Abstract
Alterations in splicing patterns of genes contribute to the regulation of gene function by generating endogenous inhibitor or activator molecules. Nucleotide-binding and oligomerization domain (NOD) 2 is an intracellular receptor for bacterial cell wall components and plays an important role in initiating immune responses against cytoinvasive pathogens. NOD2 overexpression sensitizes intestinal epithelial cells toward bacterial cell wall components, activates the proinflammatory transcription factor NF-κB, and induces the subsequent release of the chemotactic cytokine IL-8. Here, we have assessed the regulation and function of a transcript isoform of NOD2, NOD2-S, generated by the skipping of the third exon, which encodes for a protein that is truncated within the second caspase recruitment (CARD) domain. NOD2-S is preferentially expressed in the human colon and is up-regulated by the antiinflammatory cytokine IL-10. Overexpression of NOD2-S down-regulates NOD2-induced NF-κB activation and IL-8 release. Moreover, NOD2-S also interferes with the maturation and secretion of pro-IL-1β downstream of NOD2 and its adaptor molecule receptor-interacting protein kinase 2. We provide a molecular basis for these effects, as we show that NOD2-S interacts with both, NOD2 and receptor-interacting protein kinase 2 and inhibits the “nodosome” assembly by interfering with the oligomerization of NOD2. These data unveil another level of complexicity in the regulation of intracellular innate immunity and may have important implications for the molecular understanding of NOD/NALP protein-driven disease pathophysiology.
Keywords: alternative splicing, cytokine, inflammation, innate immunity, Crohn's disease
CATERPILLER [Caspase recruitment domain, transcription enhancer, R(purine)-binding, pyrin, lots of leucine repeats] genes encode for a growing family of regulatory proteins with a central nucleotide-binding and oligomerization domain (NOD), C-terminal leucine-rich repeats (LRRs), and N-terminal effector binding moieties such as pyrin or caspase recruitment domains (CARDs) (1). CARDs play an essential role as homotypic interaction modules of large protein complexes involved in apoptotic or inflammatory signaling pathways (2).
The NOD2/CARD15 gene encodes for an intracellular protein with 2 adjacent N-terminal caspase recruitment domains, a central nucleotide binding domain, and 10 C-terminal LRRs (3). The LRRs of NOD2 are homolog to those seen in resistance R proteins in plants and Toll-like receptors, which recognize pathogen-associated molecular patterns, thus enabling innate immune responses. Recently, muramyl dipeptide (MurNAc-l-Ala-d-isoGln, MDP) derived from peptidoglycan was identified as the essential structure of bacteria recognized by NOD2 (4, 5). NOD2 has been shown to engage the transcription factor nuclear factor κB (NF-κB) pathway by a recruitment of the receptor-interacting protein kinase 2 (RIP2), subsequently activating the IκB-kinase complex via induced proximity signaling (6).
A frameshift mutation in the LRR of NOD2, which leads to a partial truncation of the LRR, and several other single-nucleotide polymorphisms are associated with the manifestation of Crohn's disease, a human chronic relapsing-remitting inflammatory bowel disease (7–9). These mutations lead to a decreased MDP responsiveness and MDP-mediated NF-κB activation (10).
There is increasing evidence that alterations in splicing patterns of genes may be involved in the regulation of gene functions by generating endogenous inhibitor or activator molecules. In innate immunity signaling, a lipopolysaccharide-inducible short form of MyD88 appears to play a major role in lipopolysaccharide-tolerance induction and inhibits Toll-like receptor/IL-1 signaling (11–13). Abundancy and regulation of splice variants have also been described in other CATERPILLER genes, e.g., PYPAF1/NALP3/CIAS1, but these variants lack a detailed functional characterization (14, 15). For the CATERPILLER family, the importance of different isoforms generated by alternative splicing in humans is also emphasized by findings in R genes in plants, where the dynamic regulation of the ratio of alternative transcripts has shown to be critical for pathogen resistance from a single gene locus (16).
In this report, we describe the identification and functional characterization of a splice variant of NOD2, which encodes for a NOD2 protein isoform truncated within the second CARD domain. This variant is up-regulated by the antiinflammatory cytokine IL-10. NOD2-S interacts with RIP2 and the CARD domains of NOD2 and inhibits NOD2/RIP2-induced NF-κB activation, the expression and secretion of the proinflammatory cytokine IL-8 and NOD2/RIP2-dependent IL-1β maturation, suggesting an important role for this CARD-only containing isoform in the modulation of NOD2/RIP2-signaling events.
Results
Identification and Expression Patterns of NOD2-S.
We screened a human colon cDNA panel for NOD2 splice variants by using primers that amplify the whole ORF (exon 1 forward and exon 12 reverse priming). The amplicons were cloned and sequenced to identify potential splice variants. This approach revealed the existence of an abundant NOD2 splice variant that lacks exon 3 (Fig. 1A; AY187246 hs). The resulting frame shift causes a premature stop in exon 4 and leads to a predicted 21-kDa short NOD2 protein variant (NOD2-S) with a complete CARD1 and a truncated CARD2 domain (54 amino acids). The 10 previously undescribed C-terminal amino acids do not share a significant similarity with other protein domains (data not shown). To detect whether this splice variant is conserved across mammalian species, we amplified cDNA from mouse intestine by using primers located on exon 2 (forward) and exon 4 (reverse), and resulting amplicons were cloned and sequenced. Again, a variant with a skipped exon 3 was detected (DQ289013 mm).
