Abstract
Fluorescent proteins have proven to be excellent reporters and biochemical sensors with a wide range of applications. In a split form, they are not fluorescent, but their fluorescence can be restored by supplementary protein–protein or protein–nucleic acid interactions that reassemble the split polypeptides. However, in prior studies, it took hours to restore the fluorescence of a split fluorescent protein because the formation of the protein chromophore slowly occurred de novo concurrently with reassembly. Here we provide evidence that a fluorogenic chromophore can self-catalytically form within an isolated N-terminal fragment of the enhanced green fluorescent protein (EGFP). We show that restoration of the split protein fluorescence can be driven by nucleic acid complementary interactions. In our assay, fluorescence development is fast (within a few minutes) when complementary oligonucleotide-linked fragments of the split EGFP are combined. The ability of our EGFP system to respond quickly to DNA hybridization should be useful for detecting the kinetics of many other types of pairwise interactions both in vitro and in living cells.
Keywords: split EGFP, DNA duplex, EGFP reassembly, protein folding, DMD simulations
Split fluorescent proteins are convenient tools to detect specific protein–protein or protein–nucleic acid interactions (1–5). The approach is based on the reassembly of a fluorescent protein from two nonfluorescent fragments driven by additional biomolecular interactions, and it results in restoration of fluorescence. The development of fluorescence, however, usually takes several hours because of the requirement of the de novo formation of the chromophore within the reassembled protein (6). Because this approach provides a slow response, it would clearly be advantageous to accelerate it.
A straightforward way to do this would be to use a fragment of a split protein with a preformed chromophore that is not fluorescent per se but is capable of bright fluorescence within a full-size protein. To the best of our knowledge, such a strategy has not been previously accomplished. In this report, we demonstrate the feasibility of an alternative approach based on the nucleic acid-supported fast complementation of EGFP fragments, one of which contains a mature profluorescent chromophore.
Results and Discussion
Molecular Modeling of Protein Folding: Large EGFP Fragment Can Potentially Form a Chromophore.
In this study, we used two fragments of the EGFP, which are linked, in its native structure, by a flexible loop of nine amino acids, residues 153–161 (7, 8). The larger, N-terminal EGFP fragment is known to contain the three amino acids that form a chromophore, which is fluorescent in native, but not in denatured, protein (6, 7). It is also known that this tripeptide chromophore exhibits no fluorescence in a separate large EGFP fragment (2, 4). EGFP chromophore formation is a self-catalytic process requiring correct protein folding (6). We were curious to see whether the N-terminal EGFP fragment (approximately two-thirds of the entire EGFP) was sufficiently large to develop a compact folded structure by itself. We also wondered whether this structure might be conformationally close enough to the corresponding part of the complete EGFP so that the chromophore could spontaneously form within the folded large EGFP fragment, even though it is not fluorescent.
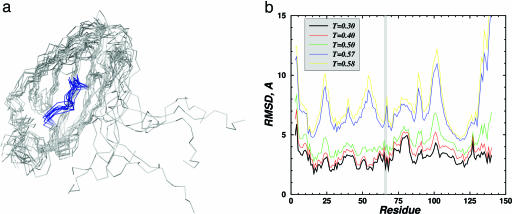
To test these hypotheses, we performed a molecular modeling analyses of EGFP and its large fragment by using the discrete molecular dynamics (DMD) approach (9, 10). The results of DMD simulations are shown in Fig. 1(also see Figs. 5–8, which are published as supporting information on the PNAS web site). Fig. 1a shows that at normal, low enough, temperatures, the large EGFP fragment is indeed folded into a compact structure, except for its dangling 20-residue-long C terminus. The arrangements of the chromophore-forming amino acids in the full-size EGFP and within its folded large fragment are essentially the same (Fig. 1b), hence making chromophore formation possible. However, as it can be seen in Fig. 1a, the chromophore-forming amino acids in the large EGFP fragment are exposed to a solvent, in contrast to the full-size EGFP, where these amino acids are buried deep inside the protein (7, 8). These amino acids also lack many important contacts with other residues of the smaller EGFP fragment, which are present in the full-size protein (11). Thus, even if the chromophore formed within the large EGFP fragment, it might not exhibit strong fluorescence.
