Abstract
Small noncoding microRNAs (miRNAs) can contribute to cancer development and progression and are differentially expressed in normal tissues and cancers. From a large-scale miRnome analysis on 540 samples including lung, breast, stomach, prostate, colon, and pancreatic tumors, we identified a solid cancer miRNA signature composed by a large portion of overexpressed miRNAs. Among these miRNAs are some with well characterized cancer association, such as miR-17-5p, miR-20a, miR-21, miR-92, miR-106a, and miR-155. The predicted targets for the differentially expressed miRNAs are significantly enriched for protein-coding tumor suppressors and oncogenes (P < 0.0001). A number of the predicted targets, including the tumor suppressors RB1 (Retinoblastoma 1) and TGFBR2 (transforming growth factor, beta receptor II) genes were confirmed experimentally. Our results indicate that miRNAs are extensively involved in cancer pathogenesis of solid tumors and support their function as either dominant or recessive cancer genes.
Keywords: microarray, transcriptome, tumorigenesis
MicroRNAs (miRNAs) are a recently discovered class of small noncoding RNAs that regulate gene expression (1). Mature miRNAs are the results of sequential processing of primary transcripts (pri-miRNAs) mediated by two RNase III enzymes, Drosha and Dicer (2). Mature 18- to 24-nt-long miRNAs negatively regulate protein expression of specific mRNA by either translational inhibition or mRNAs degradation (3). miRNAs are differentially expressed in human cancers. We have previously reported that miR-15a and miR-16-1 are located within a 30-kb region of minimal loss in chronic lymphocytic leukaemia (CLL) and that both genes are often deleted or down-regulated (4). Later, other groups reported changes of miRNA expression in colorectal neoplasia (5), paediatric Burkitt lymphoma (6), lung cancer (7), large cell lymphoma (8), glioblastoma (9), and B cell lymphomas (10). miRNAs can also contribute to tumor development (11). The miR-17/92 cluster cooperates with c-MYC to accelerate tumor development (12, 13). We described and validated a microarray platform to evaluate the global expression of miRNA (14). Bead-based flow cytometry was also recently used to classify human cancers, and a general down-regulation of miRNAs was observed, with 129 of 131 differentially regulated miRNAs underexpressed in cancer (15). We used a microarray platform containing probes specific for active as well as precursor molecules to investigate the miRNA profile in B-CLL (16, 17) as well as other cancer specific miRNA profiles, including breast carcinomas (18). Because of the many evidences that miRNAs are involved in cancer, it is of high importance to elucidate their roles in solid tumors and to further reveal common miRNA-driven pathways.
Results and Discussion
For further details, see Fig. 4 and Tables 3–14, which are published as supporting information on the PNAS web site.
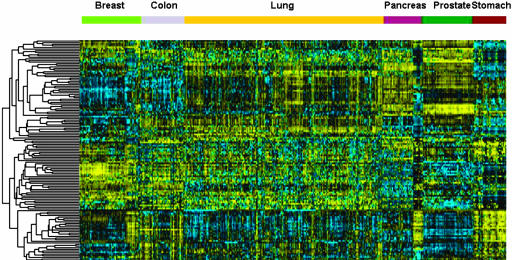
We describe here a large-scale detailed analysis of the miRNA profiles in 540 samples from six solid tumors (see Table 3). The clustering of miRNA expression profiles derived from 228 miRs in 363 solid cancer and 177 normal samples is shown in Fig. 1. The tree was constructed by using 137 different miRNAs, which are expressed in at least 90% of the samples and shows a very good separation between the different tissues. We initially compared all tumors against all normal tissues to identify 26 overexpressed and 17 underexpressed miRNAs (Table 4). These results indicated that, in solid cancers, the spectrum of expressed miRNAs is very different from that of normal cells (43 of 137 miRNAs, 31%). This analysis was performed to evaluate the results by using the same method as Lu et al. (15). The cancer versus normal tissue comparison was performed by using a reduced subset of lung samples (80 cancers and 40 normal samples), and thus numerically balance the different tissues, for a total of 404 samples. For statistical analysis, 137 miRs were retained from 228 measured, where expression values were above 256 (thresholded value) in at least 90% of samples. A t test was used to identify differentially expressed miRNAs (Table 11). The P values of the t test were corrected for multiple testing procedures and to control type I error rates. Adjusted P values were obtained by performing resampling with 500,000 permutations (19). We obtained 43 miRs with an adjusted P value <0.05. Twenty-six miRs are overexpressed and 17 underregulated when the six solid cancers (breast, colon, lung, pancreas, prostate, and stomach) are grouped together. As an alternative to t test, we used significance analysis of microarrays (SAM) for identification of differentially expressed miRNAs. This procedure allows the control of false detection rate (FDR). The delta was chosen as to result in an FDR ≤ 0.01, and 49 miRs were detected as differentially expressed, of which 34 were up-regulated (Table 12). We then identified the miRNA subset that results in the best tumor classification across all of the tissues, i.e., which best predicts the two classes (cancer and normal), by using the method of the nearest shrunken centroids, as implemented in prediction analysis of microarray. The prediction error was calculated by means of 10-fold cross-validation. The miRNAs were selected yielding the minimum misclassification error after cross-validation. Thirty six overexpressed miRNAs in cancer are associated to positive cancer scores, and 21 down-regulated miRNAs are associated to negative cancer scores (Table 13).
Fig. 1.
Clustering analysis of 540 samples representing six solid cancers and the respective normal tissues. MiRNAs were included in the tree when their expression level (background-subtracted intensity) was higher than the threshold value (256) in at least 90% of the samples. One hundred thirty-seven miRNAs were retained for clustering. Arrays were median-centered and normalized by using gene cluster 2.0. Average linkage clustering was performed by using uncentered correlation metric.
This overall classification might not be tailored to identify tissue-specific miRNA alterations that are consistently resulting in transformation, because miRNA expression is heavily tissue specific, as shown in Fig. 1. Thus, to identify the miRNAs that are really prognostic for cancer status, without incurring in the bias due to tissue specificity, we used an alternative approach. We first identified the six tissue-specific cancer signatures by performing independent prediction analysis of microarray tests (summarized in Tables 1 and 5–10), and then selected the miRNAs shared among the different histotypes miRNA signatures (Table 2). To compute the P values for this multitissue combined analysis, we performed a resampling test with 1,000,000 random permutations on the miRNA identities. A score was defined as the sum of the frequencies of the miRNAs reaching the sharing threshold (three tumors). The P value was defined as the relative frequency of simulation scores exceeding the real score. Table 2 includes 21 dysregulated miRNAs common to at least three types of solid cancers (P value = 2.5 × 10−3) and shows the frequency of each miRNA in the six solid cancers signatures.
Table 1.
miRNAs used to classify human cancers and normal tissues
| Cancer | Up-regulated miRs | Down-regulated miRs | Misclassification error after 10-fold cross validation |
|---|---|---|---|
| Breast | 15 | 12 | 0.08 |
| Colon | 21 | 1 | 0.09 |
| Lung | 35 | 3 | 0.31 |
| Pancreas | 55 | 2 | 0.02 |
| Prostate | 39 | 6 | 0.11 |
| Stomach | 22 | 6 | 0.19 |
Median normalization was performed, and the method of the nearest shrunken centroids was used to select predictive miRNAs (28).
Table 2.