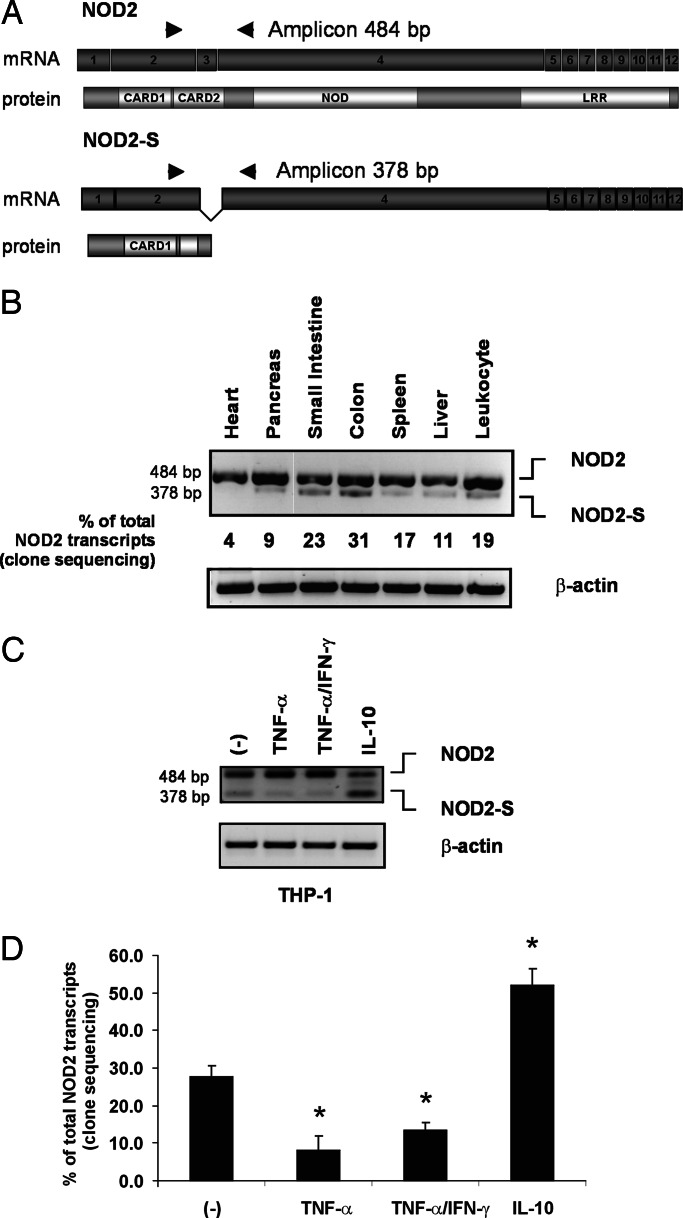
Fig. 1.
NOD2-S is preferentially expressed in the intestine and is up-regulated by the antiinflammatory cytokine IL-10. (A) Schematic cDNA/protein representations of full-length NOD2 and the NOD2-S splice variant missing the third exon. Arrowheads indicate the position of the primers used for RT-PCR expression analysis. (B) Expression of NOD2-S mRNA in adult human tissues. cDNA samples from different human tissues were amplified by RT-PCR with the indicated primers. In parallel, RT-PCR products from each sample were cloned into the pCR2.1 vector, and >200 clones were sequenced for each tissue. The relative amount of NOD2-S clones is expressed in percent of total NOD2 transcript containing clones. (C) Myelomonocytic THP-1 cells were treated with TNF-α (25 ng/ml), TNF-α (25 ng/ml)/IFN-γ (10,000 units/ml), or IL-10 (50 ng/ml) for 12 h. NOD2 was amplified as outlined and analyzed on an agarose gel. (D) PCR products were cloned, and >200 clones per experimental condition were sequenced to determine the NOD2-S:NOD2 ratio (depicted by NOD2-S containing clones in %) (∗, P < 0.05). All experiments were performed in triplicate.
To identify expression patterns of the variant, we analyzed NOD2-S mRNA expression in adult human tissues by RT-PCR methods with primers spanning exon 3 (Fig. 1A). The RT-PCR analysis showed that NOD2-S mRNA is expressed in the small intestine, colon, spleen, and in peripheral blood leukocytes (Fig. 1B). To further analyze the transcript ratio of NOD2-S and full-length NOD2 in different tissues, PCR products were cloned and >200 individual clones were sequenced for each tissue. The percentage of clones with exon 3 skipping varied from 4% in the heart to 31% in the colon (Fig. 1B). Primary intestinal epithelial cells isolated from normal individuals expressed high mRNA amounts of the short splice form of NOD2 (Fig. 7A, which is published as supporting information on the PNAS web site).
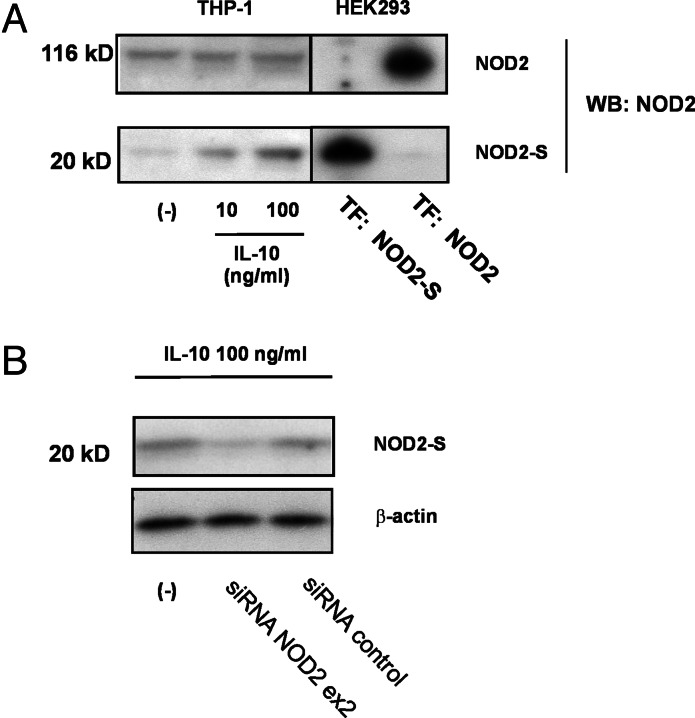
To analyze whether the NOD2-S:NOD2 mRNA ratio is regulated by cytokine stimulation, myelomonocytic THP-1 cells were stimulated for 12 h with TNF-α (25 ng/ml), TNF-α (25 ng/ml)/IFN-γ (10,000 units/ml) or IL-10 (50 ng/ml). NOD2 was amplified as outlined and analyzed on an agarose gel (Fig. 1C). In an independent approach, PCR products were ligated into a sequencing vector and >200 individually isolated clones per experimental condition were sequenced to determine the NOD2-S:NOD2 ratio. Both experiments showed a relative down-regulation of NOD2-S mRNA by TNF-α and TNF-α/IFN-γ treatment, whereas a significant increase of the short isoform could be detected in IL-10-stimulated THP-1 cells. (Fig. 1D). A similar increase of the short form of NOD2 was detectable in primary human monocytes treated with IL-10 (Fig. 7B). To provide evidence for endogenous NOD2-S protein, lysates of untreated THP-1 cells or after 12 h of IL-10 stimulation were subjected to Western blot analysis. IL-10 treated cells displayed an up-regulation of a protein band comigrating with the exogenously expressed NOD2-S (Fig. 2A), whereas the expression level of endogenous full-length NOD2 was unaffected. This band was decreased in lysates from IL-10-treated THP-1 cells, which were transfected with a NOD2-specific short-interfering RNA (siRNA) located in exon 2 (Fig. 2B).
Fig. 2.
