Abstract
The opportunistic pathogen Pseudomonas aeruginosa is responsible for systemic infections in immunocompromised individuals and chronic respiratory disease in patients with cystic fibrosis. Cyclic nucleotides are known to play a variety of roles in the regulation of virulence-related factors in pathogenic bacteria. A set of P. aeruginosa genes, encoding proteins that contain putative domains characteristic of diguanylate cyclases (DGCs) and phosphodiesterases (PDEs) that are responsible for the maintenance of cellular levels of the second messenger bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) was identified in the annotated genomes of P. aeruginosa strains PAO1 and PA14. Although the majority of these genes are components of the P. aeruginosa core genome, several are located on presumptive horizontally acquired genomic islands. A comprehensive analysis of P. aeruginosa genes encoding the enzymes of c-di-GMP metabolism (DGC- and PDE-encoding genes) was carried out to analyze the function of c-di-GMP in two disease-related phenomena, cytotoxicity and biofilm formation. Analysis of the phenotypes of DGC and PDE mutants and overexpressing clones revealed that certain virulence-associated traits are controlled by multiple DGCs and PDEs through alterations in c-di-GMP levels. A set of mutants in selected DGC- and PDE-encoding genes exhibited attenuated virulence in a mouse infection model. Given that insertions in different DGC and PDE genes result in distinct phenotypes, it seems likely that the formation or degradation of c-di-GMP by these enzymes is in highly localized and intimately linked to particular targets of c-di-GMP action.
Keywords: cytotoxicity, biofilm
The binding of cyclic nucleotides to cognate regulatory proteins is an important component of a variety of intracellular signaling pathways. In prokaryotes, cAMP is a transcriptional coregulator for a number of metabolic and virulence-related genes. The analysis of currently available prokaryotic genome sequences has identified multiple proteins with nucleotide cyclase domains, the most common of which are the class III regulated cyclases (1, 2).
Recent advances in genome sequencing have also led to the identification of families of enzymes responsible for the turnover of bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP), which appears to be a common chemical messenger present uniquely in bacteria (1). c-di-GMP was initially identified as an activator of a cellulose synthetase complex in Gluconacetobacter xylinus (3). In G. xylinus and other bacteria, c-di-GMP is synthesized by a family of enzymes called diguanyl cyclases (DGCs), which share a sequence motif (the so-called GGDEF domain) (4). The levels of c-di-GMP are also controlled by c-di-GMP phosphodiesterases (PDEs) that contain a conserved EAL domain (4). A number of proteins contain both of these domains, suggesting they may be bifunctional enzymes with opposing activities, although the EAL domains of these presumptive bifunctional proteins tend to be more divergent and perhaps inactive (5).
The significant fraction of proteins with GGDEF and/or EAL domains identified by the analysis of genome sequences show a multimodular arrangement, in which the DGC and PDE domains are fused to signal receiver or transmission domains, suggesting that the activities of many DGCs and PDEs are regulated by environmental signals (6). VieA and PvrR, regulators of virulence in Vibrio cholerae and in certain strains of Pseudomonas aeruginosa, are examples of modular proteins with a PDE domain fused to a response regulator domain (6, 7). WspR, a P. aeruginosa chemosensor with a DGC and a response regulator domain, directs formation of c-di-GMP when expressed in the absence of WspF (8). Finally, two related regulators of surface protein expression in Bordetella pertussis and in P. aeruginosa are chimeric proteins with a response regulator module fused to a PDE domain and appear to function in conjunction with a linked two-component signal transduction system (9, 10). The targets of the regulatory activities of c-di-GMP in a variety of species appear to be surface-localized adhesive structures, including exopolysaccharides or fimbrial organelles. A number of DGC and PDE proteins are anchored to the membrane via transmembrane segments (11), suggesting that this localization is important for their function.
A number of proteins with predicted or documented DGC or PDE activity have been shown to regulate virulence-related traits of diverse pathogenic bacteria, including biofilm formation. Late-stage biofilm formation by Burkholderia cepacia involves a protein with a DGC domain (12), and biofilm formation in Yersinia pestis is controlled by HmsT (DGC) and HmsP (PDE), containing the consensus DGC and PDE domains, respectively (13). In Salmonella species, GcpA, which contains a GGDEF domain, regulates the synthesis of the cellulose component in biofilm communities (14). Another Salmonella regulator of biofilm formation and motility, AdrA, has a DGC domain and is responsible for synthesis of c-di-GMP in vivo (15). In Vibrio species, DGCs or PDEs have been implicated in regulating capsular production, motility, biofilm formation, the rugose phenotype, and virulence (16–19). Finally, Arr, a P. aeruginosa PDE domain-containing protein regulates biofilm formation in response to aminoglycosides (20).