The miRNAs shared by the signatures of the six solid cancers
| miR | N | Tumor type |
|---|---|---|
| miR-21 | 6 | Breast, colon, lung, pancreas, prostate, stomach |
| miR-17-5p | 5 | Breast, colon, lung, pancreas, prostate |
| miR-191 | 5 | Colon, lung, pancreas, prostate, stomach |
| miR-29b-2 | 4 | Breast, colon, pancreas, prostate |
| miR-223 | 4 | Colon, pancreas, prostate, stomach |
| miR-128b | 3 | Colon, lung, pancreas |
| miR-199a-1 | 3 | Lung, pancreas, prostate |
| miR-24-1 | 3 | Colon, pancreas, stomach |
| miR-24-2 | 3 | Colon, pancreas, stomach |
| miR-146 | 3 | Breast, pancreas, prostate |
| miR-155 | 3 | Breast, colon, lung |
| miR-181b-1 | 3 | Breast, pancreas, prostate |
| miR-20a | 3 | Colon, pancreas, prostate |
| miR-107 | 3 | Colon, pancreas, stomach |
| miR-32 | 3 | Colon, pancreas, prostate |
| miR-92-2 | 3 | Pancreas, prostate, stomach |
| miR-214 | 3 | Pancreas, prostate, stomach |
| miR-30c | 3 | Colon, pancreas, prostate |
| miR-25 | 3 | Pancreas, prostate, stomach |
| miR-221 | 3 | Colon, pancreas, stomach |
| miR-106a | 3 | Colon, pancreas, prostate |
The list includes 21 commonly up-regulated microRNAs in 3 or more (N) types of solid cancers (P value = 2.5 × 10−3).
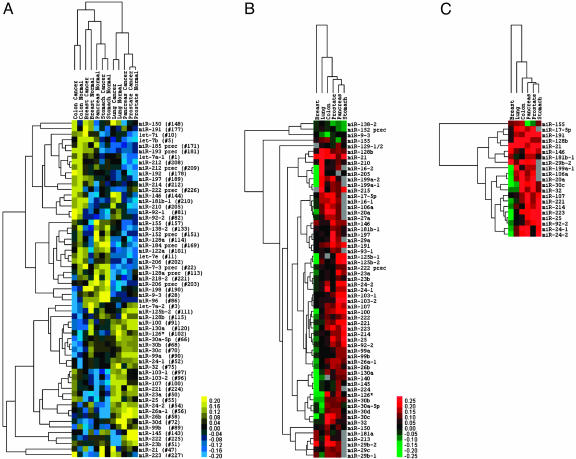
To maximize concision, we then computed the mean of the miRNA absolute expression levels for the six cancer/normal pairs. These miRNAs correctly cluster the different tissues, irrespective of the disease status (Fig. 2A). Fig. 2B shows the differential expression of the common miRNAs across the different tumors, in relation to the normal tissues. The tree displays the different cancer types according to the fold changes in the miRNA subset. Prostate, colon, stomach, and pancreas are most similar among them, whereas lung and breast are represented by a fairly different signature (Fig. 2B). Fig. 2 clearly shows which miRNAs are associated to a particular type of cancer. Strikingly, miR-21, miR-191, and miR-17-5p are significantly overexpressed in all of the tumor types we considered, or in five of six, respectively. miR-21 was reported to be overexpressed in glioblastoma and to have antiapoptotic properties (9). Lung cancer shares a portion of its signature with breast cancer and a portion with the other solid tumors, including miR-17/20/92, all three members of the miRNA cluster that actively cooperates with c-Myc to accelerate lymphomagenesis (12). The presence of these miRNAs as overexpressed is an excellent confirmation of our approach. A second miRNA group that is activated includes miR-210 and miR-213; together with miR-155, which was already reported to be amplified in large cell lymphomas (8), children with Burkitt lymphomas (6), and various B cell lymphomas (10), these miRNAs are the only ones up-regulated in breast and lung cancer. miR-218-2 is consistently down-regulated in colon, stomach, prostate, and pancreas cancers, but not in lung and breast carcinomas.
Fig. 2.
miRNA expression signature in six solid cancers. (A) Expression of the differentially regulated miRNAs across solid cancers. Sixty-one miRNAs are present in at least 90% of the tissues. The tree displays their average absolute expression values after log2 transformation. The mean was computed over all samples from the same tissue or tumor histotype. Genes and arrays were mean-centered and normalized by using gene cluster 2.0. Average linkage clustering was performed by using Euclidean distance. (B) Fold changes (cancer vs. normal) of the miRNAs present in at least 75% of the solid tumors with at least one tumor absolute value higher than 2. The tree displays the log2 transformation of the average fold changes (cancer over normal). The mean was computed over all samples from the same tissue or tumor histotype. Arrays were mean centered and normalized by using gene cluster 2.0. Average linkage clustering was performed by using uncentered correlation metric. (C) Fold changes (cancer vs. normal) of the miRNAs present in the signatures of at least 50% of the solid tumors. The tree displays the log2 transformation of the average fold changes (cancer over normal). The mean was computed over all samples from the same tissue or tumor histotype. Arrays were mean centered and normalized by using gene cluster 2.0. Average linkage clustering was performed by using uncentered correlation metric.