Expression of NOD2-S protein. (A) Twenty micrograms of cellular lysates of HEK293 cells transfected with a plasmid carrying the full-length cDNA of the NOD2-S transcript (positive control) and THP-1 cells stimulated with IL-10 were subjected to 4–12% SDS/PAGE and Western blot analysis by using an N-terminal anti-NOD2 antibody. (B) THP-1 cells were transfected with a NOD2-specific siRNA (located in exon 2) or an irrelevant siRNA as control and subjected to Western blot analysis. Representative of three independent experiments (n = 3).
NOD2-S Is a Negative Regulator of NOD2/RIP2-Induced NF-κB Activation.
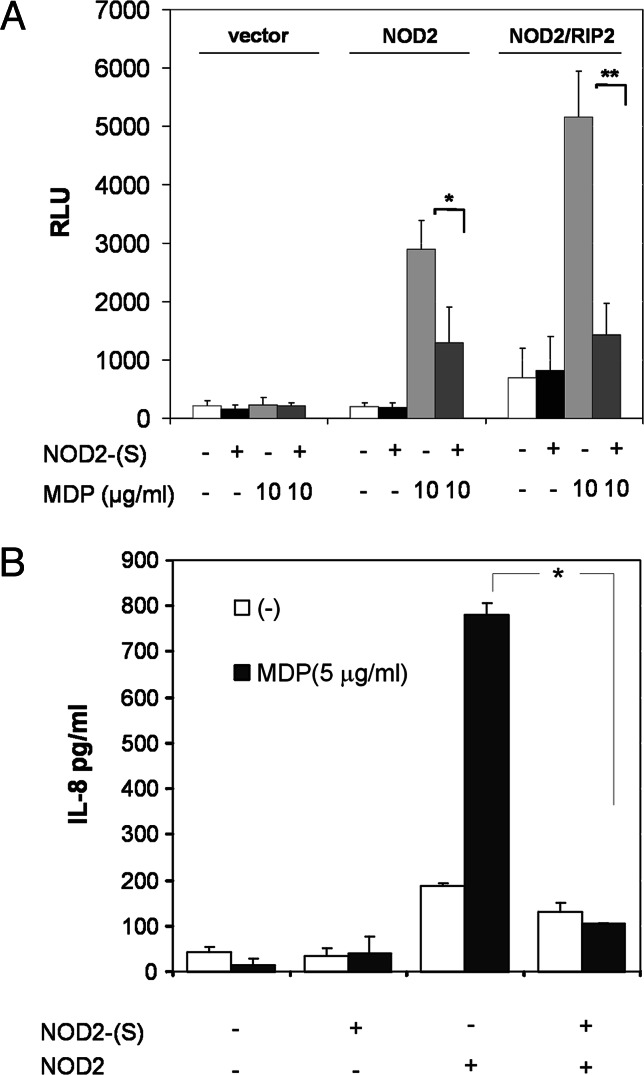
To explore the effects of NOD2-S on NOD2-induced NF-κB activation, cells were transiently transfected with the indicated amounts of expression plasmids encoding Flag-tagged NOD2-S protein, full-length NOD2, and/or RIP2 together with an NF-κB-dependent luciferase reporter plasmid. As shown in Fig. 3A, NOD2-S alone does not activate NF-κB when expressed in HEK293 cells. NOD2-S blocked muramyl-dipeptide-induced NF-κB activation in NOD2 and NOD2/RIP2-transfected cells. Higher amounts of MDP (5–10 μg/ml) were used, because stimulation was performed in the absence of transfection reagent. To address the question whether NOD2-S can interfere with the signaling of disease-relevant NOD2 variants, we cotransfected NOD2-S together with the Crohn-associated variants R702W and L1007fsinsC. The residual NF-κB activity induced upon MDP stimulation of the R702W mutant was significantly inhibited by NOD2-S coexpression (Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 3.
NOD2-S inhibits NOD2-induced NF-κB activation and IL-8 expression in HEK293 cells. (A) HEK293 cells were transfected in 96-well plates with 10 ng of plasmid encoding for NOD2 (pcDNA3.1-NOD2) or 10 ng NOD2 (pcDNA3.1-NOD2)/5 ng RIP2 (pVSV-RIP2) either in the absence or the presence of 15 ng of NOD2-S-encoding plasmid (pFlag-NOD2-S). Induction of NF-κB activity was determined by using a dual luciferase reporter assay (pNF-κB and pRL-TK) as described in Materials and Methods. Total amount of DNA was maintained at 50 ng by the addition of control plasmid DNA (pcDNA3.1-mock). Data are expressed as relative luciferase activity (mean ± SD; quadruplicate cultures; n = 3 independent experiments). (B) HEK293 cells were either transfected with an expression plasmid containing NOD2 (pcDNA3.1-NOD2) and/or NOD2-S (pFlag-NOD2-S) or the appropriate vector control. Eight hours after transfection, cells were stimulated with MDP (5 μg/ml). Secretion of IL-8 was detected by ELISA. Values represent the mean ± SD of triplicate cultures (∗, P < 0.05; ∗∗, P < 0.01).
Inhibition of IL-8 Expression and Secretion and IL-1β Processing by NOD2-S Overexpression.
Because NOD2-induced NF-κB activation has previously been shown to induce the expression and secretion of chemotactic cytokine IL-8 (17), we tested the influence of NOD2-S on IL-8 mRNA expression and secretion upon MDP stimulation in HEK293 cells transiently transfected with NOD2 or a respective control plasmid. NOD2-dependent IL-8 expression and secretion was blocked in the presence of the short NOD2 isoform (Fig. 3B and Fig. 9, which is published as supporting information on the PNAS web site).
