Abstract
Although conserved noncoding elements (CNEs) constitute the majority of sequences under purifying selection in the human genome, they remain poorly understood. CNEs seem to be largely unique, with no large families of similar elements reported to date. Here, we search for CNEs among the ancestral repeat classes in the human genome and report the discovery of a large CNE family containing >900 members. This family belongs to the MER121 class of repeats. Although the MER121 family members show considerable sequence variation among one another, the individual copies show striking conservation in orthologous locations across the human, dog, mouse, and rat genomes. The element is also present and conserved in orthologous locations in the marsupial, but its genome-wide dispersal postdates the divergence from birds. The comparative genomic data indicate that MER121 does not encode a family of either protein-coding or RNA genes. Although the precise function of these elements remains unknown, the evidence suggests that this unusual family may play a cis-regulatory or structural role in mammalian genomes.
One of most striking discoveries to arise from comparative genomic studies of the human genome is that the majority of functional sequences that have been under purifying selection during mammalian evolution do not encode proteins (1). Specifically, comparative genomics of the human, dog, mouse, and rat (HDMR) has revealed that ≈5–6% of the human genome is under purifying selection, but only 1–2% of this sequence is attributable to protein-coding sequences. The remainder consists of conserved noncoding elements (CNEs). Intense interest has focused on trying to decipher the function of these CNEs, which are likely to control gene regulation, chromosome structure, and other key functions.
Deciphering the function of the CNEs is particularly challenging because the vast majority seem to be unique in the genome; so far, no large families of similar CNEs have been discovered. For example, a study of the mammalian CNEs within a 1.8 Mb region containing the cystic fibrosis gene (CFTR) found the vast majority to be unique in the human genome (2). Similarly, a genome-wide comparison of human and pufferfish found that only 43 of the 1,373 identified CNEs showed any similarity to another CNE, with all of these cases being linked to paralogy of nearby genes (3). A recent attempt to cluster all of the CNEs identified from genome-wide alignments of human, mouse, and rat found that ≈96% of the elements were unique in the human genome (4). In this analysis, CNEs with similar human sequences were initially grouped together, and smaller clusters were then extracted by identifying highly connected subgraphs within each group. Only ≈250 of the initial groups had >10 CNEs, and these groups tended to be loosely connected. The largest group contained ≈800 CNEs, but each was similar, on average, to only two other elements within the group.
Here, we report an entire family of nearly 1,000 closely related CNEs in the human genome. This large family was previously missed because the study of CNEs has concentrated on unique sequences in the genome. Instead, the highly conserved CNE family reported here lies within the ancestral repeat (AR) sequences in the human genome.
ARs consist primarily of transposon fossils that predate the mammalian radiation (5). They cover ≈25% of the human genome and include ≈780 currently recognized classes (5). Because ARs are largely nonfunctional sequences, they have been used as a control to measure the background rate of neutral evolution against which to recognize the greater conservation of CNEs (1, 6–8). However, sporadic cases are known in which an AR element has acquired a new, useful function after insertion and has come under purifying selection. Two recent papers noted a total of three dozen clear instances (9, 10), and other papers have proposed other cases in which ARs may have been coopted (11, 12).
Against this background, we were surprised to find widespread conservation across an entire family of repetitive elements, the MER121 family of ancestral repeats. In this article, we characterize the conservation properties of this unusual family and speculate about its possible function.
MER121 Is Highly Conserved Among Mammals
Search for Perfectly Conserved Sequence Within ARs.
We sought to identify CNEs in the ARs in the human genome, by analyzing multiple sequence alignments (generated at University of California, Santa Cruz) among the HDMR genomes (6, 1, 7, 8). As an initial screen, we searched for long stretches of orthologous sequence showing perfect conservation, defined as 50 identical bases in all four species, without gaps. This is a stringent test. AR sequence is often partially or completely deleted in one or more species, and, even when an AR base has been retained in all four species, the frequency of perfect conservation across all four species is only ≈50%. Assuming uniform mutation rates at each base, the probability that a 50-mer would be retained across all four species and show perfect conservation would be <(1/2)50 ≈ 1/1015. The chance of seeing even a single such occurrence is remote.
In fact, we found 115 instances of perfect four-way conservation of at least 50 bp in the human genome. Strikingly, the majority fall into only a handful of repeat classes: MER121, L3b, L3, L2, and MIR-related (Table 1). Within these overrepresented classes, MER121 clearly stands out as the most enriched: although the MER121 class contains only 1/4,000th of AR sequence, nearly one-quarter of the 115 cases lie within this class. The MER121 class also shows similar enrichment when we repeat the analysis for perfectly conserved 30-mers (Table 1).