Several observations further strengthen our results. First, in this study, we determined the expression levels of both the precursor pre-miRNA and the mature miRNA for the majority of genes. Of note, with the exception of miR-212 and miR-128a, in all other instances the abnormally expressed region was that corresponding to the active part, the only one interacting with the target mRNA. Second, as shown in Fig. 2C, the expression variation of the solid cancer miRNA signature is often univocal (namely down- or up-regulation) across the different types of cancers, suggesting a common mechanism of involvement in human tumorigenesis. Third, the microarray data were validated by solution hybridization for 12 breast samples (18) (miR-125b, miR-145, and miR-21) and 17 endocrine pancreatic cancers and normals (miR-103, miR-155, and miR-204) (C.R. and C.M.C., unpublished data), strongly confirming the accuracy of our microarray data.
We did not observe the almost exclusively down-regulation of miRNA (129 of 131) in cancer that has been reported recently by Lu et al. (15). The difference between our work and that of Lu et al. might be due either to the difference in samples number (Lu et al. used <100 biopsies collectively derived from >10 different solid cancer types) or to the different technical platform. It is noteworthy to underline that proven oncogenic miRNAs, such as miR-155, miR-17-5p, miR-92-1, and miR-21 were not identified in Lu et al.’s paper as overexpressed. On the contrary and rather surprisingly, miR-17-5p, miR-20, and miR-92 were described as underregulated in tumors. Different analytical approaches were used; Lu et al. analyzed all of the tumor profiles versus all of the normal profiles, rather then performing separate tissue-matched analyses as we also did. In the all tumors vs. all normals comparison, we correctly detected miR-21 and miR-17-5p as overexpressed in cancer, in agreement with the published literature (9, 13).There is also a general agreement between the overall t test analysis and the tissue-specific cancer expression of the selected miRNAs. Nevertheless, occasionally this procedure can be misleading: for example, in our set of samples, miR-155 is overexpressed in breast, lung, and colon (Fig. 2C), but because it is strongly up-regulated in normal pancreas, when an all tumors vs. all normals comparison is performed, miR-155 appears as down-regulated in solid tumors, albeit with a borderline significance (P value = 0.04).
The functional significance of miRNA dysregulation in cancer we showed here needs to be understood. In solid tumors, it appears as the most common miRNA event is gain of expression, whereas loss of expression in cancer is a more limited event and more tissue specific. We used a three-step consequential approach in the following order: first, in silico prediction of targets, then luciferase assay for first validation of cancer relevant targets, and finally, ex vivo tumor correlation between miRNA expression (by microarray) and target protein expression (by Western blotting) for a specific miRNA::mRNA interactor pair. Relevant targets for cancer miRNAs can be either recessive (tumor suppressor) or dominant (oncogene) cancer genes. To test this hypothesis we first used an in silico approach to identify the predicted miRNAs targets by using TargetScan, the database of conserved 3′ UTR miRNA targets (20). TargetScan contained 5,121 predictions for 18 miRNAs (out of the differentially expressed included in Table 2) of the dysregulated miRNAs, in the total 22,402 (26.5%) predictions. A total of 115 (of 263, 44%) of well known cancer genes were predicted as targets for these 18 miRNAs (Table 14). Such a high percentage of cancer genes targeted by cancer miRNAs is very unlikely due to chance (P < 0.0001, Fisher’s exact test).