One of the remarkable features of proteins involved in the metabolism of c-di-GMP is the apparent redundancy of proteins with DGC and PDE domains (11). However, it is unclear what environmental conditions control expression of each of their respective genes, and how the activity of their products is regulated. Even more intriguing is how c-di-GMP concentrations can affect targets specific for one enzyme that produces or degrades c-di-GMP, considering that, as a small molecule, this dinucleotide is presumably freely diffusible in the cytoplasm.
Here we report the results of a systematic analysis of phenotypes of DGC and PDE domain-containing proteins in P. aeruginosa. In the annotated sequence of two P. aeruginosa strains, PAO1 and PA14, multiple coding sequences for proteins with GGDEF/EAL domains were identified. We examined a comprehensive set of mutants with transposon insertions in each of these genes generated by near-saturation mutagenesis of the genome of strain PA14, as well as of clones of the putative DGCs and PDEs overexpressed in wild-type PA14. We identified a number of mutants with distinct phenotypes: a block in type III secretion system (TTSS)-mediated cytotoxicity and inability to form biofilm or pellicle. We were also able to demonstrate formation or hydrolysis of c-di-GMP in P. aeruginosa expressing a number of DGCs and PDEs, respectively. Different PA14 mutants in DGCs and PDE genes displayed different degrees of attenuation of virulence in murine infection models, implicating c-di-GMP in virulence.
Results
Identification of Putative DGC and PDE Proteins in P. aeruginosa.
The annotated genome of P. aeruginosa PAO1 (www.pseudomonas.com) encodes 17 different proteins with a DGC domain, 5 with a PDE domain, and 16 that contain both of these domains, diguanyl cyclase-phosphoesterase fusion (DGC-PDE). The genome of strain PA14 (http://ausubellab.mgh.harvard.edu/pa14sequencing) also contains most of these same DGC- and PDE-encoding genes, with three exceptions: PA14 is missing one DGC- (PA2771) and one PDE-encoding gene (PA2818) (arr) and has one additional gene, pvrR (6), encoding a PDE domain. In the PA14 genome, the corresponding regions of the PAO1 genome that encode PA2771 and PA2818 contain a number of genes, none of which encode proteins with DGC or PDE domains (discussed in more detail below). A schematic representation of the domains within these proteins is shown in Fig. 5, which is published as supporting information on the PNAS web site. In addition to the GGDEF/EAL domains, all of the recognizable domains in these DGC and PDE domain-containing proteins are involved in signal transduction and include the PAS and PAC domains, implicated in heme binding: a CheY-like response regulator domain, a member of the CHASE family of extracellular domains; a HAMP domain, found as a linker between various receptor domains and signal-transducing protein (21); the MHYT domain, which is often found at the N terminus of proteins that are part of signal transduction pathways (22); and a protein with a 7TMR-DISMED2/7TMR-DISM_7TM (seven-transmembrane receptor with diverse intracellular signaling module) domain at its N terminus, implicated in the sensing of external carbohydrates (23). Collectively, these structural features suggest that all of these proteins are involved in signal transduction and most likely function in the synthesis or hydrolysis of c-di-GMP after reception of signals that stimulate enzymatic activity.
We determined the evolutionary relationships among the DGC (GGDEF domain, pfam PF00990) and PDE (EAL domain, pfam PF0053) (Fig. 6 A and B, which is published as supporting information on the PNAS web site). The phylogenetic trees show that the DGC modules can be grouped into three families (I–III). The proteins in group I contain almost exclusively DGC domains with a conserved GGEEF motif and are not fused to a PDE module (Fig. 6 A and C). All but two members in groups II and III are DGC-PDE proteins containing both DGC and PDE modules. This phylogenetic relationship suggests that proteins with only a DGC domain have evolved independently from those in which a DGC domain is linked to a PDE domain. Groups II and III can be distinguished based on their diminishing similarity to the consensus DGC sequence, with members of group III being most distant (Fig. 6D). When known DGCs are included in this analysis [PleD of Caulobacter crescentus (24) and the G. xylinus DGC (3, 25)], both of these proteins cluster in group I. The poor correlation of group III GGDEF domains with the consensus sequence suggests they may be enzymatically inactive.
A similar alignment and phylogenic analysis of the PDE domains are shown in Fig. 6 B and D. Three of these putative PDE modules, in PA2200, PA2818, and PA3285, appear to be more closely related and are not linked to DGCs. The other two (PvrR and PA3947) are in a separate branch of the tree. PvrR shows a significant overall similarity to V. cholerae VieS, and this close relationship is reflected in their respective PDE domains.
Genomic Analysis of Genes Encoding DGC, PDE, and DGC-PDE.