Table 1.
Conserved words within aligned ancestral repeats
| Repeat class | Human copies* | Human bases* | Perfectly conserved words of size ≥50 |
Perfectly conserved words of size ≥30 |
||
|---|---|---|---|---|---|---|
| No. | Per Mb | No. | Per Mb | |||
| MER121 | 909 | 170,592 | 26 | 152.4 | 140 | 820.7 |
| MER51-int | 115 | 47,135 | 1 | 21.2 | 1 | 21.2 |
| L3b | 7,689 | 1,059,580 | 7 | 6.6 | 58 | 54.7 |
| MER34B-int | 472 | 187,485 | 1 | 5.3 | 13 | 69.3 |
| Tigger8 | 845 | 201,639 | 1 | 5.0 | 1 | 5.0 |
| L3 | 47,395 | 9,355,520 | 30 | 3.2 | 164 | 17.5 |
| Charlie10 | 2,229 | 425,094 | 1 | 2.4 | 2 | 4.7 |
| MIRm | 32,045 | 2,854,339 | 5 | 1.8 | 25 | 8.8 |
| MARNA | 3,288 | 593,280 | 1 | 1.7 | 7 | 11.8 |
| MER103 | 6,834 | 768,897 | 1 | 1.3 | 3 | 3.9 |
| MER113 | 4,516 | 841,553 | 1 | 1.2 | 4 | 4.8 |
| MER102b | 3,975 | 843,979 | 1 | 1.2 | 2 | 2.4 |
| Charlie7 | 4,760 | 1,160,703 | 1 | 0.9 | 8 | 6.9 |
| Charlie8 | 7,617 | 1,231,496 | 1 | 0.8 | 4 | 3.2 |
| L1ME4a | 41,425 | 9,003,449 | 7 | 0.8 | 42 | 4.7 |
| L1MCa | 7,400 | 3,494,602 | 1 | 0.3 | 4 | 1.1 |
| MIRb | 279,852 | 40,575,849 | 10 | 0.2 | 91 | 2.2 |
| L1MC4 | 31,172 | 9,801,466 | 2 | 0.2 | 9 | 0.9 |
| MIR | 204,382 | 30,426,367 | 4 | 0.1 | 62 | 2.0 |
| L2 | 408,189 | 92,131,740 | 11 | 0.1 | 112 | 1.2 |
| MIR3 | 71,855 | 8,730,512 | 1 | 0.1 | 26 | 3.0 |
| L1MC4a | 30,167 | 9,732,206 | 1 | 0.1 | 4 | 1.1 |
*We count only instances on autosomes and chromosome X, not on chromosome Y or unplaced human contigs.
The MER121 class of medium-frequency repeats contains 919 copies in human. The overall consensus sequence for the family has length 412 bp, but the individual instances in the human genome have a median size of 180 bp (Table 2). The human copies display substantial sequence variation, typically containing different portions of the consensus and showing ≈26% sequence divergence from the consensus. Little has been said about these elements. The Repbase database (13) of repeat sequences characterizes MER121 as a “possible nonautonomous DNA transposon.” In contrast to most other repeat classes, the repeatmasker library (5) does not identify MER121 as a member of any larger repeat family.
Table 2.
Properties of MER121 and controls for conservation analysis
| Repeat class | Copies in human* | Size of consensus† | Median % divergence‡ | Size of human copy§ |
|||
|---|---|---|---|---|---|---|---|
| Mean | q25 | Median | q75 | ||||
| MER121 | 919 | 412 | 25.8 | 187.7 | 128 | 180 | 240 |
| MER119 | 1,168 | 586 | 23.1 | 266.4 | 115 | 210 | 433 |
| L2 | 408,478 | 2,977 | 30.5 | 227.4 | 87 | 132 | 288 |
*There are 909 MER121 copies on the autosomes and chromosome X. There are an additional seven copies on chromosome Y and three on unplaced contigs.
†Length of consensus sequence for repeat as given in repeatmasker repeat library.
‡repeatmasker reports the divergence of each copy from the element consensus.
§Size as reported by repeatmasker.
MER121 Shows Significant Conservation by Several Measures.