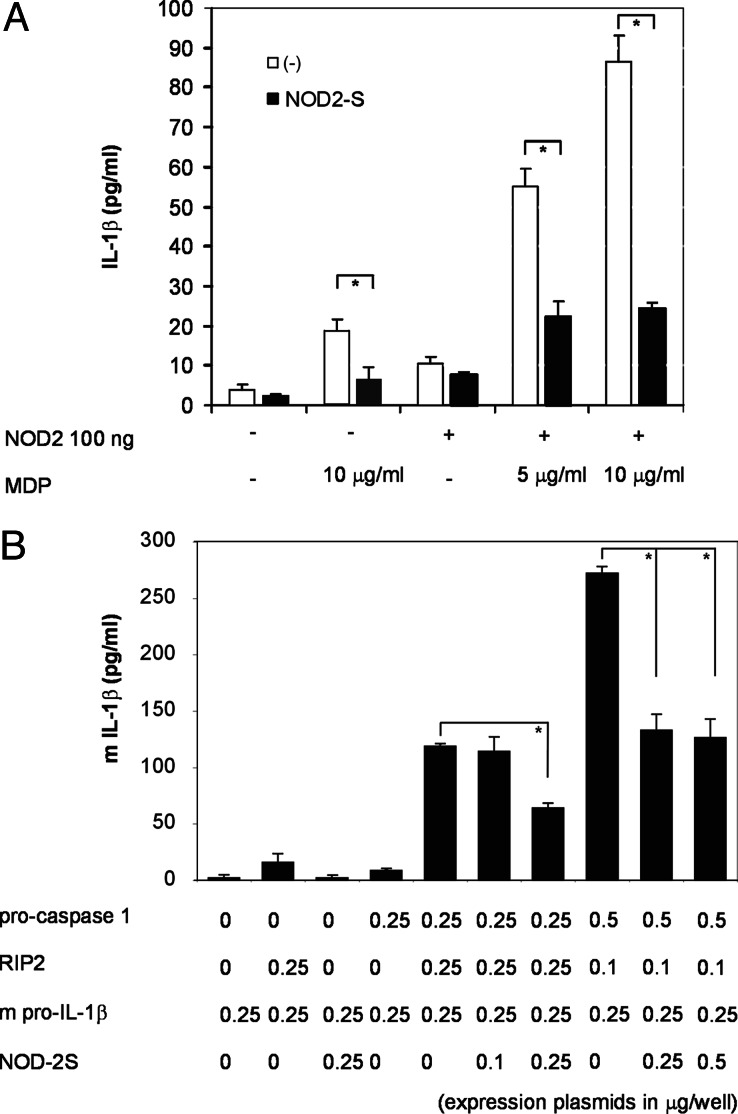
Because it had been suggested that NOD2 may also induce caspase-1 activation, we analyzed whether IL-1β secretion was inducible and whether NOD2-S might suppress IL-1β processing downstream of NOD2/RIP2 activation. THP-1 cells were transfected with an expression plasmid encoding full-length NOD2 to enhance NOD2-stimulated IL-1β secretion or the respective control plasmid and/or a NOD2-S-encoding plasmid. Cells were stimulated with MDP, and supernatants were harvested 48 h after the transfection and assayed for secreted IL-1β. Mock-transfected cells showed an increase in MDP-induced IL-1β levels (up to 28 pg/ml), which is due to activation of endogenous NOD2. In NOD2-transfected cells, MDP treatment resulted in the release of up to 90 pg/ml mature IL-1β into the medium. The increase in IL-1β concentration after MDP treatment under both conditions (NOD2 basal expression and increased NOD2 levels upon transfection) significantly decreased when NOD2-S was coexpressed (8.7 ± 1.7 and 24 ± 1.5 pg/ml, P < 0.05; Fig. 4A).
Fig. 4.
NOD2-S blocks NOD2-induced IL-1β secretion: evidence for a direct inhibition of RIP2-dependent IL-1β processing. (A) THP-1 cells were transfected with plasmids encoding 0.5 μg NOD2 and/or NOD2-S (total DNA was normalized to 1 μg with empty pcDNA3.1 vector) followed by MDP stimulation (10 μg/ml) after 18 h. After 48 h, supernatants were collected and subjected to ELISA for IL-1β. Data represent the mean ± SD (n = 3). (B) HEK293 cells were cotransfected with plasmids encoding mouse pro-IL-1β in combination with various amounts of various expression plasmids encoding pro-caspase-1, RIP2, and/or NOD2-S as indicated (total DNA normalized to 1 μg with empty pcDNA3 DNA (vector). After 24 h, supernatants were collected and subjected to ELISA for mouse IL-1β. Data represent the mean ± SD (n = 3) (P < 0.05).
To test the hypothesis that NOD2-S could block RIP2-induced activation of caspase-1 and subsequent IL-1β secretion independent from NOD2, we transiently transfected a murine pro-IL-1β expression plasmid into HEK293 cells together with the indicated combinations of plasmids encoding for human RIP2, human pro-caspase-1, and NOD2-S. Supernatants were harvested 24 h after transfection, and secreted murine IL-1β (mIL-1β) was assessed by ELISA. Consistent with other studies, expression of RIP2 and caspase-1 induced a significant release of mature mIL-1β into the medium (120–270 pg/ml depending on the amounts of plasmids used). Coexpression of NOD2-S dose-dependently decreased IL-1β secretion in this assay, in proportion to the amount of NOD2-S plasmid used (Fig. 4B).
NOD2-S Interacts with NOD2 and RIP2 and Abolishes MDP Induced NOD2 Self-Association.
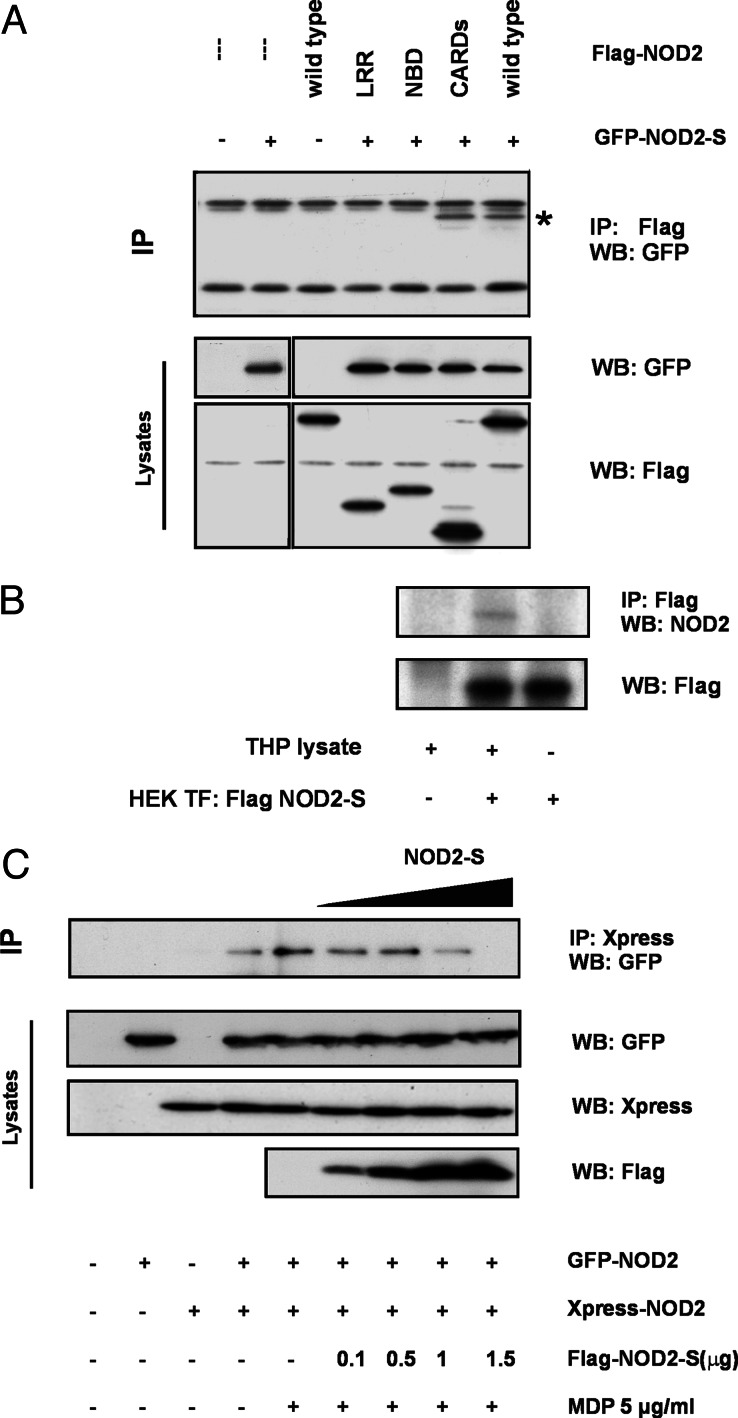
To explore whether NOD2-S associates with NOD2 and to map the interaction site, we performed coimmunoprecipitation assays in transiently transfected HEK293 cells. Twenty-four hours after transfection, NOD2 was immunoprecipitated by using an anti-FLAG monoclonal antibody. Immune complexes were then probed for the presence of coprecipitating NOD2-S GFP fusion protein. Fig. 5A illustrates that NOD2-S was detectable only in coprecipitates containing both CARD domains or full-length NOD2 but does not interact with its NBD or LRR domain.
