explain those in the inactivation profiles. For example, having the mesophilic counterweight region in the thermophilic CORE domain (AKc4 and AKc6) resulted in a decrease in Tm (Fig. 2) but barely affected the inactivation profile (Fig. 3C). On the contrary, the corresponding change in the mesophilic CORE (AKc3 and AKc5) caused both an increase in Tm (Fig. 2) and a shifting of the inactivation profile to the higher temperature (Fig. 3C). This result suggests different inactivation mechanisms between the mesophilic and thermophilic AKs (see Discussion).
Plotting together all of the activity profiles of the WT and chimeric AKs makes more apparent the exclusive control of the activity by the AMPbind and LID domains (Fig. 3E). The activation profiles of the AKs with the mesophilic AMPbind and LID domains (AKmeso, AKc2, AKc3, AKc6, and AKc7) exhibit clear separation from those of the other AKs with the thermophilic AMPbind and LID domains (AKthermo, AKc1, AKc4, AKc5, and AKc8). It is easier to discern the difference when comparing the five AKs having the AKthermo-like CORE (AKthermo, AKc2, AKc4, AKc6, and AKc8). Because their inactivations start at ≈70°C, they have enough data points in their activation profiles for comparison. The chimeras with mesophilic AMPbind and LID domains (AKc2 and AKc6) show not only increased activities but also lower slopes than the other three AKs (Fig. 3F). Among the five chimeras containing the mesophilic AMPbind and LID domains, those with the AKthermo-like CORE (AKc2 and AKc6) seem to show even higher activity than the other three chimeras with the AKmeso-like CORE (AKmeso, AKc3, and AKc7) at low temperatures. It is possible that a more robust CORE can help catalysis by retaining a higher affinity for the substrates. However, additional study with more chimeras and more data points at lower temperatures is needed to draw a more definitive conclusion on this point.
Discussion
Although chimeric proteins generated by mixing two or more natural proteins have been used in various biological studies, their construction is not always simple. Small numbers of chimeras with limited linkage points can be made relatively easily by using conventional cloning techniques with preexisting restriction sites or introduced by site-directed mutagenesis. It is also possible to generate chimeras by using hybrid primers and PCR without any restriction site (23). However, these methods are not efficient to prepare large number of chimeras with multiple linkage points. DNA shuffling techniques may be useful to generate large-number chimera libraries from homologous proteins (24), but extensive screening is required to obtain specific chimeras. The divide and swap method used in this study can be an alternative in constructing chimeric proteins. With two or more homologous proteins, it is most likely that one can find conserved amino acid residues where restriction sites can be inserted. For a given amino acid sequence the codon degeneracy allows many possible DNA sequences, one of which probably has a restriction site. Once template genes with the restriction sites are designed and synthesized, chimeric genes can be constructed by simply exchanging DNA fragments using these sites. It is also possible to substitute any region with a new DNA fragment made by fusing two complementary oligonucleotides. This approach provides additional opportunities for variations in sequences of chimeras because the nucleotides can have any sequence except the terminal regions needed for ligation. With decreasing costs for synthetic genes, this divide and swap method can become highly efficient.
The chimeric AKs produced in this study have been characterized. Thermal denaturation as measured by DSC showed that the overall stabilities of the chimeras are determined by the identities of their CORE domains. This result does not mean that the other two domains, the AMPbind and LID, cannot affect stability. In other studies, mutations in the LID domain caused considerable differences in overall stability of AKs (20, 25). The DSC results should be interpreted to mean that the stabilities of the mesophilic and thermophilic AMPbind and LID domains are similar, and thus their overall stabilities are limited by the stabilities of their CORE domains. This finding is supported by the result of the activity assay of AKc2, which comprises the thermophilic CORE and the mesophilic AMPbind and LID. As shown in Fig. 3B, the activity of AKc2 was extended up to 75°C. This result indicates that the mesophilic AMPbind and LID domains may remain stable at high temperatures because AKs are generally assumed to become inactive if the AMPbind and LID domains are denatured.