We examined the prevalence of genes encoding putative DGCs and PDEs among P. aeruginosa clinical and environmental isolates using the microarray analysis described previously (26). The results shown in Fig. 7A, which is published as supporting information on the PNAS web site, demonstrate that the majority of these genes, with the exception of PAO1 genes PA2771 and PA2818, are components of the P. aeruginosa core genome and are found in all strains tested, suggesting that the products of these genes are used by the bacteria in one or several environmental niches that this organism can occupy. The two exceptions, PA2771 and PA2818, are missing in a significant fraction of strains analyzed, including PA14. These genes are located in or near regions of high genomic plasticity and are probably components of genomic islands (26). We determined the occupancy of these sites by PCR-mediated amplification of their respective chromosomal regions using primers that were located in core genomic sequences flanking PA2771 and PA2818. All strains harboring PA2771 (13 of 20 examined) had sequences identical to those found at the corresponding location in PAO1, whereas the seven strains that lacked PA2771 matched the sequence in the PA14 genome (Fig. 7B). In the PA14 genome, the PA2771 location contains four ORFs, one encoding a putative acid phosphatase and three hypothetical proteins.
PA2818 is found only in a limited number (5 of 20 tested) of P. aeruginosa strains. In PAO1, it is located near a cluster of three t-RNA genes (PA2819.1–3), a region of the genome that is highly variable in P. aeruginosa strains of different origin. Analysis of this chromosomal region by PCR and sequencing showed that three distinct gene sequences can occupy this locus (Fig. 7C). Five strains contain a sequence corresponding to that found in the PAO1 genome, including PA2818 and PA2819. The second island is found in PA14 and 12 other strains harbor sequences that are identical to the flanks of a DNA segment specifically found in the so-called clone C strains, which is highly prevalent in European isolates (27). Last, three strains contain sequences at the 5′ end that are homologous to the DNA of P. aeruginosa Clone C and a sequence at their 3′ homologous to PAGI-2, a genomic island found in one of the P. aeruginosa clone C strains (28). Neither Clone C DNA nor PAGI-2 DNA harbors ORFs encoding DGC or PDE proteins. Thus, in addition to the sequence present in PAO1, this location can apparently be occupied by one of two different genomic islands that share similarity with the ends of genetic elements found in PA14 and certain CF strains (27, 29). Therefore, PA2818 and PA2771 are located near sites of integration for multiple genomic islands.
Phenotypes of Transposon Mutants of DGC and PDE-Encoding Genes.
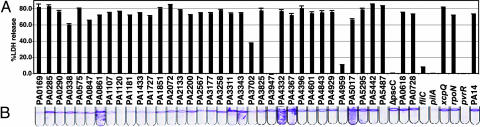
The contribution of genes encoding DGC, PDE, and DGC-PDE proteins to TTSS-mediated cytotoxicity and biofilm formation, two virulence-associated traits of P. aeruginosa, was assessed. A library of mariner transposon insertions has been generated in strain PA14 and a substantial number (24,089) of the insertions precisely mapped by sequencing the junction of the transposon and the chromosome (30). A set of mutants was selected from the PA14 mutant transposon bank that had the highest blast scores of the respective junction sequences to the target DGC and PDE coding genes and the proximity of the insertions to the first codon. Table 2, which is published as supporting information on the PNAS web site, provides additional information about the PA14 genes, the identifiers of the insertion mutants, and a summary of the phenotypes examined in this study. This set of mutants, corresponding to insertions in all of the DGC, PDE, and DGC-PDE genes in the PA14 genome, was tested for their ability to abrogate TTSS-cytotoxicity. Insertions in PA3947 (rocR), PA4959 (fimX), and pvrR were defective for cytotoxicity, and insertion in PA3702 (wspR) caused a partial defect (Fig. 1A). These genes have been implicated in TTSS (10, 28, 31, 32). With several mutants (rocR, fimX, and pvrR), we observed that the phenotypes of corresponding deletion mutants did not agree with those generated by transposon insertions. A more detailed analysis of the cytotoxicity phenotypes of fimX and rocR is described in Supporting Text and Fig. 8, which are published as supporting information on the PNAS web site.
Fig. 1.
Phenotypic analysis of P. aeruginosa PA14 transposon mutants in genes encoding the DGC, PDE, and DGC-PDE domain proteins. (A) Cytotoxic effect of P. aeruginosa PA14 transposon mutants on CHO cells after infection, monitored by the release of LDH. (B) Biofilm phenotypes were measured as the attachment of P. aeruginosa to tubes and formation of the biofilm rings and pellicles after 20-h static incubation at 30°C.