Fig. 5.
NOD2-S interacts with NOD2 and inhibits the self-association of NOD2 upon muramyl-dipeptide stimulation. (A) Coimmunoprecipitation of NOD2-S with full-length NOD2 and the CARD domains. 293 cells (1.5 × 106) were cotransfected with GFP-NOD2-S and the indicated Flag-NOD2 expression constructs encoding for wild-type NOD2 (wild type), LRR, NBD, or the CARDs. Twenty-four hours after transfection, extracts were prepared and immunoprecipitated with α-Flag antibody. Coprecipitating GFP-NOD2-S was detected by immunoblotting with anti-GFP monoclonal antibody (specific band depicted by asterisk). Expression of NOD2-S-GFP and Flag-NOD2 constructs is shown in the lower insets. (B) NOD2-S associates with endogenous levels of NOD2. Native lysates of 293 cells expressing Flag-tagged NOD2-S were mixed with cellular extracts of THP1 cells known to endogenously express NOD2 at low levels. Endogenous NOD2 is coprecipitating with Flag-NOD2-S in the absence of MDP. (C) NOD2-S interferes with MDP-induced NOD2 oligomerization. HEK293 cells were cotransfected with GFP- and Xpress-tagged full-length NOD2 and varying amounts of NOD2-S. Self-association of NOD2 was determined by immunoprecipitation with an α-Xpress mAb. Association of NOD2 was detected by using an α-GFP mAb. Total lysates were immunoblotted with α-Xpress, α-GFP, and α-FLAG antibodies (lower three blots). No crossreactivity of the α-Flag and α-Xpress Tag antibodies was observed. Each gel is representative of at least three independent experiments.
To show that NOD2-S can interact with endogenous NOD2, we immunoprecipitated FLAG-tagged NOD2-S from native lysates of 293 cells, which were mixed with cellular extracts of THP1 cells known to endogenously express NOD2. Fig. 5B demonstrates that NOD2-S coprecipitates endogenous NOD2 expressed at low levels, thus ruling out CARD/CARD interactions induced by overexpression of both protein isoforms. This interaction is detectable without MDP stimulation and points to a constitutive binding of NOD2-S to the CARDs of NOD2.
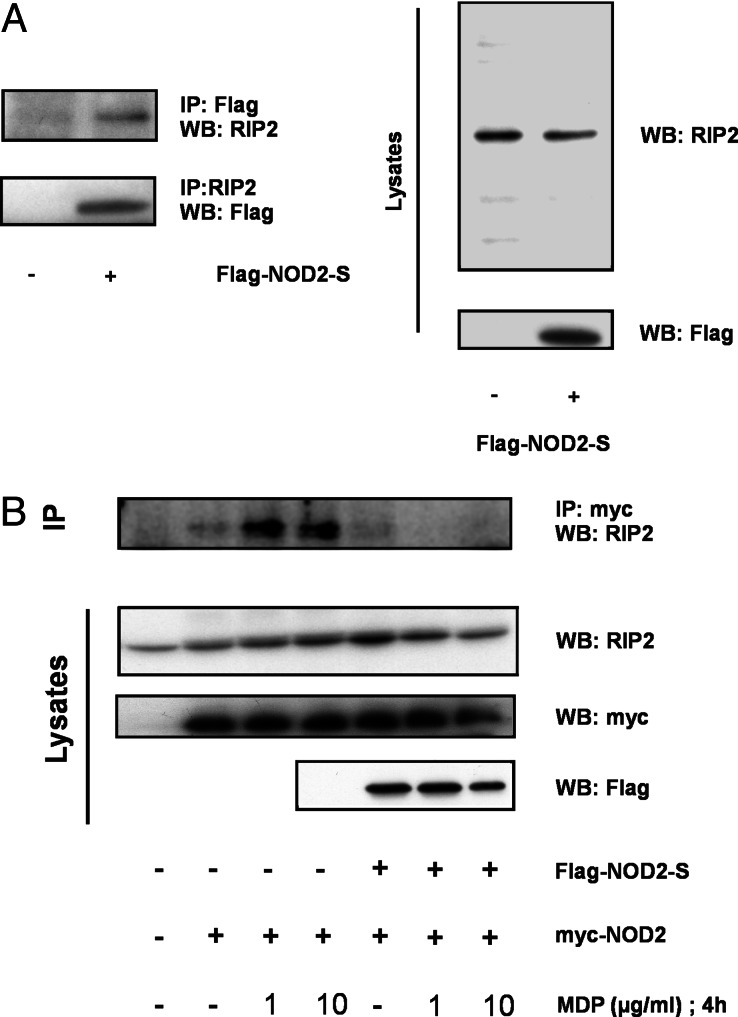
To determine the effect of NOD2-S on the self-association of NOD2 upon MDP stimulation, HEK293 cells were transiently transfected with Xpress-tagged and GFP-tagged NOD2 with or without FLAG-tagged NOD2-S. As shown in Fig. 5C, a weak self-association of NOD2 is readily detectable under basal conditions, which is increased after 15 min of MDP stimulation in the presence of transfection reagent. The self-association of NOD2 can be dose-dependently inhibited by cotransfection of NOD2-S. Concurring with other studies (18), no crossreactivity between Flag and Xpress antibody was detected despite the sequence similarity of the tags. Albeit the short variant does not interact with several CARD-containing caspases caspase-1, -2, -5, and -9, as well as the CARD-containing protein NOD1 (data not shown), NOD2-S coprecipitates with endogenous RIP2 in the absence of MDP (Fig. 6A). To demonstrate whether NOD2-S also affects RIP2 recruitment to the NOD2 complex formed upon MDP stimulation, further immunoprecipitation experiments were carried out. Fig. 6B depicts that NOD2-S expression, in addition to inhibiting the self-oligomerization of NOD2, inhibits the MDP-induced recruitment of RIP2 to the NOD2 complex. Taken together, these results suggest that NOD2-S interferes with the molecular assembly of the “nodosome” and induced proximity signaling via RIP2, possibly by preventing its oligomerization and/or its association with RIP2.
