Abstract
The Mdm2 and Mdm4 oncoproteins are key negative regulators of the p53 tumor suppressor. However, their physiological contributions to the regulation of p53 stability and activity remain highly controversial. Here, we combined a p53 knock-in allele, in which p53 is silenced by a transcriptional stop element flanked by loxP sites, with the mdm2- and mdm4-null alleles. This approach allows Cre-mediated conditional p53 expression in tissues in vivo and cells in vitro lacking Mdm2, Mdm4, or both. Using this strategy, we show that Mdm2 and Mdm4 are essential in a nonredundant manner for preventing p53 activity in the same cell type, irrespective of the proliferation/differentiation status of the cells. Although Mdm2 prevents accumulation of the p53 protein, Mdm4 contributes to the overall inhibition of p53 activity independent of Mdm2. We propose a model in which Mdm2 is critical for the regulation of p53 levels and Mdm4 is critical for the fine-tuning of p53 transcriptional activity, both proteins acting synergistically to keep p53 in check.
Keywords: cre/lox
The p53 tumor suppressor activity is induced by stress signals, such as DNA damage, oxidative stress, or oncogene activation (1). This induction is mainly due to p53 posttranslational modifications that lead to accumulation of the protein and increased transcriptional activities. p53 downstream responses includes cell cycle arrest and apoptosis and are mediated by its multiple target genes, such as bax and PUMA (2), p21 (3), and Ptprv (4).
Because of its growth-inhibitory activities, p53 function and stability are tightly regulated under most physiological conditions. Of critical importance for this regulation are the two related proteins Mdm2 and Mdm4. Mdm2 is a binding partner and negative regulator of p53 (5). Mdm2 binds p53 in its transactivation domain, suggesting that Mdm2 interferes with p53 function by disrupting its interaction with the basal transcription machinery and/or essential coactivator(s) (6). Interestingly, because mdm2 is a transcription target of p53, a feedback loop exists in which p53 drives expression of the protein that down-regulates its activity (7, 8). Biochemically, Mdm2 acts as an E3 ubiquitin ligase responsible for the ubiquitination of p53 and itself (9–12). However, whether low/physiological levels of Mdm2 are necessary and sufficient to induce proteasome-dependent p53 degradation in vivo remains controversial (13). Genetic experiments have demonstrated the importance of the Mdm2/p53 interaction both during embryonic development and in adult homeostatic tissues. mdm2-deficient mice die at the preimplantation stage, whereas mice lacking both p53 and Mdm2 are viable (14, 15). Moreover, in mice with a hypomorphic mdm2 allele both the transcriptional activity and apoptotic functions of p53 are increased (16). Intriguingly, increased frequency of spontaneous apoptosis was only observed in a subset of actively dividing cells, suggesting that Mdm2 function might be dispensable in quiescent cells in vivo.
Mdm4 was identified as a p53 binding partner (17), and its ability to heterodimerize with Mdm2 was later reported (18). Importantly, germ-line inactivation of mdm4 causes midgestation embryonic lethality that can be completely rescued on a p53-deficient background (19, 20). These data demonstrate that Mdm4 regulates the Mdm2–p53 network in vivo; however, the molecular mechanisms underlining this regulation are yet to be clarified (21).
Therefore, although genetic studies highlight the importance of the p53/Mdm2 and p53/Mdm4 interactions, a clear understanding of the physiological contributions of Mdm2 and Mdm4 to the regulation of p53 is lacking. We therefore sought to gain mechanistic insights on the Mdm2–Mdm4–p53 interplay using sophisticated mouse models.
Results
We used for this study mice carrying a transcriptional stop element flanked by loxP recombination sites [loxP-stop-loxP (LSL)] upstream of the coding region of the p53 gene (p53LSL/LSL) (Fig. 1A). These mice were recently generated by D. Tuveson and T. Jacks (A. Ventura, D. Tuveson, and T. Jacks, unpublished data). To confirm that this p53 allele is silenced by the LSL element and expressed specifically upon Cre introduction, we generated mouse embryonic fibroblasts (MEFs) carrying one p53-null allele (22) and one p53LSL allele (p53LSL/−). We used Cre-expressing adenovirus (Ad-Cre-GFP) to efficiently introduce Cre recombinase. Control cells were infected with GFP-expressing adenovirus (Ad-GFP). No p53 mRNA or protein expression could be detected in noninfected or GFP-expressing cells, indicating that the stop element silences gene transcription effectively (data not shown). Introduction of Cre into the cells leads to efficient excision of the LSL element as determined by PCR on genomic DNA, up-regulation of p53 transcription as measured by quantitative RT-PCR, and detection of the p53 protein by Western blotting (data not shown).