Fig. 1.
Structure of the large EGFP fragment (1–158 N-terminal amino acids) analyzed by DMD simulations. (a) Backbone representation of 10 folded and aligned structures of the large EGFP fragment obtained in DMD simulations at T = 0.3 (T is measured in ε/KB units). The segment from 62 to 70 amino acids, containing the chromophore-forming amino acids (T66, Y67, and G68), is colored blue. The C terminus of this polypeptide is very flexible because of a small number of contacts with the rest of the molecule, so the alignment was made by omitting these amino acids. (b) The root-mean-square deviation (RMSD) of each residue in the folded large EGFP fragment relative to the intact EGFP structure as a function of temperature. The chromophore-forming residues are in the shaded region, and their spatial arrangement at lower temperatures is essentially fixed, with deviation ≤2Å.
The small, C-terminal EGFP fragment consists of the two β-hairpins, which do not interact with each other, so that, in contrast to the larger EGFP fragment, this polypeptide cannot form a well defined compact structure by itself. However, DMD simulations of EGFP folding (see Figs. 5–7) suggest that once the small EGFP fragment binds to its larger counterpart, it can find the correct position to become a part of a united compact protein structure with a buried chromophore, and the dangling part of the large EGFP fragment also folds during the reassembly. These simulations suggest that a mature profluorescent chromophore can form within the separate N-terminal fragment of the split EGFP. If this were the case, the protein fluorescence could be rapidly be restored when the large EGFP fragment is complemented with the C-terminal fragment of EGFP.
Isolation of EGFP Fragments, One of Which Contains a Profluorescent Chromophore.
Based on the molecular modeling analyses, we suspected that the large EGFP fragment might be isolated in vitro with a preformed chromophore. We genetically dissected EGFP between amino acids 158 and 159 by cloning and isolating two separate protein fragments corresponding to those tested in DMD simulations. The EGFP fragments were overexpressed in E. coli as fusions with small self-splitting SspDNAB intein (12) to facilitate protein purification (13). These polypeptides were isolated from inclusion bodies after refolding (see Materials and Methods for details). It has been shown that intein fusions with green fluorescent protein do not affect its proper folding (13). Fig. 2a shows that both EGFP fragments were obtained with high enough purity. Refolded protein samples contained ≥70% of the large and ≈90% of the small EGFP fragments.
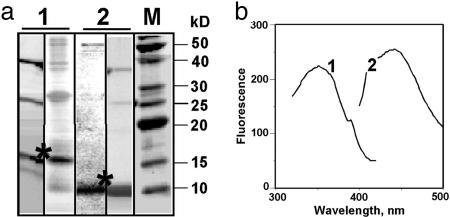
Fig. 2.
Characteristics of EGFP fragments overexpressed in E. coli and isolated by using the intein self-splicing technology. (a) Fifteen percent SDS/PAGE analysis of protein samples containing the large (lanes 1) and small (lanes 2) EGFP fragments (two samples of each fragment from different protein preparations are shown as examples). Lane M corresponds to a molecular mass protein ladder. Large and small EGFP fragments are seen as ≈15 kDa and ≈10 kDa bands, respectively (marked with asterisks). Although the small EGFP fragment is practically pure, the large EGFP fragment is somewhat contaminated by intein (≈25 kDa) and unsplit fusion (≈40 kDa). (b) Fluorescence excitation (curve 1) and fluorescence emission (curve 2) spectra of the large EGFP fragment (2 μM in PBS buffer, pH 7.4).
It was found that the absorption spectrum of the large EGFP fragment features significant absorbance in the range 300–400 nm (data not shown), which is characteristic for the chromophore of denatured EGFP (6) and which was also observed for other photoactive split EGFP variants (14). Note that, as expected, such long-wavelength absorbance was absent in the absorption spectrum of the small, C-terminal EGFP fragment.