Having established that MER121 has by far the highest incidence of large conserved words (that is, n-mers), we proceeded to better characterize its conservation properties. As controls, we used two other repeat classes: MER119, a known nonautonomous DNA transposon, and L2, a LINE element (Table 2). Both classes are ancient and show divergence levels from their consensus that is comparable with MER121, but they serve as complementary controls in other ways. MER119 has a similar number of copies (1,168) and consensus size (586 bp) as MER121, and it contains no instances of perfectly conserved 30- or 50-mers. In contrast, L2 has many more copies (≈400,000) and a much larger consensus sequence (2,977 bp), and the family has a number of instances of large conserved words. In our analysis, we used all copies of MER119 and randomly selected 1,000 copies of L2.
We first determined the fraction of human copies that are present in all four species (Table 3). We required that at least 50 bp be present in each species, thereby allowing partial deletion. The proportion of MER121 retained across all four species (82%) is far higher than for the two controls (28% and 37%). (The difference between the controls likely reflects the larger size of L2, making it less likely to be completely deleted.)
Table 3.
Conservation of MER121, relative to typical ancestral repeat elements
| MER121 | MER119 | L2 | Significance* | |
|---|---|---|---|---|
| Retention across HDMR† | 82% | 28% | 37% | P < 10−100 |
| Length ratio relative to human‡ | 1.00 (0.05) | 0.89 (0.22) | 0.91 (0.22) | P < 10−27 |
| Four-way aligned bases§ | 96% | 73% | 69% | P < 10−100 |
| Four-way sequence identity¶ | 72% | 49% | 48% | P < 10−100 |
*Significance levels based on binomial test for proportions; Wilcoxon test for the length ratio. Significance levels below 10−100 are capped at this value.
†Proportion of human copies with orthologous copies in each of dog, mouse, and rat.
‡Median length ratio for mouse repeat relative to human repeat, for orthologous copies. Interquartile range is shown in parentheses.
§Proportion of human bases aligning to (ungapped) bases in each of dog, mouse, and rat. The values for MER119 and L2 are similar to the average across all ARs.
¶Proportion of four-way aligned bases that are identical across all four species. The values for MER119 and L2 are similar to the average across all ARs.
We next characterized the variation in the length of retained repeats at orthologous sites. Because rodents are especially prone to delete neutral sequences (1), we measured size variation as the ratio of the mouse and human size for each repeat instance. For both control classes, the retained copies are typically 10% smaller in mouse, but also show wide variability in the size ratio within the class. In sharp contrast, deletion and insertions (indels) are much rarer within the MER121 copies: the orthologous copies typically have nearly identical sizes.
Another test also confirms the relative lack of indels in MER121. For all retained repeats, we calculated the fraction of human bases that are aligned to bases in all other species (that is, ungapped columns in the multiple alignment, regardless of whether the sequence is conserved). As expected based on the conservation of overall length, nearly all MER121 bases lie within such four-way alignments (96%), whereas the proportion for the controls is only ≈70%.
Finally, we determined how often a four-way aligned base is identical in all four species. The rate of perfect conservation for MER121 is extremely high (72%), far above the controls (≈50%) and only slightly lower than is seen for coding exons (78%).
By all of these measures, the individual copies of MER121 show remarkable cross-species conservation. This conservation contrasts with the LINE L2 class, which contains a few examples of extreme conservation of large words, but does not show significant conservation across the class. The cross-species conservation of the individual MER121 copies is all the more remarkable given the great variability among the various human copies. The contrast between intraspecies variation and interspecies conservation can be appreciated in Fig. 1.
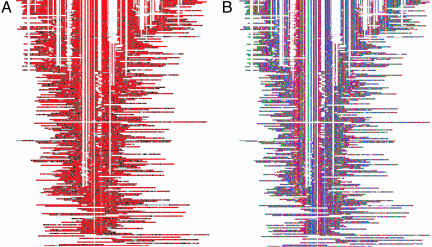
Fig. 1.
Conservation and orthologous bases of the top 200 human MER121 instances most similar to the consensus. Human instances are aligned to one another, starting from the copy most similar to consensus and following a progressive alignment strategy. This approach results in a multiple alignment with 200 rows, one for each human instance. To incorporate information about orthologous sequence in the other species, we replace each human base with its corresponding four-way HDMR multiple alignment column, thereby enlarging the alignment to 800 rows (= 200 × 4). (A) Positions are colored according to four-way conservation (red, bases with perfect four-way conservation; black, bases with four-way alignment but not perfect conservation; gray, human base lacks an orthologous base in at least one other species. (B) Positions are colored according to their DNA base (blue, A; green, G; orange, C; red, T).