If the CORE domain governs overall stability, which part(s) of the enzyme is responsible for the temperature dependence of the catalytic activity? Although it has been suggested that dynamics of the AMPbind and LID domains were closely related to catalysis (8, 9, 11), little is known about the mechanism of the movement. Do the AMPbind and LID domains trigger their own motions? Or are other parts, such as the hinge regions or the counterweight loops, important for the dynamics? Studying the temperature dependence of activity of the chimeric AKs helps answer these questions. The results of the activity assays clearly show that the two mobile domains themselves (the AMPbind and LID domains), not their hinges or the counterweight loops, control the temperature dependence of the catalytic activity. This finding should be carefully interpreted. The results do not mean that the hinge and counterweight regions are not important for catalysis. Rather, they imply that the effects of the hinges and the counterweight loops on activity are similar in AKmeso and AKthermo, but their AMPbind and LID domains can cause differences in activity. Because the opening of the AMPbind and LID domains is suggested to be the rate limiting step for catalysis of AK (11), the results also indicate that the AMPbind and LID domains may control their own functional dynamics. Because almost all of the contacts between the two mobile domains and other parts (the CORE domain and the substrate analog) are conserved between AKmeso and AKthermo, intrinsic properties of the two domains, not the interactions with others, may control the movement. Although their stabilities may be similar, the two mobile domains in AKmeso can be more flexible than those in AKthermo, which may help the opening by relieving constraints of the motion in critical areas.
Any differences in flexibility must be because of sequence changes between AKmeso and AKthermo. It would require extensive additional study using more chimeras in which smaller areas are exchanged and/or point mutations to determine specific residue substitutions responsible for the differences. However, it is possible to suggest potentially important residues for the difference with the available information. One of the residues is Arg-131 of AKthermo. Arg-131 forms a salt bridge with the conserved Glu-156 within the LID domain in the crystal structure (18) and the molecular dynamics simulation (26). Because AKmeso has Ser-131 at the position, AKmeso cannot have the salt bridge. Thus, the Arg-131–Glu-156 salt bridge may provide additional rigidity only to the LID of AKthermo.
The results from the activity assays also suggest a difference in the inactivation mechanism of AKmeso and AKthermo. In their activity profiles (Fig. 3A), AKmeso showed its maximum activity above its Tm (47.6°C), whereas the inactivation of AKthermo started below its Tm (74.5°C). Because the denaturation of AKmeso may begin with the CORE domain, AKmeso can be active at higher temperatures than its Tm due to the increased stability of the CORE domain caused by binding of the substrates during the assay. Conversely, in AKthermo we propose that one or both of the AMPbind and LID domains begins to denature around the temperature at which its CORE starts to unfold. Thus, the substrate binding may not be as effective as in AKmeso. Even if it is possible, the stabilization of the CORE by the substrate binding cannot prevent inactivation, which is presumably mediated by the unfolding of the AMPbind and/or LID domains. The similarity of the inactivation profiles of AKthermo, AKc4, and AKc6 (Fig. 3C) can be explained in the same way. Although they have different counterweight loops in their CORE domains resulting in the different Tm values, their activities are limited by the denaturation of their AMPbind and/or LID domains as well as that of their CORE domains, and thus the inactivation profiles are almost identical because of the similar stabilities of their AMPbind and LID domains.
This work also provides insight into temperature adaptation of proteins. Since Somero (27) proposed the “corresponding state” hypothesis postulating that homologs exhibit comparable flexibilities to perform catalysis at their physiologically relevant temperatures, it has remained unclear whether the difference in stability between the homologs is a consequence of adjusting flexibility for catalysis or simply a result of a lack of selective pressure (28). In this study, we were able to show that the increased activity at low temperatures can be achieved without giving up stability by generating the chimeras (AKc2 and AKc6) that are as stable as AKthermo at high temperatures and as active as AKmeso at low temperatures. Thus, it seems that high activity does not have to be related to low stability and can accompany high stability by adjusting the flexibility of regions that are less important for stability. Uncoupling between global flexibility and stability also has been suggested in other systems using different experimental techniques such as NMR relaxation (16) and H/D exchange (17).
The strategy used in this study can be generally applied. If a thermophilic homolog of a target protein is available, one can use the divide and swap method to produce a series of chimeras in which the desired region(s) is exchanged. Then, temperature dependence of any properties of the chimeras can be analyzed and compared with those of the target protein and its thermophilic homolog to determine important region(s) for the properties. One of the advantages of this strategy is that consequence of the substitutions can be double checked. One can swap specific region(s) resulting in two chimeras, a mesophile with thermophilic part(s) and a thermophile with mesophilic part(s), and test effect of the substitution in two directions. This approach is not limited to a pair of mesophilic and thermophilic homologs but can be applied to any set of homologous proteins with any distinct properties such as different activities, binding kinetics, or optimal functional conditions (pH, pressure, salinity, etc.). Thus, with increasing phylogenic knowledge of proteins and decreasing cost for synthetic genes, the strategy used in this study can be more applicable to many other studies.