The presence of a chromophore in the large EGFP fragment is more evident in its fluorescence spectrum (Fig. 2b). This polypeptide exhibits fluorescence with distinct maxima near 360 nm in the excitation spectrum and near 460 nm in the emission spectrum, whereas intensity is ≈100 times weaker than the peak fluorescence of intact EGFP. Although these spectra are quite different from those of the intact, full-length EGFP with peak excitation at 488 nm (7) and peak emission at 507 nm (see Fig. 4a), they correspond well to fluorescence spectra of the synthetic chromophore and to the spectra of a short, chromophore-containing peptide isolated from the intact fluorescent protein by partial proteolysis (15). Thus, these spectral data indicate that the large EGFP fragment isolated from the fusion to intein that was refolded from inclusion bodies does contain a preformed chromophore.
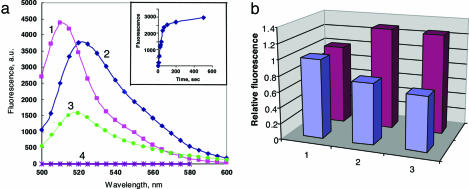
Fig. 4.
Fluorescent responses of the split EGFP system upon DNA hybridization. (a) Fluorescence spectra of intact EGFP (1) and of the split EGFP-based protein complex reassembled by DNA hybridization from the tripartite molecular constructions (2), each taken at ≈200 nM concentrations in PBS buffer at pH 7.4 (spectra recorded 20 min after mixing) (3), the same as sample 2 plus 100-fold excess of one of the two complementary oligonucleotides (nonbiotinylated oligo 1) (4), and control containing both EGFP fragments coupled to streptavidin but without oligonucleotides. (Inset) The time course of the fluorescence development in sample 2 was recorded at 524 nm. (b) Effect of Mg2+ cations on intact EGFP (blue) and on the reassembled split EGFP complex containing duplex DNA (purple). Column 1, no Mg2+; columns 2 and 3: 2 min and 3 h, respectively, after addition of 2 mM Mg2+.
Design and Implementation of the Split EGFP Reassembly with Quick Response to DNA Hybridization.
In our design (Fig. 3a), two EGFP fragments are coupled with complementary oligonucleotides by using biotin-streptavidin chemistry. The larger polypeptide contains a chromophore that becomes fluorescent only in a full-size protein. Indeed, the chromophore is not fluorescent in the EGFP fragment because it is exposed to and quenched by the solvent, and it also lacks necessary contacts with amino acids of the C-terminal fragment. However, if the two EGFP fragments are brought close to each other by nucleic acid complementary interactions, the C-terminal EGFP fragment should restore missing amino acid contacts and shield the chromophore from solution, which could result in the development of fluorescence.
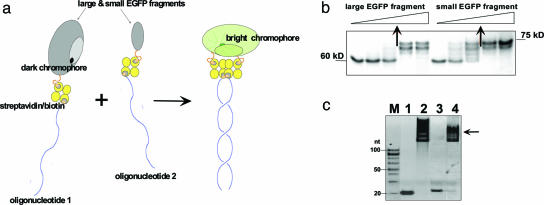
Fig. 3.