Conservation Profile from HDMR Alignments.
We next examined variations in the cross-species conservation rate within all MER121 instances with the goal of identifying the best conserved portions shared between copies. To do this, we mapped the four-way alignments for each orthologous copy to the MER121 consensus sequence (thereby providing a common coordinate system)
Most copies have retained a central region of ≈150 bp. Fig. 2A shows the probability that each position in the consensus is covered by a four-way aligned column. Interestingly, although the flanking regions are less frequently retained than the central region, they show approximately the same conservation rate when they are present (Fig. 2B). The entire element is thus likely to be functional, although only portions are present in any given instance. Not surprisingly, we also observed that the rate of perfect four-way conservation is higher at the subset of aligned bases that can be mapped to the consensus sequence (88%) than for all aligned bases (72%) (Table 3).
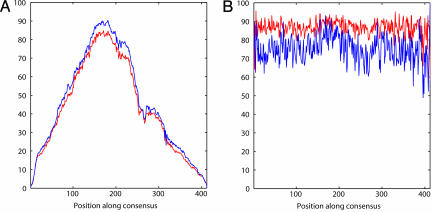
Fig. 2.
Conservation profile along the MER121 consensus. (A) Percentage of aligned human MER121 elements with an aligned orthologous base in the other species at the indicated position. (B) Percentage of these aligned bases showing perfect conservation across the species. Red curves indicate four-way comparison of HDMR; blue curves indicate two-way comparison of human and marsupial.
MER121 Dispersal Preceded Divergence from Marsupials.
We searched for instances of MER121 in the high-quality draft genome sequence of the opossum, Monodelphis domestica (≈7.2-fold redundancy, N50 supercontig size of 4.0 Mb). Using the repeatmasker program and the human MER121 consensus, we identified 1,064 copies. At least 620 can be reliably aligned to an orthologous copy in the human genome. This is likely an underestimate, due to the difficulties in establishing clear orthology when working with a draft assembly of an evolutionarily distant genome.
The pairwise human–marsupial alignments were also mapped to positions along the consensus (see Methods). The human–marsupial conservation landscape shows striking similarity to that seen for the HDMR comparison (Fig. 2 A and B). Although the absolute levels of sequence identity cannot be directly compared (one is a four-way analysis, the other a pairwise analysis), the overall shapes are strongly preserved.
MER121 Dispersal Postdated Divergence from Birds.
We followed the same procedure to search for instances of MER121 in the chicken genome (14). The repeatmasker program identified only three short instances of sequence similar to MER121 (lengths 69, 70, and 96 bases). The largest instance (96 bases) was highly diverged from consensus and did not align as credibly to the consensus as the other two instances. More sensitive analysis using Smith–Waterman alignments of both the MER121 consensus and a randomized control to the entire chicken genome confirmed that the two smaller instances are statistically significant, whereas the larger instance is not. To address the possibility that the chicken genome might contain additional MER121 elements that are too diverged to be detected with the mammalian consensus, we also searched the chicken genome using the 69- and 70-bp sequences identified as similar to MER121 (which we reasoned might be closer to additional elements in chicken). However, this strategy uncovered no additional copies in chicken.
We conclude that only two short sequences with similarity to the mammalian consensus for MER121 can be reliably identified in the current chicken genome sequence (chr1:91985730–91985798 and chr13:6024059–6024122 in the galGal2 assembly). Both share similarity with the same central portion of the repeat consensus (positions 176–239). Interestingly, one of the chicken sequences is orthologous to a short (72 bp) human MER121 element (downstream of the gene SAMSN1) and is surrounded by an orthologous sequence, some of which shows significant conservation across all four mammals. It is conceivable that this sequence could represent an ancestral source for the central region of the MER 121 element.
MER121 Encodes Noncoding Elements
Given the exceptional conservation properties of MER121, it is clear that it must have an important function that has been under purifying selection for 200 million years. We sought to show that MER121 encodes a noncoding element, by demonstrating that it does not have the properties expected of a family of protein-coding or RNA genes.
Conservation Pattern of MER121 Differs from Protein-Coding Genes.