The same comprehensive set of DGC, PDE, and DGC-PDE mutants was also tested for their ability to form biofilm in a static culture (33). It has been shown that in many bacteria, biofilm formation is enhanced by a DGC-dependent increase in c-di-GMP and reduced by a PDE-mediated decrease in c-di-GMP concentrations (13, 15). Insertions in PA0169, PA1120, PA4959, and PA5487 abolished biofilm formation, and insertion in PA1107, PA1181, PA1433, PA1727, and PA3702 resulted in biofilm reduction. In contrast, pellicle formation (hyperbiofilm) was observed for insertions in PA0861, PA3311, PA3343, PA4332, PA4367, and PA5017 (Fig. 1B). Thus, insertions in genes encoding DGC or DGC-PDEs affected biofilm formation, whereas insertion in genes encoding only PDE domains, with the exception of pvrR, did not cause a biofilm-related phenotype in our assay. These results suggested that P. aeruginosa regulation of biofilm is affected by a number of DGC and DGC-PDE proteins.
Analysis of Overexpression of Genes Encoding Proteins with DGC and PDE Domains.
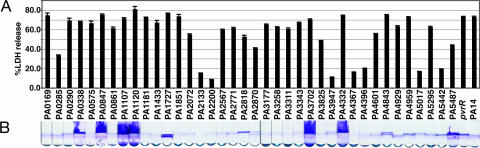
Proteins with DGC and PDE domains were overexpressed in P. aeruginosa PA14 by using vectors with the inducible tac promoter to assess the consequences of alterations in c-di-GMP levels. After the growth of these strains in isopropyl β-d-thiogalactoside-containing media, the ability to kill CHO cells and form biofilm was determined (Fig. 2A). A number of overexpressing clones showed a significant impairment of cytotoxicity, without interfering with the ability of these bacteria to form biofilm. PA14, overexpressing PA2133, PA2200, PA3947, PA4367, PA4396, PA5017, and PA5442, were significantly impaired in their ability to kill CHO cells. All of these genes, with the exception of PA4396, encode proteins with a single PDE domain or DGC-PDE. Thus, the reduced cytotoxicity of these overexpressing clones is most likely a consequence of reduced c-di-GMP levels.
Fig. 2.
Phenotypic analysis of P. aeruginosa PA14 overexpressing genes encoding the DGC, PDE, and DGC-PDE domain proteins. (A) Cytotoxic effect of PA14 transposon mutants on CHO cells after infection, monitored by the release of LDH. (B) Biofilm phenotypes were measured as the attachment of P. aeruginosa to glass tubes and the formation of the biofilm rings and pellicles after 12-h static incubation at 30°C.
The overexpression of a number of genes encoding DGC domains enhanced pellicle formation without affecting CHO cell cytotoxicity (Fig. 2B). Specifically, P. aeruginosa PA14 overexpressing PA0338, PA0847, PA1107, PA1120, PA3702, PA4332, and PA5487 formed pronounced pellicles, whereas overexpression of PA1727 resulted in the formation of a thinner pellicle. All of the hyperbiofilm pellicle-forming clones encoded proteins with a single DGC domain, with the exception of PA1727. PA1727 contains both a DGC and a PDE domain and had the weakest hyperbiofilm phenotype. These findings suggest that an increase in the levels of DGCs and presumably in c-di-GMP results in enhanced production of factors that promote bacterial binding to abiotic surfaces and interbacterial adherence. Moreover, these results indicated that changes associated with the surface of these bacteria that affect biofilm formation do not interfere with the activity of the TTSS.
Enzymatic Activities of Proteins with DGC and PDE Domains.
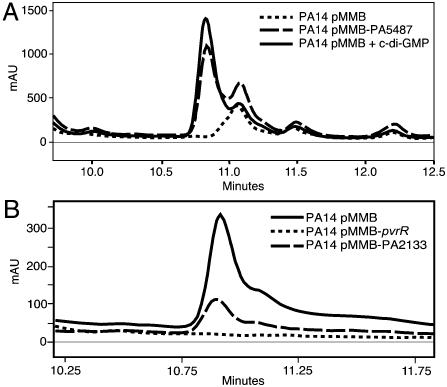
We used reverse-phase HPLC to assess whether proteins with predicted DGC and PDE domains specify enzymes capable of formation or hydrolysis of c-di-GMP, respectively (Fig. 3, Table 1). To detect DGC activity, cultures overexpressing genes that encode DGC-containing proteins were extracted and analyzed for c-di-GMP content (Fig. 3A). We detected c-di-GMP in extracts of 7 of 17 strains expressing predicted DGCs and in one strain expressing a DGC-PDE protein (PA1727) with both a DGC and a PDE domain. The ability of the PDE-containing proteins to hydrolyze c-di-GMP was tested by adding synthetic c-di-GMP to lysates of strains overexpressing these proteins and measuring the disappearance of the dinucleotide from the reaction mixture (Fig. 3B, Table 1). c-di-GMP hydrolysis was detected in all but one strain expressing genes encoding PDE enzymes and none in genes encoding DGC-PDE.
Fig. 3.