Design and assembly of a nucleic acid-supported EGFP complementation system with rapid signal response. (a) Schematics of the complementation of split fluorescent protein by DNA hybridization. Fluorescent protein (EGFP) is dissected into two nonfluorescent fragments, one of which contains preformed chromophore capable of bright fluorescence within a full-size protein. Both protein fragments are linked to complementary oligonucleotides via biotin–streptavidin interactions. In our protocol, we endeavor to create a 1:1:1 ratio of protein/streptavidin/oligonucleotide complex. In a mixture, the two nucleoprotein constructs associate by sequence-specific duplex DNA formation, which triggers complementation of the large and small EGFP fragments, resulting in fast development of fluorescence. (b) Gel-shift assay (10% SDS/PAGE) showing binding of increased amounts of biotinylated EGFP fragments with a fixed amount of streptavidin (2 μg; 60-kDa band). Arrows indicate the protein amounts resulting in 1:1 complexes (70- to 75-kDa bands), which correspond to ≥70% yield of biotinylation. (c) Gel-shift assay (10% PAGE) demonstrating the formation of 1:1:1 tripartite molecular constructions depicted in Fig. 1a and comprising the large or small EGFP fragment, streptavidin, and a corresponding oligonucleotide (see Table 1, which is published as supporting information on the PNAS web site, for their sequences). Lanes 1 and 2, biotinylated oligo 1 in the absence (1) or presence (2) of the large EGFP fragment coupled to streptavidin; lanes 3 and 4, biotinylated oligonucleotide 2 in the absence (3) or presence (4) of the small EGFP fragment coupled to streptavidin; M, 20-bp size marker. Arrow marks the position of the required oligonucleotide–protein complexes that are strongly shifted upward as expected.
The large and small EGFP fragments were expressed with extra cysteine residues at the C- and N-termini, respectively, for biotinylation with the sulfhydryl-reactive reagent, N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (biotin-HPDP) via S-S bond formation. The C- and N-terminally biotinylated polypeptides then can be coupled with biotinylated oligonucleotides via the biotin binding of streptavidin, which acts as a linker. We assumed that the terminal Cys in the A fragment of EGFP will be the major target site for biotinylation, whereas internal Cys-49 and Cys-71, which are buried to some extent inside the polypeptide (as supported by the DMD structure in Fig. 1) will be much less reactive.
We chose the noncovalent biotin/streptavidin-based coupling of proteins to oligonucleotides because it allows modular design (16, 17), which can be advantageous when different protein-oligonucleotide constructs are prepared for multiplex detection. Also note that the link formed between the protein and biotin-HPDP via S-S bonding can be readily cleaved with reducing agents, if subsequent disassembly is necessary. In planning this design, we assumed that the spatial arrangement would simultaneously allow the oligonucleotides to form duplexes and the EGFP fragments to come close to each other. Indeed, when two streptavidin molecules are located side by side, their centers are separated by ≈60 Å (18). Given that the biotin-binding site is located near the middle of each streptavidin subunit (19), one can estimate the smallest distance between the two sites in the contacting proteins as ≈30 Å. The length of biotin linkers in biotin-HPDP reagent and in the oligonucleotides was ≥25 Å, sufficient for all corresponding partners of the assembly to associate.
The biotinylated EGFP fragments were attached to streptavidin at a 1:1 ratio (Fig. 3b) and then coupled with the corresponding oligonucleotides bearing biotin at the 5′ or 3′ end (Fig. 3c). When these 1:1:1 tripartite molecular constructions were combined in equimolar amounts, a strong increase in fluorescence was detected with excitation/emission spectra resembling EGFP (Fig. 4a). In contrast, control experiments, i.e., mixing streptavidin-bound protein fragments without complementary oligonucleotides, did not show any appreciable fluorescence increase. The kinetics of the DNA-templated EGFP reassembly was fast with a t1/2 ≤ 1 min (Fig. 4a Inset). This time is close to the kinetics of renaturation of EGFP from denatured protein with mature chromophore (6, 7), and it agrees well with essentially immediate formation of DNA duplexes (20–22). The fluorescence intensity of the reassembled complexes varied from experiment to experiment with maximal response close to that of intact EGFP.