Fig. 6.
NOD2-S interacts with RIP2 and inhibits NOD2/RIP2 complex formation upon MDP stimulation. (A) 293 cells (1.5 × 106) were transfected with a plasmid encoding for Flag-NOD2-S. (A Left) Twenty-four hours after transfection, extracts were prepared and immunoprecipitated with α-RIP2-Ab or an α-Flag-mAb. Coprecipitating NOD2-S or RIP2 were detected by immunoblotting with the reciprocal antibodies (α-Flag-mAb or α-RIP2-Ab). (A Right) Expression of RIP2 protein and NOD2-S was confirmed by immunoblotting of the lysates. (B) NOD2-S interferes with MDP-induced NOD2/RIP2 complex formation. HEK293 cells were cotransfected with myc-tagged full-length NOD2 and Flag-tagged NOD2-S as indicated. Association of NOD2 and endogenous RIP2 was determined by immunoprecipitation with an α-myc mAb, followed by immunodetection with an α-RIP2 antibody. Total lysates were immunoblotted with α-myc, α-RIP2, and α-FLAG antibodies (lower three blots). Each gel is representative of at least three independent experiments.
Discussion
Although an abundancy of splice variants has been reported for genes of the CATERPILLER family, e.g., PYPAF1/NALP3/CIAS1 (14, 15), a detailed functional characterization of CATERPILLER variants in the context of inflammatory signaling pathways is still missing. The salient finding of the present study is the identification of a previously undescribed isoform of the NOD2/CARD15 gene, NOD2-S, that serves as an endogenous inhibitor of NOD2/RIP2-dependent signaling. NOD2-S comprises the complete CARD1 and a part of CARD2 of the full-length NOD2 protein. The CARD2 is truncated at the end of exon 2 due to the frameshift generated by the skipping of exon 3. The 10 previously undescribed C-terminal amino acids of NOD2-S (encoded by exon 4) do not share known sequence homologies. Formally, the alternatively spliced variant appears as a putative target for nonsense-mediated mRNA decay (NMD) as it harbors a premature stop codon within exon 4 followed by functional introns (19). The abundant levels of detectable transcript and the evidence for NOD2-S protein translation indicate that NMD or other mechanisms in transcripts with premature stop codons (e.g., negative regulation of nuclear export or translation; ref. 20) may not play a functional role. Interestingly, corresponding truncated variants of R genes in plants also seem to be resistant to NMD (21). The study of this NOD2 isoform may therefore be useful in further elucidating the regulation of NMD mechanisms in more detail.
A number of CARD proteins are characterized by single CARD moieties without any other recognizable motifs (“CARD-only proteins”), e.g., ICEBERG (22) and PseudoICE/COP (23, 24). It has been suggested that such molecules may exert their function as decoy-CARD proteins competing with other CARD proteins in homotypic protein–protein interactions. For NOD2, it has been shown that overexpression of both CARDs is able to induce NF-κB signaling in the absence of MDP, whereas neither CARD1 nor CARD2 alone have this effect (3). CARD-only proteins may thus act as positive or negative regulators of CARD-dependent caspase- or NF-κB-signaling pathways. We show that NOD2-S inhibits NOD2-induced NF-κB activation and NF-κB-dependent target gene expression (Fig. 2). This interference may be mediated in a dual way by both association with RIP2 and interaction with full-length NOD2 inhibiting self-assembly of NOD2 upon MDP stimulation (Fig. 4). These findings support the hypothesis of NOD2-S as a competitive inhibitor in nodosome assembly and downstream signaling. Interestingly, the finding that NOD2-S is capable of binding to RIP2 also sheds new light on the initial study of Ogura and colleagues (3), which described that both CARDs of NOD2 are essential for RIP2 recruitment during induced proximity signaling. This unique feature of the NOD2/RIP2 interaction has not yet been fully resolved on the structural level, because other RIP2-interacting proteins only possess a single CARD domain (e.g., NOD1, bcl10, and caspase-1). Our results further dissect the molecular requirements of this interaction because the first 54 of the 93 amino acids of CARD2, which are still enclosed in NOD2-S, are sufficient to mediate the NOD2-RIP2 association.
Coexpression of NOD2-S inhibits NOD2-induced IL-1β release into the supernatant. The two other known CARD-only proteins ICEBERG and pseudoICE/COP have been reported to antagonize IL-1β processing by association with the prodomain of caspase-1 (22–24). Although NOD2-S also significantly down-regulated RIP2-induced IL-1β release, we did not detect a direct physical interaction of NOD2-S with caspase-1 in coimmunoprecipitation experiments (data not shown). However, a strong interaction of NOD2-S with both NOD2 and RIP2 may be indicative for a role of NOD2-S as a competitive inhibitor of both NOD2- and RIP2-dependent signaling. A reduced availability of RIP2 by complexation with NOD2-S might not only inhibit NOD2-dependent signaling pathways, but also contribute to a NOD2-independent disturbance of innate immune signaling, because it was recently shown that depletion of RIP2 resulted in diminished Toll-like receptor-mediated cellular responses (25, 26).
NOD2-S expression varies considerably among different tissues (Fig. 1 A and B). In the myelomonocytic cell line THP-1, which was chosen for further characterization of NOD2-S:NOD-2 ratio changes, NOD2-S comprises ≈30% of the total NOD2 transcripts at the basal state. The ratio is dramatically changed upon cytokine stimulation. The antiinflammatory cytokine IL-10 up-regulates the inhibitory NOD2-S splice variant (>50% of the total transcripts), whereas it is down-regulated by the proinflammatory stimuli TNF-α and IFN-γ in vitro (Fig. 1 C and D). Interestingly, in contrast to our findings in intestinal epithelial cells (17), there was no evidence for an up-regulation of the full-length transcript by TNF-α or IFN-γ in THP-1 cells. This observation has been confirmed in primary monocytes (D.S., unpublished observations). A systematic study on the expression of NOD2 in myelomonocytic cell lines (27) has demonstrated that NOD2 is only weakly expressed in HL-60 cells under basal conditions but can be strongly induced by TNF-α treatment. In THP-1 cells, however, a strong expression already under basal, unstimulated conditions was seen, which reflected the findings in the isolated primary monocytes in the same study. It will be interesting to see whether the basal expression level in monocytes also depends on NF-κB activation or whether other mechanisms are used to maintain high levels of NOD2 expression.