Analysis of enzymatic activities of proteins with DGC and PDE domains. Extracts of P. aeruginosa PA14, overexpressing genes with DGC or DGC-PDE domains, were fractionated by reverse-phase HPLC, and levels of c-di-GMP were quantified. For the assessment of PDE activity, synthetic c-di-GMP was added to lysates of cultures overexpressing proteins with PDE and DGC-PDE domains, and loss of the c-di-GMP peak in the HPLC analysis was quantified. (A) Representative traces from the HPLC analysis of the PA14 extract with the pMMB67EHGent vector or overexpressing PA5487. Trace of synthetic c-di-GMP. (B) Traces of extracts of reaction mixtures containing synthetic c-di-GMP and lysates of PA14 with pMMB67EHGent vector, overexpressing pvrR, or PA2133.
Table 1.
Enzymatic activities of the comprehensive set of P. aeruginosa DGCs and PDEs
| Gene | Conserved motif | Activity |
|||
|---|---|---|---|---|---|
| DGC* | PDE-A† | ||||
| None | — | — | |||
| PA0169 | GGEEF | — | |||
| PA0285 | GGDEF | ESL | — | — | |
| PA0290 | GGEEF | — | |||
| PA0338 | GGEEF | — | |||
| PA0575 | GGDEF | EAL | — | — | |
| PA0847 | GGDEF | +(16) | |||
| PA0861 | GGDEF | ELL | — | — | |
| PA1107 | GGEEF | +(35) | |||
| PA1120 | GGDEF | +(134) | |||
| PA1181 | GGDEF | ELL | — | — | |
| PA1433 | RGGEF | KVL | — | — | |
| PA1727 | GGDEF | EAL | +(24) | — | |
| PA1851 | GGEEF | — | |||
| PA2072 | GGDEF | EAL | — | — | |
| PA2133 | ETL | — | +(71) | ||
| PA2200 | EAL | +(61) | |||
| PA2567 | SPTRF | EAL | — | — | |
| PA2771 | GGEEF | — | |||
| PA2818 | EAL | — | |||
| PA2870 | GGEEF | +(96) | |||
| PA3177 | GGEEF | — | |||
| PA3258 | GGDDF | EAL | — | — | |
| PA3311 | AGDEF | EAL | — | — | |
| PA3343 | GGEEF | +(22) | |||
| PA3702 | GGEEF | +(445) | |||
| PA3825 | EVL | +(27) | |||
| PA3947 | EVL | +(100) | |||
| PA4332 | GGEEF | — | |||
| PA4367 | GGDQF | EAL | — | — | |
| PA4396 | DEQHF | — | |||
| PA4601 | GGDEF | EAL | — | — | |
| PA4843 | GGEEF | — | |||
| PA4929 | GGEEF | — | |||
| PA4959 | GDSIF | EVL | — | — | |
| PA5017 | ASNEF | EAL | — | — | |
| PA5295 | GSDEF | EAL | — | — | |
| PA5442 | AGDEF | EAL | — | — | |
| PA5487 | GGEEF | +(109) | |||
| pvrR | EAL | +(100) | |||
*DGC activity is measured as pmol of c-diGMP produced per mg wet cell weight.
†PDE activity is measured as percent of the added synthetic c-diGMP degraded.
Given the limits of sensitivity of the HPLC assay (12 pmol/mg cell wet weight), we cannot conclude that any particular DGC or PDE domain does not have the corresponding enzymatic activity in vivo. However, our inability to detect activity in all but one of the enzymes containing both DGC and PDEs may be significant. This may reflect some form of reciprocal inhibition of enzymatic activity by the two modules. Alternatively, for some of these enzymes, a high level of activity may require activating signals not present during the assay.
Contribution to Virulence During Acute Infection.
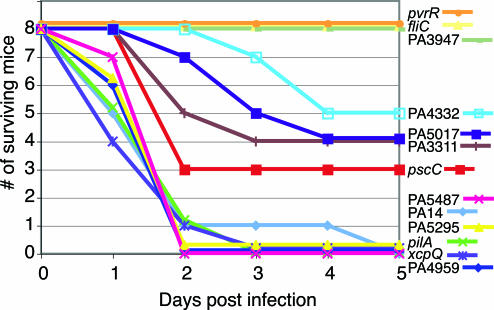
To assess whether mutations in genes encoding DGC and PDE influence virulence, we tested several of the mutants with biofilm or cytotoxicity phenotypes in a murine thermal injury model (34). For comparison, we also included mutants in various virulence factors (fliC, pscC, pilA, and xcpQ) and selected an additional transposon mutant (PA5295) that lacks a phenotype in the biofilm and cytotoxicity assays. When thermally injured mice were infected with wild-type PA14, almost 80% died by the second day after infection (Fig. 4). No attenuation of virulence was seen in the mutants that lacked the major subunit of the type IV pilus adhesin (pilA) or were defective in type II secretion (xcpQ). Insertions into PA3947 (rocR), pvrR, and fliC completely abolished the ability of PA14 to cause lethal infection. Similarly, the virulence phenotypes of mutants in PA5295 or the biofilm-defective PA5487 was identical to wild type. Mutations in PA4332, PA3311, and PA5017 all exhibited an intermediate virulence phenotype similar to the TTSS mutant in pscC, even in the presence of demonstrable CHO cell cytotoxicity. The virulence phenotypes of mutants in PA4959 (fimX) and in PA3702 (wspR) were identical to wild type, despite a strong or partial defect in CHO cell cytotoxicity, respectively. The lack of correlation between cytotoxicity and virulence in mice suggests that some of the genes encoding DGCs and PDEs play a broader role in acute infections that may not involve TTSS.