Two differences between the fluorescence spectra of the intact EGFP and reassembled protein should be noted. First, the excitation/emission maxima for reassembled protein were red-shifted to 490/524 nm, as compared to 488/507 nm for EGFP. The spectral changes may be explained by somewhat different arrangement of amino acids surrounding the chromophore within the reassembled protein and by the presence of streptavidin and/or negatively charged DNA within the complex. The second difference becomes apparent upon the addition of Mg2+ ions. The fluorescence of native EGFP gradually decreases after the addition of 2 mM MgSO4 and reaches ≈70% of its initial value in 3 h after, in accordance with the known quenching effect of bivalent cations on EGFP fluorescence (7). In contrast, the fluorescence of the reassembled complex increased ≈30% within a few minutes upon the addition of Mg2+ and remained essentially unchanged (Fig. 4b). This difference can be explained by a stabilizing effect of Mg2+ on duplex DNA, which is playing a major role in the reassembly of EGFP within the DNA–protein complex.
Restored Protein Fluorescence Can Be Quenched by Dissociating the DNA Duplex.
Finally, we examined the possibility of quenching the fluorescence of restored split EGFP by dissociating the assembled multicomponent complex by using competitive DNA hybridization. When one of the two complementary oligonucleotides was added in excess to the fluorescent complex, an essentially instant drop in fluorescence was detected (Fig. 4a). The competing hybridization of a nontagged oligonucleotide presumably displaces its protein-tagged equivalent and, as a result, splits the complemented protein complex.
Note that competitive hybridization was able to shut down only ≈50% of the restored fluorescence; increasing amounts of competitor had only little effect. The incomplete quenching may simply represent a higher stability of the complex of the two 1:1:1 conjugates, as compared to free DNA duplexes [this could occur because intact EGFP is more stable than the two separated fragments (see Fig. 6)]. Alternatively, it is conceivable that when the two biotinylated protein fragments are brought together to generate fluorescence, one of them may move to the neighboring streptavidin molecule and become linked to the same streptavidin molecule as the other, complementing protein fragment. This rearrangement would yield a reconstituted protein that generates fluorescence independent of hybridization state. Such an unwanted complexation could be avoided with covalent conjugation chemistry. It is also possible that some fraction of the restored EGFP molecules has a spatial arrangement that allows them to form very stable, essentially undissociatable, complexes. The irreversibility of protein reassembly has recently been observed, although with somewhat different variant of split green fluorescent protein (4).
Concluding Remarks.
It should be emphasized that in all prior reports, EGFP reassembly in vitro was most likely performed with protein lacking a mature chromophore, which formed only during or subsequent to reassembly (1–5). Because de novo formation of the mature profluorescent EGFP chromophore requires hours (6), the fluorescence development in these studies was very slow. In contrast, our split EGFP-based system responds very rapidly to assembly via nucleic acid complementary interactions, which are known, themselves, to be fast (20–26). This fast response occurs because the large EGFP fragment we isolated in vitro already contains the mature chromophore. Our data also show that the restored protein fluorescence is rapidly quenched upon dissociating the DNA duplex. Given that DNA hybridization-dehybridization events can be remotely controlled by local heating and/or electrical fields (20, 21, 27–29), it is possible to perform multiple on-off cycles of the optical signal generated by the system.
We believe that the nucleoprotein assemblies with DNA hybridization-controllable optical response will find applications in various biomolecular devices (20,30–33) by exploiting the modular design of the system and wide spectral range covered by the family of EGFP-like fluorescent proteins (34, 35), allowing multiplex probe detection. It is also clear that the fast response properties of our EGFP-based system should be useful for detecting many other types of pairwise interactions or promoter activities in living cells by using more conventional fusions of the EGFP fragments to interacting proteins (36, 37).
Materials and Methods
Molecular Modeling.
Modeling of EGFP and its fragments was performed by using a string of beads method (9). Each amino acid of a polypeptide is represented by two beads corresponding to the Cα and Cβ positions. Neighboring beads are constrained to mimic the backbone geometry and flexibility. The interactions between amino acids are simulated by a Gō-like structure-based potential (38). In such a model, two amino acids are assigned an attractive or repulsive potential depending on whether they form a contact in the native protein state. The conformation of native EGFP was taken from the Protein Data Bank (x-ray structure; PDB ID code 1c4f). To choose the contact potential for amino acids in EGFP fragments we used native structures of the full-size protein. Protein folding thermodynamics and kinetics were analyzed by the DMD approach (9, 10) starting from completely unfolded conformations.