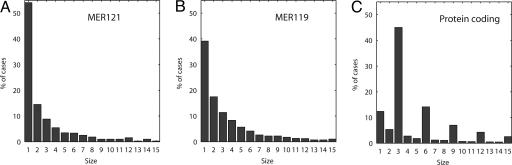
The high intraspecies variability of MER121 strongly suggests that MER121 does not encode a family of protein-coding genes. As more direct evidence, we examined the size distribution of indels in the four-way alignments. We saw no enrichment for indel sizes that are multiples of three, as typically observed in coding regions. Fig. 3 shows the distribution of gap sizes in four-way alignments of MER121 and MER119 and for a set of well-studied coding exons. Within the coding regions of a set of 5,430 well studied genes, 73% of the indels have sizes that are multiples of 3. For MER121 and MER119, the proportions are 14% and 19%.
Fig. 3.
Frequency of insertions and deletions of various lengths within aligned regions for MER121 (A), MER119 (B), and protein-coding regions (C). Graphs show gap events in human–dog sequence comparison within four-way alignments of HDMR. Gap events are human bases that align to a “-” character in dog.
No Significant Evidence of Transcription of MER121 in Public Databases.
We next searched for evidence that MER121 is expressed at the level of RNA. We searched for matches between the MER121 consensus sequence (412 bp) and cDNAs deposited in available mammalian cDNA databases, including the Mammalian Gene Collection (15) and the Fantom3 database (16). These databases together contain 149,645 sequences totaling 276 Mb. We considered only alignments with significance scores of E ≤ 10−3 with the blastn program (17). We found only a single transcript (AK051942) containing a significant alignment (E = 10−9, identity 60/69). The transcript has length 2,718 bp but contains only 69 bases that align to MER121. We also examined the entire collection of human and mouse ESTs deposited in GenBank (≈11 million sequences, totaling 5.53 Gb). Only 19 ESTs were found that contain >50 bp with similarity (E ≤ 10−3) to the MER121 consensus. In all cases, the region of shared similarity accounts for less than half of the total EST length.
These few cases of larger transcripts containing small segments of MER121-related sequence do not constitute biologically significant evidence of expression. The lack of MER121 transcripts in available databases does not formally rule out that the element encodes an RNA transcript [inasmuch as the available databases are enriched for transcripts with poly(A) tails]. However, we note that many known RNA genes do show significant evidence of expression in the same databases (Table 4, which is published as supporting information on the PNAS web site). The evidence thus argues against MER121 encoding a family of RNA genes.
No Significant Evidence of Conserved RNA Secondary Structure.
As further evidence, we considered whether the conservation pattern of MER121 elements is indicative of conserved RNA secondary structure, as seen in most gene families that encode structural RNAs. Toward this end, we compared all human instances of MER121 with recent genome-wide computational predictions of conserved folding structure from two different sources (18, 19).
Washietl et al. (18) identified regions of conserved RNA folding structure based on structural conservation and evidence of thermodynamic stability within conserved regions of the human genome. Within their set of high-confidence (P > 0.9) set of 35,843 predictions, there is only a single case of trivial overlap (6 bp) with a MER121 copy. Within their more permissive set (P > 0.5) with 91,368 predictions, there are five cases of overlap: three are trivial (<10 bp) and two are large (261 bp, 320 bp). Pedersen et al. (J. S. Pedersen, G. Bejerano, A. Siepel, K. Lindblad-Toh, E.S.L., J. Rogers, J. Kent, W. Miller, and D. Haussler, unpublished results) (Evofold predictions) identified regions by using a probabilistic model that incorporates a model of evolution together with one RNA secondary structure. Within their set of 48,479 predictions, there are 51 cases of overlap. All are small, with a median size of 24 bases (range 2–49 bases).
The tiny overlap between the MER121 elements and these computationally derived sets argues against conserved folding structure across the family of MER121 elements.
Further Characterization of MER121
Because the evidence indicates that MER121 does not encode a family of protein-coding or RNA genes, it seems likely that it encodes a cis-acting regulatory or structural element. Formal proof will require demonstrating a specific function, which we cannot yet do. However, we sought to characterize properties relevant to such DNA elements, such as highly conserved motifs and local clustering.
Conservation Profile of 6-Mer Motifs in MER121.