Fig. 4.
Virulence of P. aeruginosa PA14 strains with mutations in genes encoding the DGC, PDE, and DGC-PDE domain proteins in a murine model of acute infection. Survival of mice after thermal injury and infection by 2 × 106 wild-type and various P. aeruginosa mutants was monitored daily.
Discussion
The cellular levels of a newly recognized second-messenger c-di-GMP appear to be involved in regulating the expression of a number of prokaryotic surface proteins and carbohydrates. The abundance of genes with predicted DGC and PDE domains in prokaryotic genomes raises the questions of how a presumably freely diffusible small molecule can function specifically in different regulatory pathways, and whether these enzymes affect the overall (as opposed to localized) concentration of c-di-GMP in the cell. We have undertaken a systemic analysis of P. aeruginosa DGCs and PDEs to assess whether individual mutants in the corresponding genes or strains overexpressing the corresponding gene products show distinct virulence-related phenotypes. We analyzed genome sequences of P. aeruginosa PAO1 and PA14 for genes encoding proteins with DGC and PDE domains and identified a total of 39 genes that specify proteins with either one or both of these domains. In virtually all cases, the DGC and PDE domains are found either in proteins that also contain known signal-receiving modules or in proteins containing multiple hydrophobic segments, suggesting membrane localization. Although most of the DGC and PDE genes are components of the core genome, two (PA2771 and PA2818) in strain PAO1 and one (pvrR) in PA14 are on genomic islands and therefore were likely horizontally acquired. Interestingly, the distribution of PA2771 in P. aeruginosa strains is the same as the type III-delivered cytotoxin-encoding exoS (26). The significance of the coevolution of ExoS and PA2771 or ExoU with corresponding PA14 genomic island is not apparent. Another interesting evolutionary feature of the DGC family is that the proteins with a DGC domain but lacking PDE domain appear to be phylogenetically related in a single class, suggesting a common ancestry or convergent evolution of sequences related to their activity.
Examination of phenotypes of DGC, PDE, and DGC-PDE transposon insertion mutants revealed that several insertions have biofilm-related phenotypes, either a loss of the ability to form biofilm on solid surfaces or a so-called hyperbiofilm phenotype (pellicle producing). Other transposon insertions altered the ability of P. aeruginosa to intoxicate mammalian cells via the TTSS pathway. Mutation of pvrR, encoding a putative PDE, or fimX, which encodes a protein with relatively degenerate DGC and PDE domains, also compromised biofilm formation and the ability to kill CHO cells, a phenotype similar to a pilA mutant (lacking type IV pili). FimX-mediated twitching motility appears to be required for biofilm formation but is dispensable for cytotoxicity (Figs. 1B and 8).
We have also performed a comprehensive analysis of phenotypes of P. aeruginosa overexpressing each PDE and DGC domain and attempted to correlate these with measurements of c-di-GMP synthesis or degradation. In many instances, we observed multilayer pellicles that were associated with overexpression of enzymes containing a DGC module but not a PDE domain, although overexpression of PA1727, containing both PDE and DGC domains, consistently showed an enhanced ability to form biofilm. In general, we were able to correlate pellicle formation with high levels of c-di-GMP. PA1727 was the only bimodular (DGC-PDE) enzyme that showed elevated levels of c-di-GMP. In all, these data suggest that surface changes associated with enhanced ability to form biofilm depend at least in part on an increase in levels of c-di-GMP. Moreover, the ability of P. aeruginosa to form pellicles, presumably a consequence of production of surface components that promote interbacterial adherence, does not interfere with cytotoxicity, which requires attachment via the type IV pili and TTSS injection of effectors.
One of the more striking outcomes of the comprehensive analysis of DGCs and PDEs encoded in the P. aeruginosa genome is that they do not appear to be redundant, because related mutants have different phenotypes when grown under identical conditions. Although all of these enzymes presumably regulate cellular levels of c-di-GMP, the phenotypes associated with mutating particular DGC or PDE genes do not strictly correlate with alterations in the levels of this dinucleotide. For example, P. aeruginosa expressing PA2870 and PA3343, two DGCs that produce high levels of c-di-GMP in our assay (Table 1), do not cause an alteration in the biofilm phenotype. Therefore, simple changes in c-di-GMP levels cannot explain the observed effect on biofilm, cytotoxicity, or any other biological function in which this dinucleotide participates. The distinct specificity of each enzyme may be manifested locally, where the production or hydrolysis of c-di-GMP is intimately related to its site of activity.