Cloning, Expression, and Refolding of Polypeptides.
A plasmid containing the EGFP-1 gene (Clontech) was used as a template for PCR amplification of DNA sequences coding for the large (A) and small (B) EGFP fragments. Sequences of primers for PCR amplification are given in Table 1. The large fragment contained 158 N-terminal amino acids plus a C-terminal cysteine, and the small fragment contained the remaining C-terminal 81 amino acids plus an N-terminal cysteine. To isolate the protein fragments by using intein self-splitting chemistry (12, 13), PCR products were cloned in the TWIN-1 vector (New England Biolabs) to yield the C-terminal fusions to the SspDNAB intein that were expressed in BL21(DE3) pLys competent Escherichia coli cells (Stratagene). The structure of all constructs was verified by sequencing.
Cells were grown overnight to A600 = 0.6 and induced with 0.35 mM IPTG overnight at 25°C. Cells were precipitated by centrifugation, washed with a buffer containing 50 mM Tris·HCl (pH 8.5), 25% sucrose, 1 mM EDTA, 10 mM DTT, then frozen (−70°C for 10 min) and thawed (37°C for 5 min) three times. Cells were kept on ice and were lysed by sonication with three 30-sec bursts, each followed by 30-sec intervals (Sonifier cell disrupter W185c, Branson). The resulting mixture was centrifuged at 1,000 × gfor 5 min at 4°C; the pellets with inclusion bodies were resuspended in the same buffer and sonicated again for an additional three 30-sec bursts. Pellets were washed three times and then resuspended in buffer containing 25 mM Mes (pH 8.5), 8 M urea, 10 mM NaEDTA, 0.1 mM DTT and left at room temperature for 1 h. The solubilized proteins were centrifuged at 1,000 × g for 5 min, and the supernatant was then refolded by adding drop by drop to the refolding buffer (50 mM Tris, pH 8.5/500 mM NaCl/1 mM DTT) at a final dilution ratio of 1:100.
Purification of EGFP Fragments.
The refolded proteins were purified by using chitin columns (New England Biolabs), as adapted from the manufacturer’s instruction manual. Specifically, a solution of refolded protein (40 ml of SspDNAB intein-EGFP fragment fusion) was loaded on a column (0.8 × 4 cm Bio-Rad) containing 2 ml of chitin bead suspension. The column was preequilibrated with 10 ml of buffer containing 50 mM Tris·HCl (pH 8.5), 500 mM NaCl, 1 mM EDTA, 1 mM PMSF, and 0.1% Triton X-100 and washed with 20 ml of the same buffer. Next, the column was washed with 10 ml of the cleavage buffer (the same composition as before but at pH 7.0), then loaded with 10 ml of this buffer and left at room temperature overnight for intein self-cleavage. After that, the buffer was allowed to run through the column; 1.5-ml fractions were collected and then analyzed by SDS/PAGE (Fig. 2a). Protein absorption spectra were recorded on a Hitachi U-3010 spectrophotometer.
Coupling of Proteins with Oligonucleotides, Protein Complementation, and Fluorescence Measurements.
The EGFP protein fragments were transferred into PBS-EDTA buffer at pH 7.5 by using G-25 microspin columns (Amersham Pharmacia Biosciences). Then these solutions were mixed at a 10:1 volume ratio with 10 mM biotin-HPDP (Pierce) in dimethylformamide and incubated 2 h at room temperature to reach ≥70% biotinylation. Unreacted biotin-HPDP was removed from biotinylated proteins by gel filtration. Next, solutions that consisted mostly of 1:1 complexes of biotinylated EGFP fragments with streptavidin were obtained by incubating these fragments with equimolar amounts of streptavidin (as determined by titration experiments; see Fig. 3b) for 15 min at 37°C in PBS-EDTA buffer. Finally, an equimolar amount of the corresponding biotinylated oligonucleotide (for sequences, see Table 1) was added to each binary complex to get mostly 1:1:1 tripartite molecular constructions (see Fig. 3c). These constructions were mixed at a 1:1 molar ratio in the PBS-EDTA buffer to a final concentration of ≈200 nM. Fluorescence was monitored on a Hitachi F-2500 spectrofluorometer. To dissociate the reassembled oligonucleotide-supported protein constructs, a 100-fold excess of nonbiotinylated oligonucleotide (with the same sequence as the biotinylated oligomer used for coupling with the large EGFP fragment) was added, and the resulting fluorescence changes were recorded.