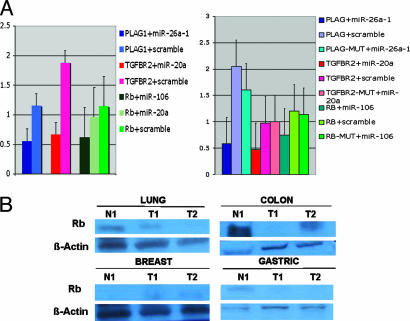
We then experimentally confirmed the in-silico predictions for three different cancer genes, Retinoblastoma (RB1), TGF-β-2 receptor (TGFBR2), and PLAG1. Three of four miRNA::mRNA predicted interactions (75%) were found to be positive in a luciferase assay by showing a significant reduction of protein translation in respect to the scrambled control oligoRNAs (Fig. 3A). Retinoblastoma 3′UTR, for example, was found to be functionally interacting strongly with miR-106a. The biological significance of the proven miRNA::mRNA interactions is reinforced by previous reports showing that Rb1 gene is normally transcribed in colon cancers, whereas various fractions of cells do not express RB1 protein (21). This finding suggests a posttranscriptional mechanism for regulation of RB1 that could be well explained by the concomitant miR-106a overexpression we detected in colon carcinoma (Fig. 2B). Furthermore, miR-20a is down-regulated in breast cancer (Fig. 2C) and, in fact, TGFBR2 protein is expressed in the epithelium of breast cancer (22). In addition, recently it has been shown that restoration of TGF-β signaling by introduction of exogenous TGFBR2 in lung cells lacking this protein reduces tumorigenicity in human lung cancers (23). Interestingly, miR-20a, which we proved to interact with TGFBR2 in vitro, is overexpressed in the lung cancer samples that we analyzed.
Fig. 3.
Protein-coding cancer genes as targets of solid cancer miRNA signature components. (A) The 3′ UTR of different cancer protein coding genes enable cancer miRNA regulation. The data present the relative repression of firefly luciferase expression standardized to a transfection control, renilla luciferase. The miRNAs were selected from those differentially regulated in solid cancers as shown in Fig. 2 and Table 2. PLAG1, pleiomorphic adenoma gene 1; TGFBR2, transforming growth factor, beta receptor II, Rb, retinoblastoma gene. pGL-3 (Promega) was used as the empty vector. miR-20a, miR-26a-1, and miR-106 oligoRNAs (sense and scrambled) were used for transfections. A second experiment using as control a mutated version of each target mRNA lacking the 5′ miRNA-end complementarity site (MUT) is shown at Right. All of the experiments were performed twice in triplicate (n = 6). (B) In cancer patients, the levels of RB1 protein correspond to an inversely correlation with miR-106a expression. For normalization, we used β actin.
Finally, we tested a set of patient samples to verify whether RB1 protein expression correlates with miR-106a expression (Fig. 3B). As expected, in gastric, prostate, and lung tumor samples, RB1 was down-regulated (in respect to the paired normal) and miR-106a was found to be overexpressed, whereas in breast tumor samples, where miR-106a is slightly down-regulated (Fig. 2C), RB1 is expressed at slightly higher levels than in the paired normal control.
These experimental proofs reinforce the hypothesis that key cancer genes are regulated by aberrant expression of miRNAs in solid cancers (24). These data add examples to the list of miRNA with important cancer gene targets as previously shown by Johnson et al. (25) (let-7::Ras interaction), O’Donnell et al. (13) (miR-17-5p::E2F1) and Cimmino et al. (26) (miR-16::Bcl2). Of note, the last two mentioned miRNAs are members of the described miRNA solid cancer signature we identified here. In conclusion, our data indicate that miRNAs are involved in cancer pathogenesis of solid tumors and support their function in either dominant or recessive fashion, by controlling the expression of protein-coding tumor suppressors and oncogenes.
Materials and Methods
Samples.
Five hundred and forty samples, including a total of 363 primary tumors and 177 normal tissues, were used in this study (Table 3). The solid cancers represented were lung carcinoma, breast carcinoma, prostate carcinoma, stomach carcinoma, and pancreatic endocrine tumor. All of the samples were obtained with patient’s informed consent and were histologically confirmed. The prostate samples have been received from The National Cancer Institute (NCI) Cooperative Prostate Cancer Tissue Resource (CPCTR). The normal samples were paired from the affected individual for lung and stomach and from noncancer individuals for the remaining tissues. All normal breast samples were obtained by pooling five unrelated normal tissues each. Total RNA from tissues was isolated by TRIzol (Invitrogen) according to manufacturer’s instructions.