Finally, we determined that mutations in genes encoding DGC and PDE result in distinct phenotypes in a P. aeruginosa murine model of burn-wound infection. In this acute infection model, initiation of biofilm formation is apparently not important, as indicated by the lack of attenuation of the pilA mutant. In contrast, hyperbiofilm formation (pellicle) may reduce the virulence capacity of P. aeruginosa. It is also conceivable that, in tissues of infected animals, these proteins control novel virulence traits. The next challenge is to elucidate the precise sites of activity of c-di-GMP, which would provide clues about the function of DGCs and PDEs in bacterial physiology, adaptation to environmental niches, and virulence.
Materials and Methods
Biofilm Assay.
Log-phase-grown bacteria were diluted to OD600 nm = 0.002 in LB broth, and 3 ml was incubated at 30°C for 20 h in 14-ml borosilicate tubes. Bacterial pellicles were stained by gently pouring off the media, washing twice with water, and staining with 0.1% crystal violet. Tubes were washed and rinsed with water until all unbound dye was removed (33). Three independent assays were carried out for each strain.
CHO Cell Cytotoxicity Assay.
The cytotoxic activity of P. aeruginosa transposon mutants was measured by release of lactate dehydrogenase (LDH) from CHO after exposure to bacteria, as described (35). Briefly, CHO cells were washed and covered with F-12 media containing 1% FBS and 2 mM glutamine. CHO cells were infected with mid-log P. aeruginosa at an initial multiplicity of infection of 10. Culture supernatants were collected at the indicated times, and LDH released from cells was measured by using a Roche Applied Science LDH kit, per the manufacturer’s instructions. Three independent assays were carried out for each strain.
Murine Model of Burn-Wound Infection.
Thermal injury and infection were carried out as described (36, 37). Briefly, mice were infected with 2 × 106 wild-type and mutant P. aeruginosa, and the surviving mice were counted daily. At this dose, 10% of mice infected with wild-type P. aeruginosa PA14 survived at day 5.
Supplementary Material
Acknowledgments
We thank Jason Gardner for assistance with mouse virulence studies. This work was supported by National Institutes of Health Grant AI21451 (to S.L.). The work in F.M.A.’s laboratory was funded by National Heart, Lung, and Blood Institute Grant U01 HL66678 and by a grant from the Cystic Fibrosis Foundation. Work in A.N.N′s laboratory is supported by the Shriners of North America. V.L. was supported by postdoctoral fellowships from the National Institutes of Health and the Cystic Fibrosis Foundation.
Abbreviations
- c-di-GMP
bis-(3′-5′)-cyclic dimeric GMP
- DGC
diguanyl cyclase
- PDE
phosphodiesterase
- TTSS
type III secretion system
- CHO
Chinese hamster ovary
- LDH
lactate dehydrogenase
- DGC-PDE
diguanyl cyclase-phosphoesterase fusion.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Galperin M. Y. Environ. Microbiol. 2004;6:552–567. doi: 10.1111/j.1462-2920.2004.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shenroy A. R., Visweswariah S. S. FEBS Lett. 2004;561:11–21. doi: 10.1016/s0014-5793(04)00128-0. [DOI] [PubMed] [Google Scholar]
- 3.Ross P., Weinhouse H., Aloni Y., Michaeli D., Ohana P., Mayer R., Braun S., de Vroom G. E., van der Marel A., van Boom J. H., et al. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 4.Ross P., Mayer R., Benziman M. Microbiol. Rev. 1991;55:35–58. doi: 10.1128/mr.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt A. J., Ryjenkov D. A., Gomelsky M. J. Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drenkard E., Ausubel F. M. Nature. 2002;416:740–743. doi: 10.1038/416740a. [DOI] [PubMed] [Google Scholar]
- 7.Tischler A. D., Lee S. H., Camilli A. J. Bacteriol. 2002;184:4104–4113. doi: 10.1128/JB.184.15.4104-4113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hickman J. W., Tifrea D. F., Harwood C. S. Proc. Natl. Acad. Sci. USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merkel T. J., Barros C., Stibitz S. J. Bacteriol. 1998;180:1682–1690. doi: 10.1128/jb.180.7.1682-1690.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulasekara H. D., Ventre I., Kulasekara B. R., Lazdunski A., Filloux A., Lory S. Mol. Microbiol. 2005;55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- 11.Galperin M. Y. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber B., Riedel K., Kothe M., Givskov M., Molin S., Eberl L. Mol. Microbiol. 2002;46:411–426. doi: 10.1046/j.1365-2958.2002.03182.x. [DOI] [PubMed] [Google Scholar]