We show that increased levels of NOD2-S can abolish residual NOD2 signaling by the Crohn's disease-associated R702W variant. IL-10, which is demonstrated to up-regulate the inhibitory short NOD2 isoform, failed as an experimental therapy in Crohn's disease despite its strong antiinflammatory properties. Thus, it is tempting to speculate that interference with the delicate balance of NOD2 signaling in inflammatory bowel disease, e.g., by changing the ratio of NOD2 splice variants, may also have detrimental effects on the course of disease. Although current treatments for Crohn's disease are immunosuppressive and act via inhibiting the secondary inflammation, future therapeutic approaches should actually aim at strengthening the immunological normal barrier function, e.g., by restoring physiological NOD2 signaling.
The ratio between NOD2 and NOD2-S appears to be critical for the decision whether the sensing of the ligand MDP via the LRR might elicit signals via the CARDs. The findings emphasize the importance of the previously unstudied role of splice variants in the modulation of inflammatory signaling via human CATERPILLER proteins. The mechanism seems to be phylogenetically conserved from plants to mammals because truncated splice variants of R gene transcripts in plants are pivotal in the adequate induction of a hypersensitivity reaction in response to pathogens (16, 28, 29). Further work will be necessary to unveil the exact physiological roles of CATERPILLER splice variants in innate immune responses in vitro and in vivo. A comprehensive inventory of splice variants and their regulation may be pivotal to understand the proteome plasticity of intracellular innate immune responses.
Materials and Methods
Cell Culture, Transfection, and Reagents.
THP1 myelomonocytic and HEK293 cells were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). All cells were cultured in RPMI medium 1640 plus 10% FCS. One day before transfection, the cells were seeded at a density of 5 × 105 cells/2 ml on six-well plates. Transfections were performed with FuGENE 6 (Roche, Basel, Switzerland). Medium was changed 4 h after the transfection to avoid transfection reagent presence at the time of MDP stimulation. TNF-α was purchased from R & D Systems. MDP was from Bachem.
Plasmid Constructs.
cDNA amplification and ligation into expression plasmids was performed by using standard protocols described in detail in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Plasmids for pcDNA3-NOD2, VSV-RIP2, pro-caspase-1, and murine pro-IL-1β are kind gifts from J. Tschopp (University of Lausanne, Epalinges, Switzerland), G. Nunez (University of Michigan, Ann Arbor) and J. Yuan (Harvard Medical School, Boston) and have been described in refs. 3, 30, and 31.
mRNA Isolation and RT-PCR.
Experiments were carried out as described in ref. 32. For a detailed description see Supporting Materials and Methods.
Determination of the NOD2-S:NOD2 Ratio by Clone Sequencing.
To determine the exact ratio of transcripts with an exon 3 skipping, we used a clone sequencing method that has been described to unveil genotype-splicing effects (33, 34). PCR products from 30 rounds of PCR (exponential phase) were ligated into a TA-cloning vector (Invitrogen), and the indicated numbers of individual clones were sequenced by using Dye terminator chemistry (Applied Biosystems) on a 3730xL DNA Analyzer (Applied Biosystems).
siRNA Transfection.
Cells were transfected by using NOD2-specific siRNA duplex (exon 2), sense: 5′-GUGAAAUGUGCUCGCAGGAGGCUUU-3′ or an irrelevant control siRNA duplex 5′- GUGUGUACUCGGACGGGAGCAAUUU-3′ by using Lipofectamine 2000 (Invitrogen). Protein lysates were subjected to Western blot analysis by using a polyclonal anti-NOD2-antibody (Cayman Chemical, Ann Arbor, MI).
Dual Luciferase Reportergene Assay.
NF-κB luciferase activity was determined with a Dual Luciferase Reporter gene kit from Promega by using the pNF-κB Luc plasmid (Stratagene) and the plasmid pRL-TK. Cell lysates were analyzed with a MicroLumatPlus LB96V microplate luminometer (EG & G Berthold, Wellesley, MA). The results for firefly luciferase activity were normalized to renilla luciferase activity.
ELISA.
Supernatants of HEK293 or THP cells transfected with the indicated amounts of plasmids were collected after 24 h (HEK293) or 48 h (THP-1). Human IL-8 (hIL-8), hIL-1β, and murine IL-1β were measured by enzyme-linked immunosorbent assay (R & D Systems).
Immunoprecipitation and Western Blots.
The cells were lysed in native lysis buffer (50 mM Tris, pH 7.6/150 mM NaCl/1% Nonidet P-40/1 mM sodium orthovanadate/10 mM sodium fluoride/5 mM sodium pyrophosphate/10 mM β-glycerophosphate) in the presence of protease inhibitors. Immunoprecipitation and Western blots were performed according to standard protocols (see ref. 33 and Supporting Materials and Methods).
Supplementary Material
Acknowledgments
We thank G. Nunez, J. Tschopp, C. Stehlik, and J. Yuan for valuable reagents and Tanja Kaacksteen and Ilka Ocker for their expert technical assistance. This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 415, a University of Kiel Internal Grant (to P.R.), the National Genome Research Network, and through a Competence Network from the German Ministry for Education and Research.