MiRNA Microarrays.
Microarray analysis was performed as described (14). Briefly, 5 μg of total RNA was used for hybridization on miRNA microarray chips. These chips contain gene-specific 40-mer oligonucleotide probes, spotted by contacting technologies and covalently attached to a polymeric matrix. The microarrays were hybridized in 6× SSPE (0.9 M NaCl/60 mM NaH2PO4·H2O/8 mM EDTA, pH 7.4)/30% formamide at 25°C for 18 h, washed in 0.75× TNT (Tris·HCl/NaCl/Tween 20) at 37°C for 40 min, and processed by using a method of direct detection of the biotin-containing transcripts by streptavidin-Alexa Fluor 647 conjugate. Processed slides were scanned by using a microarray scanner, with the laser set to 635 nm, at fixed PMT setting, and a scan resolution of 10 mm. The data were confirmed by Northern blotting as described in refs. 16, 18, and 24.
Computational Analysis.
Microarray images were analyzed by using genepix pro. Average values of the replicate spots of each miRNA were background subtracted, normalized, and subjected to further analysis. Normalization was performed by using per chip median normalization method and the median array as ref. 14. Finally, we selected the miRNAs measured as present in at least the smallest of the two classes in a data set. Absent calls were thresholded to 4.5 before statistical analysis. This level is the average minimum intensity level detected in the experiments. miRNA nomenclature was according to the Genome Browser (http://genome.ucsc.edu) and the miRNA database at Sanger Center (27); in case of discrepancies we followed the miRNA database. Differentially expressed miRNAs were identified by using the t test procedure within significance analysis of microarrays (SAM) (28). SAM calculates a score for each gene on the basis of the change in expression relative to the standard deviation of all measurements. The miRNA signatures were determined by applying nearest shrunken centroids (29). This method identifies a subgroup of genes that best characterizes each solid cancer from its respective normal counterpart. The prediction error was calculated by means of 10-fold cross validation, and for each cancer we retained the miRNA signature that resulted in the minimal prediction error. A resampling test was performed by random permutation analysis to compute the P value of the shared signature. All miRNAs identified by the statistical analysis and the cancer signatures are listed in supporting information.
Tumor Suppressor and Oncogene Target Predictions.
The most recent TargetScan predictions (April 2005) were used to identify the putative miRNA targets. They include essentially the 3′ UTR targets reported by Lewis et al. (20) with a few changes arising from updated gene boundary definitions from the April 2005 UCSC Genome Browser mapping of RefSeq mRNAs to the hg17 human genome assembly. Among the putative targets, we specified known cancer genes (tumor suppressors and oncogenes) as identified in the Cancer Gene Census at www.sanger.ac.uk/genetics/CGP/Census or reported by OMIM at www.ncbi.nlm.nih.gov.
Target in Vitro Assays.
For luciferase reporter experiments, the 3′ UTR segments of Rb1, TGFBR2, and Plag1 predicted to interact with specific cancer miRNAs were amplified by PCR from human genomic DNA and inserted into the pGL3 control vector (Promega), using the XbaI site immediately downstream from the stop codon of luciferase. The human megakaryocytic cell line MEG-01 was grown in 10% FBS in RPMI medium 1640, supplemented with 1x nonessential amino acid and 1 mmol sodium pyruvate at 37°C in a humidified atmosphere of 5% CO2. The cells were cotransfected in 12-well plates by using siPORT neoFX (Ambion, Austin, TX) according to the manufacturer’s protocol with 0.4 μg of the firefly luciferase report vector and 0.08 μg of the control vector containing Renilla luciferase, pRL-TK (Promega). For each well, 10 nM miRNA oligonucleotides (Dharmacon Research, Lafayette, CO) or scrambled oligonucleotides (Ambion) were used. Firefly and Renilla luciferase activities were measured consecutively by using dual-luciferase assays (Promega) 24 h after transfection.