- 13.Kirillina O., Fetherston J. D., Bobrov A. G., Abney J., Perry R. D. Mol. Microbiol. 2004;54:75–88. doi: 10.1111/j.1365-2958.2004.04253.x. [DOI] [PubMed] [Google Scholar]
- 14.Garcia B., Latasa C., Solano C., Garcia-del Portillo F., Gamazo C., Lasa I. Mol. Microbiol. 2004;54:264–277. doi: 10.1111/j.1365-2958.2004.04269.x. [DOI] [PubMed] [Google Scholar]
- 15.Simm R., Morr M., Kader A., Nimtz M., Romling U. Mol. Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 16.Boles B. R., McCarter L. L. J. Bacteriol. 2002;184:5946–5954. doi: 10.1128/JB.184.21.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bomchil N., Watnick P., Kolter R. J. Bacteriol. 2003;185:1384–1390. doi: 10.1128/JB.185.4.1384-1390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rashid M. H., Rajanna C., Ali A., Karaolis D. K. FEMS Microbiol. Lett. 2003;227:113–119. doi: 10.1016/S0378-1097(03)00657-8. [DOI] [PubMed] [Google Scholar]
- 19.Rashid M. H., Rajanna C., Zhang D., Pasquale V., Magder L. S., Ali A., Dumontet S., Karaolis D. K. FEMS Microbiol. Lett. 2004;230:105–113. doi: 10.1016/S0378-1097(03)00879-6. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman L. R., D’Argenio D. A., MacCoss M. J., Zhang Z., Jones R. A., Miller S. I. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 21.Williams S. B., Stewart V. Mol. Microbiol. 1999;33:1093–1102. doi: 10.1046/j.1365-2958.1999.01562.x. [DOI] [PubMed] [Google Scholar]
- 22.Galperin M. Y., Gaidenko T. A., Mulkidjanian A. Y., Nakano M., Price C. W. FEMS Microbiol. Lett. 2001;205:17–23. doi: 10.1111/j.1574-6968.2001.tb10919.x. [DOI] [PubMed] [Google Scholar]
- 23.Anantharaman V., Aravind L. BMC Genomics. 2003;4:34. doi: 10.1186/1471-2164-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aldridge P., Paul R., Goymer P., Rainey P., Jenal U. Mol. Microbiol. 2003;47:1695–1708. doi: 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- 25.Tal R., Wong H. C., Calhoon R., Gelfand D., Fear A. L., Volman G., Mayer R., Ross P., Amikam D., Weinhouse H., et al. J. Bacteriol. 1998;180:4416–4425. doi: 10.1128/jb.180.17.4416-4425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfgang M. C., Kulasekara B. R., Liang X., Boyd D., Wu K., Yang Q., Miyada C. G., Lory S. Proc. Natl. Acad. Sci. USA. 2003;100:8484–8489. doi: 10.1073/pnas.0832438100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larbig K. D., Christmann A., Johann A., Klockgether J., Hartsch T., Merkl R., Wiehlmann L., Fritz H. J., Tummler B. J. Bacteriol. 2002;184:6665–6680. doi: 10.1128/JB.184.23.6665-6680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klockgether J., Reva O., Larbig K., Tummler B. J. Bacteriol. 2004;186:518–534. doi: 10.1128/JB.186.2.518-534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuchma S. L., Connolly J. P., O’Toole G. A. J. Bacteriol. 2005;187:1441–1454. doi: 10.1128/JB.187.4.1441-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liberati N. T., Urbach J. M., Miyata S., Lee D. G., Drenkard E., Wu G., Villanueva J., Wei T., Ausubel F. M. Proc. Natl. Acad. Sci. USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang B., Whitchurch C. B., Mattick J. S. J. Bacteriol. 2003;185:7068–7076. doi: 10.1128/JB.185.24.7068-7076.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Argenio D. A., Calfee M. W., Rainey P. B., Pesci E. C. J. Bacteriol. 2002;184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedman L., Kolter R. Mol. Microbiol. 2004;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 34.Holder I. A., Neely A. N., Frank D. W. Burns. 2001;27:129–130. doi: 10.1016/s0305-4179(00)00142-x. [DOI] [PubMed] [Google Scholar]
- 35.Lee V. T., Smith R. S., Tummler B., Lory S. Infect. Immun. 2005;73:1695–1705. doi: 10.1128/IAI.73.3.1695-1705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stieritz D. D., Holder I. A. J. Infect. Dis. 1975;131:688–691. doi: 10.1093/infdis/131.6.688. [DOI] [PubMed] [Google Scholar]
- 37.Neely A. N., Law E. J., Holder I. A. Infect. Immun. 1986;52:200–204. doi: 10.1128/iai.52.1.200-204.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.