Supplementary Material
Acknowledgments
We thank Irina Smolina and Alexei Belenky for participating in early experiments on oligonucleotide–protein coupling; Maria Valencia-Burton for helpful discussions; Heiko Kuhn for drawing our attention to ref. 5; and Konstantin A. Lukyanov (Moscow Institute of Bioorganic Chemistry) and Bruno Samorì (University of Bologna), the first readers of this manuscript, for their encouraging comments and helpful suggestions. This work was sponsored by Hamilton Thorne Biosciences and Goldman Philanthropic Partnership.
Glossary
Abbreviations:
- DMD
discrete molecular dynamics
- biotin-HPDP
N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Ozawa T., Sako Y., Sato M., Kitamura T., Umezawa Y. Nat. Biotechnol. 2003;21:287–293. doi: 10.1038/nbt791. [DOI] [PubMed] [Google Scholar]
- 2.Hu C. D., Kerppola T. K. Nat. Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Remy I., Michnick S. W. Methods. 2004;32:381–388. doi: 10.1016/j.ymeth.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Magliery T. J., Wilson C. G., Pan W., Mishler D., Ghosh I., Hamilton A. D., Regan L. J. Am. Chem. Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 5.Stains C. I., Porter J. R., Ooi A. T., Segal D. J., Ghosh I. J. Am. Chem. Soc. 2005;127:10782–10783. doi: 10.1021/ja051969w. [DOI] [PubMed] [Google Scholar]
- 6.Reid B. G., Flynn G. C. Biochemistry. 1997;36:6786–6791. doi: 10.1021/bi970281w. [DOI] [PubMed] [Google Scholar]
- 7.Zimmer M. Chem. Rev. 2002;102:759–781. doi: 10.1021/cr010142r. [DOI] [PubMed] [Google Scholar]
- 8.Ormo M., Cubitt A. B., Kallio K., Gross L. A., Tsien R. Y, Remington S. J. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 9.Ding F., Buldyrev S. V., Dokholyan N. V. Biophys. J. 2005;88:147–155. doi: 10.1529/biophysj.104.046375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding F., Dokholyan N. V. Trends Biotechnol. 2005;23:450–455. doi: 10.1016/j.tibtech.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Jung G., Wiehler J, Zumbusch A. Biophys. J. 2005;88:1932–1947. doi: 10.1529/biophysj.104.044412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans T. C., Jr., Benner J., Xu M.-Q. J. Biol. Chem. 1999;274:18359–18363. doi: 10.1074/jbc.274.26.18359. [DOI] [PubMed] [Google Scholar]
- 13.Wang H., Chong S. Proc. Natl. Acad. Sci. USA. 2003;100:478–483. doi: 10.1073/pnas.0236088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akemann W., Raj C. D., Knopfel T. Photochem. Photobiol. 2001;74:356–363. doi: 10.1562/0031-8655(2001)074<0356:fcopeg>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Niwa H., Inouye S., Hirano T., Matsuno T., Kojima S., Kubota M., Ohashi M., Tsuji F. I. Proc. Natl. Acad. Sci. USA. 1996;93:13617–13622. doi: 10.1073/pnas.93.24.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gothelf K. V., Brown R. S. Chemistry. 2005;11:1062–1069. doi: 10.1002/chem.200400646. [DOI] [PubMed] [Google Scholar]