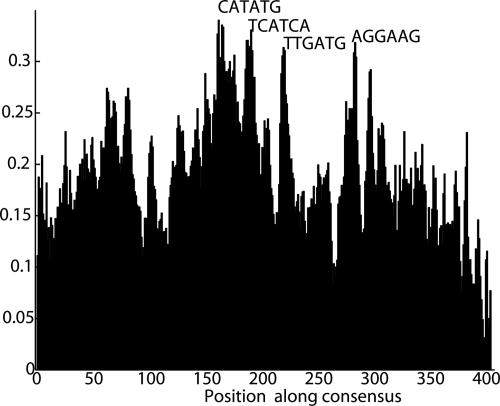
We searched for short, well conserved words within MER121 copies that might represent protein-binding sites. Although the conservation rate for individual nucleotides is relatively constant across the consensus sequence (Fig. 2B), the conservation rate for 6-mers shows a much more uneven distribution with multiple distinct peaks (Fig. 4). These peaks are suggestive of potential binding sites. It is difficult to draw strong inferences from the 6-mer motifs, but some are associated with known transcription factor-binding sites. For example, the most highly conserved 6-mer, CATATG, is a palindromic consensus for E-box motif bound by transcription factor USF. Another highly conserved 6-mer, AGGAAG, is a common motif recognized by many ETS-family transcription factors. Clearly, direct experimental evidence will be required to draw conclusions concerning a possible function for MER121 as a DNA element.
Fig. 4.
Conservation rate of 6-mers along the MER121 consensus. Histogram shows the probability that the 6-mer at the indicated position along the consensus shows perfect four-way conservation across HDMR. The 6-mer profile is notably more peaked than the single base conservation rate shown in Fig. 2B.
Genome-Wide Distribution of MER121 Elements.
Having characterized the conservation properties of MER121 elements, we also studied their distribution across the genome, in the hope of finding clues to function based on genomic context. Specifically, we asked whether these elements were typically found in nearby genes and whether they tended to cluster in the genome.
MER121 copies tend to occur in gene-poor regions, based on two different measures. For each AR class containing between 500 and 4,000 copies in the human genome, we determined the median distance between occurrences of the repeat and the nearby gene starts indicated in the Ensembl database. Of the 241 such AR classes, MER121 ranks 8th highest, with a median distance of 138 kb to the nearest gene start. By contrast, MER119 ranks 130th, with a median distance of 59 kb.
Copies of MER121 are also found preferentially in regions of low exon density. We covered the human genome with 500-kb sliding windows (offset by 50 kb) and determined the density of exonic bases. For a given repeat class, we recorded the density in the closest window surrounding each copy and calculated the median across all copies. When the 241 AR classes are ranked from lowest to highest exonic density, MER121 ranks 20th (0.4%). By contrast, MER119 ranks 175th with a density that is close to the genome-wide median (0.8%).
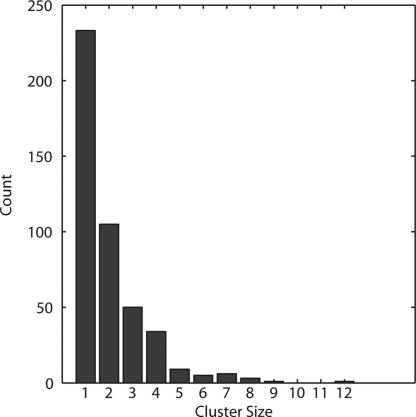
We next looked for clusters of nearby MER121 elements. The average distance D between MER121 in the human genome is ≈3.3 Mb. We defined clusters as collections of elements lying within a region of length D/2. In this process, we ignored copies that lie within 1 kb, because these are likely to represent single copies that have been disrupted in some manner, for example, by an insertion (there are 18 such cases). The distribution of cluster sizes for MER121 copies is shown in Fig. 5. Although the clusters may simply reflect the mechanism of MER121 insertion, it is possible that they may be related to the biological function of these elements. The largest cluster, with 12 copies of MER121, surrounds the inhibin beta A gene and spans ≈1.25 Mb. There are four additional clusters with 8 or more elements: they are associated with the LYPAL1 gene, the TBX3/TBX5 T-box transcription factors, and regions on chromosome 11 and 17, respectively, that contain several genes. Visual representation of the top 5 largest clusters and their genomic neighborhoods are shown in Figs. 6–10, which are published as supporting information on the PNAS web site.
Fig. 5.
Size distribution of MER121 clusters. Human copies of MER121 found within a region of size 1.65 Mb, half the expected distance if copies were distributed uniformly across the genome.