Materials and Methods
Generation of AK Chimeras.
AKmeso and AKthermo genes were commercially synthesized by Geneart (Regensburg, Germany) according to the following design. Six conserved areas in the amino acid sequences of the two AKs were selected as boundaries dividing the sequences into seven regions. Within each boundary, it was possible to find residues where a restriction site could be introduced without altering the amino acid sequence by using different codons. Two more restriction sites were inserted at each end of the genes to facilitate subcloning of the whole genes. The genes then were cloned into a modified pT7 vector (pNS008b) provided by Nayoung Suh (University of Wisconsin, Madison, WI). The restriction sites of the synthetic genes are unique in the pNS008b because the vector lacks the same restriction sites outside of its multiple cloning site. To construct chimeric genes, single or multiple regions were swapped between the synthetic AKmeso and AKthermo genes by cutting and ligating DNA fragments at the unique restriction sites. The final chimeric genes were cloned into the pET11 vector for expression. The chimeric and WT proteins were overexpressed in Escherichia coli and purified by a two-step procedure involving affinity chromatography and gel filtration as described in ref. 18.
Tm Measurement.
Thermal stabilities of the chimeric AKs were measured by DSC as described in ref. 18. The scans were performed from 5°C to 95°C. The DSC data clearly showed one major peak for each scan and could be well approximated by a two-state transition model to determine Tm values.
Activity Assay.
The enzymatic activity of the chimeric and WT AKs was determined at various temperatures in the direction of ATP formation as described in ref. 18 with minor modifications. Briefly, the enzyme reaction at each temperature was started by adding AK to reaction buffer containing ADP and stopped by adding inhibitor P1,P5-di(adenosine-5′) pentaphosphate. The amount of ATP produced by the reaction was determined using ATP-dependent reduction of NADP+ to NADPH by coupling enzymes at the room temperature. The assays were repeated three times at each temperature, and the average values were reported.
Supplementary Material
Acknowledgments
We thank N. Suh for providing the pNS008b vector and critical advice on DNA cloning, N. Matilla for assistance in chimera production, D. McCaslin for help in DSC experiments, D. Kondrashov for comments on the manuscript, and W. Cleland for helpful discussion. This work was supported by a Vilas Associate Award (to G.N.P.) and the Wisconsin Alumni Research Foundation. DSC data were obtained at the University of Wisconsin–Madison Biophysics Instrumentation Facility.
Glossary
Abbreviations:
- AK
adenylate kinase
- AKmeso
AK from Bacillus subtilis
- AKthermo
AK from Bacillus stearothermophilus
- DSC
differential scanning calorimetry
- Tm
thermal denaturation midpoint.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Schotte F., Lim M., Jackson T. A., Smirnov A. V., Soman J., Olson J. S., Phillips G. N., Jr, Wulff M., Anfinrud P. A. Science. 2003;300:1944–1947. doi: 10.1126/science.1078797. [DOI] [PubMed] [Google Scholar]
- 2.Eisenmesser E. Z., Bosco D. A., Akke M., Kern D. Science. 2002;295:1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 3.Volkman B. F., Lipson D., Wemmer D. E., Kern D. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 4.Ota N., Agard D. A. Protein Sci. 2001;10:1403–1414. doi: 10.1110/ps.800101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller D. W., Agard D. A. J. Mol. Biol. 1999;286:267–278. doi: 10.1006/jmbi.1998.2445. [DOI] [PubMed] [Google Scholar]