Abbreviations
- CARD
caspase recruitment domain
- CATERPILLER
Caspase recruitment domain, transcription enhancer, R(purine)-binding, pyrin, lots of leucine repeats
- LRR
leucine-rich repeat
- MDP
muramyl dipeptide
- NOD
nucleotide-binding and oligomerization domain
- RIP2
receptor-interacting protein kinase 2
- siRNA
short-interfering RNA
Footnotes
References
- 1.Harton J. A., Linhoff M. W., Zhang J., Ting J. P. J. Immunol. 2002;169:4088–4093. doi: 10.4049/jimmunol.169.8.4088. [DOI] [PubMed] [Google Scholar]
- 2.Bouchier-Hayes L., Martin S. J. EMBO Rep. 2002;3:616–621. doi: 10.1093/embo-reports/kvf139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogura Y., Inohara N., Benito A., Chen F. F., Yamaoka S., Nunez G. J. Biol. Chem. 2001;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 4.Inohara N., Ogura Y., Fontalba A., Gutierrez O., Pons F., Crespo J., Fukase K., Inamura S., Kusumoto S., Hashimoto M., et al. J. Biol. Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 5.Girardin S. E., Boneca I. G., Viala J., Chamaillard M., Labigne A., Thomas G., Philpott D. J., Sansonetti P. J. J. Biol. Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 6.Inohara N., Koseki T., Lin J., del Peso L., Lucas P. C., Chen F. F., Ogura Y., Nunez G. J. Biol. Chem. 2000;275:27823–27831. doi: 10.1074/jbc.M003415200. [DOI] [PubMed] [Google Scholar]
- 7.Hugot J. P., Chamaillard M., Zouali H., Lesage S., Cezard J. P., Belaiche J., Almer S., Tysk C., O’Morain C. A., Gassull M., et al. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 8.Ogura Y., Bonen D. K., Inohara N., Nicolae D. L., Chen F. F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R. H., et al. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 9.Hampe J., Cuthbert A., Croucher P. J., Mirza M. M., Mascheretti S., Fisher S., Frenzel H., King K., Hasselmeyer A., MacPherson A. J., et al. Lancet. 2001;357:1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 10.Bonen D. K., Ogura Y., Nicolae D. L., Inohara N., Saab L., Tanabe T., Chen F. F., Foster S. J., Duerr R. H., Brant S. R., et al. Gastroenterology. 2003;124:140–146. doi: 10.1053/gast.2003.50019. [DOI] [PubMed] [Google Scholar]
- 11.Janssens S., Burns K., Tschopp J., Beyaert R. Curr. Biol. 2002;12:467–471. doi: 10.1016/s0960-9822(02)00712-1. [DOI] [PubMed] [Google Scholar]
- 12.Burns K., Janssens S., Brissoni B., Olivos N., Beyaert R., Tschopp J. J. Exp. Med. 2003;197:263–268. doi: 10.1084/jem.20021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janssens S., Burns K., Vercammen E., Tschopp J., Beyaert R. FEBS Lett. 2003;548:103–107. doi: 10.1016/s0014-5793(03)00747-6. [DOI] [PubMed] [Google Scholar]
- 14.Kikuchi-Yanoshita R., Taketomi Y., Koga K., Sugiki T., Atsumi Y., Saito T., Ishii S., Hisada M., Suzuki-Nishimura T., Uchida M. K., et al. J. Biochem. (Tokyo) 2003;134:699–709. doi: 10.1093/jb/mvg195. [DOI] [PubMed] [Google Scholar]
- 15.O’Connor W., Jr., Harton J. A., Zhu X., Linhoff M. W., Ting J. P. J. Immunol. 2003;171:6329–6333. doi: 10.4049/jimmunol.171.12.6329. [DOI] [PubMed] [Google Scholar]
- 16.Dinesh-Kumar S. P., Baker B. J. Proc. Natl. Acad. Sci. USA. 2000;97:1908–1913. doi: 10.1073/pnas.020367497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenstiel P., Fantini M., Brautigam K., Kuhbacher T., Waetzig G. H., Seegert D., Schreiber S. Gastroenterology. 2003;124:1001–1009. doi: 10.1053/gast.2003.50157. [DOI] [PubMed] [Google Scholar]
- 18.Barnich N., Hisamatsu T., Aguirre J. E., Xavier R., Reinecker H. C., Podolsky D. K. J. Biol. Chem. 2005;280:19021–19026. doi: 10.1074/jbc.M413776200. [DOI] [PubMed] [Google Scholar]
- 19.Carter M. S., Li S., Wilkinson M. F. EMBO J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- 20.Lee M. H., Schedl T. Genes Dev. 2004;18:1047–1059. doi: 10.1101/gad.1188404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X. C., Gassmann W. Plant Cell. 2003;15:2333–2342. doi: 10.1105/tpc.013474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humke E. W., Shriver S. K., Starovasnik M. A., Fairbrother W. J., Dixit V. M. Cell. 2000;103:99–111. doi: 10.1016/s0092-8674(00)00108-2. [DOI] [PubMed] [Google Scholar]
- 23.Lee S. H., Stehlik C., Reed J. C. J. Biol. Chem. 2001;276:34495–34500. doi: 10.1074/jbc.M101415200. [DOI] [PubMed] [Google Scholar]
- 24.Druilhe A., Srinivasula S. M., Razmara M., Ahmad M., Alnemri E. S. Cell Death Differ. 2001;8:649–657. doi: 10.1038/sj.cdd.4400881. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi K., Inohara N., Hernandez L. D., Galan J. E., Nunez G., Janeway C. A., Medzhitov R., Flavell R. A. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 26.Chin A. I., Dempsey P. W., Bruhn K., Miller J. F., Xu Y., Cheng G. Nature. 2002;416:190–194. doi: 10.1038/416190a. [DOI] [PubMed] [Google Scholar]
- 27.Gutierrez O., Pipaon C., Inohara N., Fontalba A., Ogura Y., Prosper F., Nunez G., Fernandez-Luna J. L. J. Biol. Chem. 2002;277:41701–41705. doi: 10.1074/jbc.M206473200. [DOI] [PubMed] [Google Scholar]
- 28.Mindrinos M., Katagiri F., Yu G. L., Ausubel F. M. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- 29.Ausubel F. M., Katagiri F., Mindrinos M., Glazebrook J. Proc. Natl. Acad. Sci. USA. 1995;92:4189–4196. doi: 10.1073/pnas.92.10.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thome M., Hofmann K., Burns K., Martinon F., Bodmer J. L., Mattmann C., Tschopp J. Curr. Biol. 1998;8:885–888. doi: 10.1016/s0960-9822(07)00352-1. [DOI] [PubMed] [Google Scholar]
- 31.Wang S., Miura M., Jung Y., Zhu H., Gagliardini V., Shi L., Greenberg A. H., Yuan J. J. Biol. Chem. 1996;271:20580–20587. doi: 10.1074/jbc.271.34.20580. [DOI] [PubMed] [Google Scholar]
- 32.Waetzig G. H., Seegert D., Rosenstiel P., Nikolaus S., Schreiber S. J. Immunol. 2002;168:5342–5351. doi: 10.4049/jimmunol.168.10.5342. [DOI] [PubMed] [Google Scholar]
- 33.Valentonyte R., Hampe J., Huse K., Rosenstiel P., Albrecht M., Stenzel A., Nagy M., Gaede K. I., Franke A., Haesler R., et al. Nat. Genet. 2005;37:357–364. doi: 10.1038/ng1519. [DOI] [PubMed] [Google Scholar]
- 34.Hiller M., Huse K., Szafranski K., Jahn N., Hampe J., Schreiber S., Backofen R., Platzer M. Nat. Genet. 2004;36:1255–1257. doi: 10.1038/ng1469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.