Discussion
To search for large CNE families, we systematically explored the conservation properties of orthologous Ancestral Repeats aligned across the HDMR genomes. We discovered that MER121, with >900 copies in the human genome, is not only the best conserved family of mammalian-wide repetitive elements but is also the largest CNE family reported to date. Intriguingly, the family members vary substantially from one another, but the individual copies typically show strong cross-species similarity across HDMR. The MER121 family is also present and conserved in marsupials, with approximately the same number of copies found in human. However, it is barely detectable in the chicken genome, with only two short sequences detectable having similarity to the mammalian consensus.
The function of MER121 remains a mystery. MER121 is clearly not a protein-coding gene family. The evidence also indicates that it does not encode a family of RNA genes, based on the lack transcripts in RNA databases and the absence of compensating mutations indicative of conserved folding structures. Rather, MER121 seems likely to encode cis-acting regulatory or structural elements. Its genomic distribution may provide some clues to its function: the elements are preferentially found away from gene starts and in gene poor regions, and they occasionally cooccur in large clusters.
These observations led us to speculate that MER121 may have originated as a unique sequence with a structural or regulatory role that was picked up by a transposable element perhaps 200 million years ago, disseminated throughout the genome, retained in places where it was advantageous, and fine-tuned locally to produce copies that have been subsequently preserved in a faithful manner. There is a precedent for an active transposon harboring and mobilizing a regulatory element: the gypsy LTR retrovirus in Drosophila carries an insulator within its 5′ untranslated region (19).
Whatever the function of MER121, the results make clear that ARs may play important functional roles. Although the ≈900 copies of MER121 (171 kb in all) together constitute only a tiny fraction of the human CNE sequence (0.2%), they suggest that other AR families may harbor large families of functional elements. Indeed, we found evidence that other AR families (L3b, L3, L2) show conservation that is far above background, although no other repeat family shows such striking conservation as seen for MER121. Clearly, there exist some treasures hidden among the supposed junk in the human genome.
Methods
Multiple Alignments and Conservation of Ancient Repeats.
We identified instances of repetitive elements in the human genome (build 35, hg17) using the repeatmasker computer program (5). The Ancient Repeat classes were the same as studied in the initial analysis of the mouse genome (1). We analyzed four-way alignment of genome sequence from human (hg17), mouse (mm5), dog (dog1), and rat (rn3), as provided by the University of California, Santa Cruz (UCSC) (http://genome.ucsc.edu) and based on blastz/multiz alignments (20, 21). Alignments between human (hg17) and Monodelphis (monDom1) were also downloaded from UCSC. In the rare cases when more than one multiple alignment overlapped a human repeat annotation, we used the longest alignment based on extent in human. We identified repeats retained across HDMR by requiring that at least 50 bp be present in each of the four species, to allow for partial deletion. For retained repeats, we determined the number of bases that align within each of the three other species. We report the length ratio as the ratio of the number of aligned bases in mouse divided by the number of bases in the human copy.
We defined the four-way alignment rate as the proportion of these human bases that align to bases in all four species (a human base that aligns to a gap character in any species does not count as a four-way aligned base). The rate of perfect four-way conservation is defined as the proportion of four-way aligned bases that is identical across all four species.
Projection of Cross-Species Alignments onto the Repeat Consensus.
Each human MER121 instance aligned four-way (HDMR) or pairwise (human–opossum) was projected on the MER121 consensus by realigning the orthologous sequences and the consensus by using clustalw 1.83 (22) (default settings), resulting in either five-way or three-way multiple alignments.
Conservation Profile of Instances Most Similar to Consensus.
The conservation profile of the top 200 human instances most similar to the MER121 consensus, shown in Fig. 1, was generated as follows. First, we ranked all aligned human instances in decreasing order of similarity to the repeatmasker consensus, using the alignment score computed by a modification of a standard Needleman–Wunsch global alignment algorithm that does not penalize terminal gaps. Next, we followed a simple progressive alignment strategy to align, in decreasing order of similarity to consensus, each of the HDMR multiple alignments to a profile built from the preceding alignment steps.
Gene Set Used for Proximity and Conservation Analysis.
For the comparison to human genes, we used the Ensembl gene predictions for human build hg17 from UCSC (http://genome.ucsc.edu). The rate of perfect four-way conservation within coding regions was based on alignment of Ensembl coding exons.
Indels within Coding Regions.
For the analysis of indels within well studied genes, we used the full set of Ensembl gene predictions that are associated with a gene in the RefSeq database (www.ncbi.nlm.nih.gov/RefSeq/) that is cited at least five times in PubMed. A total of 5,430 genes met these conditions.