- 17.Niemeyer C. M., Sano T., Smith C. L., Cantor C. R. Nucleic Acids Res. 1994;22:5530–5539. doi: 10.1093/nar/22.25.5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coussaert T., Volkel A. R., Noolandi J., Gast A. P. Biophys. J. 2001;80:2004–2010. doi: 10.1016/S0006-3495(01)76170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freitag S., Le Trong I., Klumb L., Stayton P. S., Stenkamp R. E. Protein Sci. 1997;6:1157–1166. doi: 10.1002/pro.5560060604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamad-Schifferli K., Schwartz J. J., Santos A. T., Zhang S., Jacobson J. M. Nature. 2002;415:152–155. doi: 10.1038/415152a. [DOI] [PubMed] [Google Scholar]
- 21.Williams A. P., Longfellow C. E., Freier S. M., Kierzek R., Turner D. H. Biochemistry. 1989;28:4283–4291. doi: 10.1021/bi00436a025. [DOI] [PubMed] [Google Scholar]
- 22.Sekar M. M., Bloch W., St. John P. M. Nucleic Acids Res. 2005;33:366–375. doi: 10.1093/nar/gki163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pörschke D., Eigen M. J. Mol. Biol. 1971;62:361–381. doi: 10.1016/0022-2836(71)90433-5. [DOI] [PubMed] [Google Scholar]
- 24.Craig M. E., Crothers D. M., Doty P. J. Mol. Biol. 1971;62:383–401. doi: 10.1016/0022-2836(71)90434-7. [DOI] [PubMed] [Google Scholar]
- 25.Cohen R. J., Crothers D. M. J. Mol. Biol. 1971;61:525–542. doi: 10.1016/0022-2836(71)90063-5. [DOI] [PubMed] [Google Scholar]
- 26.Brucale M., Zuccheri G., Samorì B. Org. Biomol. Chem. 2005;3:575–577. doi: 10.1039/b418353n. [DOI] [PubMed] [Google Scholar]
- 27.Fixe F., Branz H. M., Louro N., Chu V., Prazeres D. M., Conde J. P. Biosens. Bioelectron. 2004;19:1591–1597. doi: 10.1016/j.bios.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 28.Gurtner C., Tu E., Jamshidi N., Haigis R. W., Onofrey T. J., Edman C. F., Sosnowski R., Wallace B., Heller M. J. Electrophoresis. 2002;23:1543–1550. doi: 10.1002/1522-2683(200205)23:10<1543::AID-ELPS1543>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 29.Su H.-J., Surrey S., McKenzie S. E., Fortina P., Graves D. J. Electrophoresis. 2002;23:1551–1557. doi: 10.1002/1522-2683(200205)23:10<1551::AID-ELPS1551>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 30.Dickson R. M., Cubitt A. B., Tsien R. Y., Moerner W. E. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 31.Choi B., Zocchi G., Wu Y., Chan S., Perry L. J. Phys. Rev. Lett. 2005;95:078102. doi: 10.1103/PhysRevLett.95.078102. [DOI] [PubMed] [Google Scholar]
- 32.Samori B., Zuccheri G. Angew. Chem. Int. Ed. Engl. 2005;44:1166–1181. doi: 10.1002/anie.200400652. [DOI] [PubMed] [Google Scholar]
- 33.Seeman N. C. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 34.Tsien R. Y. FEBS Lett. 2005;579:927–932. doi: 10.1016/j.febslet.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 35.Chudakov D. M., Lukyanov S., Lukyanov K. A. Trends Biotechnol. 2005;23:605–613. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Zhang S., Ma C., Chalfie M. Cell. 2004;119:137–144. doi: 10.1016/j.cell.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Cabantous S., Terwilliger T. C., Waldo G. S. Nat. Biotechnol. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 38.Gō N., Abe H. Biopolymers. 1981;20:991–1011. doi: 10.1002/bip.1981.360200511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.