Western Blotting for RB1.
The levels of RB1 protein were quantified by using the mouse monoclonal anti-RB1 antibody (Santa Cruz Biotechnology) using standard procedures for Western blotting. The normalization was performed with mouse monoclonal anti-actin antibody (Sigma).
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute Program Project Grants P01CA76259, P01CA81534, and P30CA56036, a Kimmel Scholar award (to G.A.C.), and grants from Italian Ministry of Public Health, Italian Ministry of University Research, Telethon, and Italian Association for Cancer Research (AIRC) (to M.N., G.L., A.S. and S.V.), Progetto CAN2005 Comitato dei Sostenitori (2005) and Fondazione Cariverona and Fundacion Zanotto, Italy (AS).
Glossary
Abbreviation:
- miRNA
microRNA.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Ambros V. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 2.Cullen B. R. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Bartel D. P. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Calin G. A., Dumitru C. D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., et al. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michael M. Z., SM O. C., van Holst Pellekaan N. G., Young G. P., James R. J. Mol. Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 6.Metzler M., Wilda M., Busch K., Viehmann S., Borkhardt A. Genes Chromosomes Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 7.Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., et al. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 8.Eis P. S., Tam W., Sun L., Chadburn A., Li Z., Gomez M. F., Lund E., Dahlberg J. E. Proc. Natl. Acad. Sci. USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan J. A., Krichevsky A. M., Kosik K. S. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 10.Kluiver J., Poppema S., de Jong D., Blokzijl T., Harms G., Jacobs S., Kroesen B. J., van den Berg A. J. Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 11.Croce C. M., Calin G. A. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 12.He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S. W., Hannon G. J., Hammond S. M. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., Mendell J. T. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 14.Liu C.-G., Calin G. A., Meloon B., Gamliel N., Sevignani C., Ferracin M., Dumitru D. C., Shimizu M., Zupo S., Dono M., et al. Proc. Natl. Acad. Sci. USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., et al. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Calin G. A., Liu C. G., Sevignani C., Ferracin M., Felli N., Dumitru C. D., Shimizu M., Cimmino A., Zupo S., Dono M., et al. Proc. Natl. Acad. Sci. USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calin G. A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S., Iorio M. V., Visone R., Sever N. I., Fabbri M., et al. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 18.Iorio M. V., Ferracin M., Liu C. G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., et al. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 19.Jung S. H., Bang H., Young S. Biostatistics. 2005;6:157–169. doi: 10.1093/biostatistics/kxh026. [DOI] [PubMed] [Google Scholar]
- 20.Lewis B. P., Burge C. B., Bartel D. P. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 21.Ali A. A., Marcus J. N., Harvey J. P., Roll R., Hodgson C. P., Wildrick D. M., Chakraborty A., Boman B. M. FASEB J. 1993;7:931–937. doi: 10.1096/fasebj.7.10.8344490. [DOI] [PubMed] [Google Scholar]
- 22.Buck M. B., Fritz P., Dippon J., Zugmaier G., Knabbe C. Clin. Cancer Res. 2004;10:491–498. doi: 10.1158/1078-0432.ccr-0320-03. [DOI] [PubMed] [Google Scholar]
- 23.Anumanthan G., Halder S. K., Osada H., Takahashi T., Massion P. P., Carbone D. P., Datta P. K. Br. J. Cancer. 2005;93:1157–1167. doi: 10.1038/sj.bjc.6602831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R. M., Okamoto A., Yokota J., Tanaka T., et al. Cancer Cell. 2006 doi: 10.1016/j.ccr.2006.01.025. in press. [DOI] [PubMed] [Google Scholar]
- 25.Johnson S. M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K. L., Brown D., Slack F. J. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Cimmino A., Calin G. A., Fabbri M., Iorio M. V., Ferracin M., Shimizu M., Wojcik S. E., Aqeilan R. I., Zupo S., Dono M., et al. Proc. Natl. Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tusher V. G., Tibshirani R., Chu G. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tibshirani R., Hastie T., Narasimhan B., Chu G. Proc. Natl. Acad. Sci. USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.