Alignment to ESTs and cDNAs.
We analyzed the MGC and Fantom3 collections of cDNA by using the online blast services available at http://mgc.nci.nih.gov/Reagents/MGCBlast and http://fantom3.gsc.riken.jp/blast/.
We downloaded all human ESTs (6,287,602 sequences, 3.35 Gb) and mouse ESTs (4,688,039 sequences, 2.18 Gb) deposited in GenBank from www.ncbi.nlm.nih.gov/blast and aligned by using blastn (17) with default parameters.
Predictions of Conserved Folding Structure.
We compared the overlap of the human instances of MER121 with two sets of predictions of conserved RNA structure (on human build hg17): RNAz (www.tbi.univie.ac.at/papers/SUPPLEMENTS/ncRNA) and EvoFold (http://genome.ucsc.edu).
Alignments of Consensus to Chicken Genome.
We aligned both the MER121 repeat consensus and a reshuffled consensus as a control to the full chicken assembly (galGal2) by using the standard Smith–Waterman algorithm. Alignments of the consensus that scored higher than the largest score observed for the randomized control were considered significant. (We also verified that the distribution of top scores obtained by aligning the randomized control to every 10-kb interval of the chicken assembly fits an extreme value distribution.)
Supplementary Material
Acknowledgments
We thank Michele Clamp for assistance with the indel analysis of coding regions, James Cuff for helping with repeatmasker, and Ben Fry for advice on visualization. We thank colleagues at the Broad Institute for helpful discussion. This work was supported in part by grants from the National Human Genome Research Institute (to E.S.L.).
Abbreviations
- CNE
conserved noncoding element
- AR
ancient repeat
- HDMR
human, dog, mouse, and rat.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., Abril J. F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 2.Margulies E. H., Blanchette M., Haussler D., Green E. D., NISC Comparative Sequencing Program Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woolfe A., Goodson M., Goode D. K., Snell P., McEwen G. K., Vavouri T., Smith S. F., North P., Callaway H., Kelly K., et al. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bejerano G., Haussler D., Blanchette M. Bioinformatics. 2004;20(Suppl. 1):I40–I48. doi: 10.1093/bioinformatics/bth946. [DOI] [PubMed] [Google Scholar]
- 5.Smit A. F. A., Hubley R., Green P. (1996–2004) repeatmasker Open-3.0, available at www.repeatmasker.org.
- 6.Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Gibbs R. A., Weinstock G. M., Metzker M. L., Muzny D. M., Sodergren E. J., Scherer S., Scott G., Steffen D., Worley K. C., Burch P. E., et al. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 8.Lindblad-Toh K., Wade C. M., Mikkelsen T. S., Karlsson E. K., Jaffe D. B., Kamal M., Clamp M., Chang J. L., Kulbokas E. J., Zody M. C., et al. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 9.Britten R. J. Gene. 1997;205:177–182. doi: 10.1016/s0378-1119(97)00399-5. [DOI] [PubMed] [Google Scholar]
- 10.Cooper G. M., Stone E. A., Asimenos G., Green E. D., Batzoglou S., Sidow A. G. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordan I. K., Rogozin I. B., Glazko G. V., Koonin E. V. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 12.Smalheiser N. R., Torvik V. I. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 13.Jurka J. Trends Genet. 2000;9:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 14.Hillier L. W., Miller W., Birney E., Warren W., Hardison R. C., Ponting C. P., Bork P., Burt D. W., Groenen M. A., Delany M. E., et al. Nature. 2004;432:695–716. [Google Scholar]
- 15.Gerhard D. S., Wagner L., Feingold E. A., Shenmen C. M., Grouse L. H., Schuler G., Klein S. L., Old S., Rasooly R., Good P., et al. Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 17.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Washietl S., Hofacker I. L., Lukasser M., Huttenhofer A., Stadler P. F. Nat. Biotechnol. 2005;23:1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- 19.Gdula D. A., Gerasimova T. I., Corces V. G. Proc. Natl. Acad. Sci. USA. 1996;93:9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz S., Kent W. J., Smit A., Zhang Z., Baertsch R., Hardison R. C., Haussler D., Miller W. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanchette M., Kent W. J., Riemer C., Elnitski L., Smit A. F., Roskin K. M., Baertsch R., Rosenbloom K., Clawson H., Green E. D., et al. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson J. D., Higgins D. G., Gibson T. J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.