- 6.Balabin I. A., Onuchic J. N. Science. 2000;290:114–117. doi: 10.1126/science.290.5489.114. [DOI] [PubMed] [Google Scholar]
- 7.Noda L. In: The Enzymes. Boyer P. D., editor. Vol. 8. New York: Academic; 1973. pp. 279–305. [Google Scholar]
- 8.Vonrhein C., Schlauderer G., Schulz G. E. Structure (London) 1995;3:483–490. doi: 10.1016/s0969-2126(01)00181-2. [DOI] [PubMed] [Google Scholar]
- 9.Müller C. W., Schlauderer G. J., Reinstein J., Schulz G. E. Structure (London) 1996;4:147–156. doi: 10.1016/s0969-2126(96)00018-4. [DOI] [PubMed] [Google Scholar]
- 10.Schulz G. E., Müller C. W., Diederichs K. J. Mol. Biol. 1990;213:627–630. doi: 10.1016/S0022-2836(05)80250-5. [DOI] [PubMed] [Google Scholar]
- 11.Wolf-Watz M., Thai V., Henzler-Wildman K., Hadjipavlou G., Eisenmesser E. Z., Kern D. Nat. Struct. Mol. Biol. 2004;11:945–949. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- 12.Miyashita O., Onuchic J. N., Wolynes P. G. Proc. Natl. Acad. Sci. USA. 2003;100:12570–12575. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H. J., Sheng X. R., Pan X. M., Zhou J. M. Biochem. Biophys. Res. Commun. 1997;238:382–386. doi: 10.1006/bbrc.1997.7301. [DOI] [PubMed] [Google Scholar]
- 14.Shapiro Y. E., Kahana E., Tugarinov V., Liang Z., Freed J. H., Meirovitch E. Biochemistry. 2002;41:6271–6281. doi: 10.1021/bi012132q. [DOI] [PubMed] [Google Scholar]
- 15.Závodszky P., Kardos J., Svingor Á., Petsko G. A. Proc. Natl. Acad. Sci. USA. 1998;95:7406–7411. doi: 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butterwick J. A., Patrick Loria J., Astrof N. S., Kroenke C. D., Cole R., Rance M., Palmer A. G., III J. Mol. Biol. 2004;339:855–871. doi: 10.1016/j.jmb.2004.03.055. [DOI] [PubMed] [Google Scholar]
- 17.Liang Z. X., Tsigos I., Lee T., Bouriotis V., Resing K. A., Ahn N. G., Klinman J. P. Biochemistry. 2004;43:14676–14683. doi: 10.1021/bi049004x. [DOI] [PubMed] [Google Scholar]
- 18.Bae E., Phillips G. N., Jr J. Biol. Chem. 2004;279:28202–28208. doi: 10.1074/jbc.M401865200. [DOI] [PubMed] [Google Scholar]
- 19.Haney P. J., Stees M., Konisky J. J. Biol. Chem. 1999;274:28453–28458. doi: 10.1074/jbc.274.40.28453. [DOI] [PubMed] [Google Scholar]
- 20.Perrier V., Surewicz W. K., Glaser P., Martineau L., Craescu C. T., Fabian H., Mantsch H. H., Barzu O., Gilles A. M. Biochemistry. 1994;33:9960–9967. doi: 10.1021/bi00199a019. [DOI] [PubMed] [Google Scholar]
- 21.Glaser P., Presecan E., Delepierre M., Surewicz W. K., Mantsch H. H., Barzu O., Gilles A. M. Biochemistry. 1992;31:3038–3043. doi: 10.1021/bi00127a002. [DOI] [PubMed] [Google Scholar]
- 22.Hayward S. J. Mol. Biol. 2004;339:1001–1021. doi: 10.1016/j.jmb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Yon J., Fried M. Nucleic Acids Res. 1989;17:4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stemmer W. P. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 25.Perrier V., Burlacu-Miron S., Bourgeois S., Surewicz W. K., Gilles A.-M. J. Biol. Chem. 1998;273:19097–19101. doi: 10.1074/jbc.273.30.19097. [DOI] [PubMed] [Google Scholar]
- 26.Bae E., Phillips G. N., Jr J. Biol. Chem. 2005;280:30943–30948. doi: 10.1074/jbc.M504216200. [DOI] [PubMed] [Google Scholar]
- 27.Somero G. N. Annu. Rev. Ecol. Syst. 1978;9:1–29. [Google Scholar]
- 28.D’Amico S., Claverie P., Collins T., Georlette D., Gratia E., Hoyoux A., Meuwis M. A., Feller G., Gerday C. Philos. Trans. R. Soc. London Ser. B. 2002;357:917–925. doi: 10.1098/rstb.2002.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.