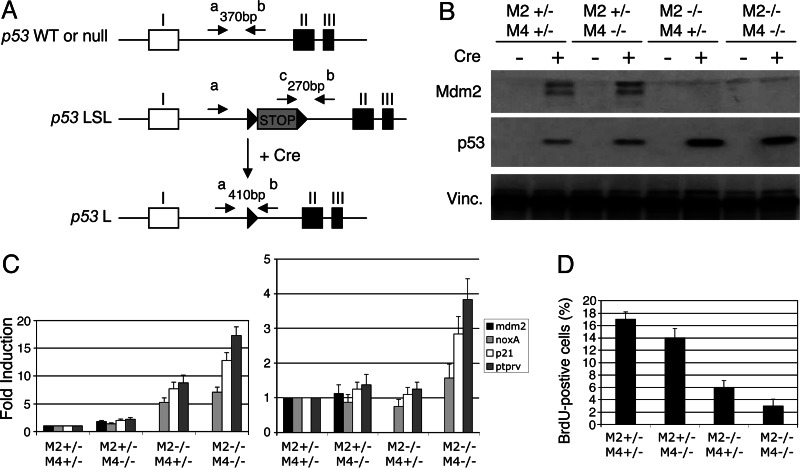
Fig. 1.
Conditional expression of p53 in vitro. (A) Schematic representation of the 5′ end of the mouse p53 LSL and loxP (L) alleles. Exons I–III are shown with coding sequences in black and the untranslated sequences in white. The transcription stop element, flanked by loxP sites (black arrowheads), is shown in light gray. Upon Cre expression, the LSL element is excised and the p53 gene is transcribed. Primers a, b, and c were used to discriminate between the different p53 alleles as detailed in Materials and Methods. Note that this diagram is not to scale. (B) Western blot analysis of early passage p53LSL/− MEFs with the indicated genotypes infected with adeno-Cre or adeno-GFP. Vinculin (Vinc.) served as a loading control. Analyses were carried out 24 h after infection. (C) (Left) Quantitative RT-PCR analysis shows induction of expression of various p53 target genes in cells lacking Mdm4 and/or Mdm2 and infected with adeno-Cre. (Right) Data were normalized to the total amount of p53 present in each condition. Analyses were carried out 24 h after infection. The data were normalized to the level of expression in Cre-infected control cells (M2+/− M4+/−) and represent the mean (± SD) of three independent experiments. (D) Graphical representation showing percentages of cells positive for GFP and BrdUrd (BrdU) from immunofluorescence analysis. The analyses were carried out 24 h after adenoviral infection. The data represent the mean (± SD) of three independent experiments. M2, mdm2; M4, mdm4.
We next generated p53LSL/− mdm2+/− mdm4+/−, p53LSL/− mdm2+/− mdm4−/−, p53LSL/− mdm2−/− mdm4+/−, and p53LSL/− mdm2−/− mdm4−/− MEFs from littermate embryos. The cells were infected with GFP- or Cre-GFP-expressing adenovirus. The data from this analysis are summarized in Fig. 1. The results obtained by using p53LSL/− MEFs in which only the mdm2 alleles or the mdm4 alleles were manipulated or MEFs harboring conditional mdm2 or mdm4 alleles (23, 24) are presented Fig. 6, which is published as supporting information on the PNAS web site. In the absence of Mdm4, p53 levels reached were no higher or only moderately higher than the levels in Mdm4 competent cells (Figs. 1B and 6 A and D). In contrast, loss of Mdm2 leads to a dramatic accumulation of p53 (Figs. 1B and 6 A and D). Importantly, the p53 levels reached in the absence of both Mdm2 and Mdm4 were comparable with the levels reached in the absence of Mdm2 alone (Fig. 1B). Together, these observations indicate that, whereas Mdm2 is essential for maintaining p53 at low levels, Mdm4 does not contribute to the regulation of p53 stability independent of Mdm2. Interestingly, the half-life of Mdm2 was identical in Cre-expressing p53LSL/− mdm2+/− mdm4+/+, p53LSL/− mdm2+/− mdm4+/−, and p53LSL/− mdm2+/− mdm4−/− MEFs, indicating that Mdm4 does not modulate Mdm2 stability, at least in these experimental conditions (data not shown). To determine the transactivation capacity of p53, we examined by quantitative RT-PCR analysis its ability to stimulate expression of a variety of known p53 target genes, including Ptprv, a p53 target gene identified because it is induced in mdm4-null embryos (Figs. 1C and 6 B and E) (4). Loss of Mdm4 alone leads only to a mild but reproducible increase in p53 transcriptional activity, whereas loss of Mdm2 results in a significant increase in p53 target gene transcription. Importantly, in cells deficient in both Mdm2 and Mdm4, p53 transcriptional activity is more pronounced compared with its activity in cells lacking only Mdm2. Strikingly, when the activity of p53 is normalized to the amount of p53 present, a significant increase is observed only when both mdm2 and mdm4 are inactivated (Fig. 1C Right). These findings suggest that, whereas Mdm2 does not interfere with p53 transcriptional activity per se, Mdm4 regulates p53 transcriptional activity in an Mdm2-independent manner. Activation of p53 function in MEFs causes cell proliferation arrest in the G1 phase of the cell cycle (25). We next examined the cell cycle arrest response by determining the percentages of GFP-positive cells in S phase, through BrdUrd labeling (Figs. 1D and 6 C and F). As expected, cells infected with adeno-GFP, which are functionally p53-null, showed high levels of BrdUrd incorporation. In p53LSL/− mdm2+/− mdm4+/− MEFs infected with adeno-Cre-GFP, BrdUrd incorporation was reduced significantly. This decrease was mildly but consistently accentuated in mdm4-negative cells and more robust in the absence of Mdm2. Furthermore, we observed an almost complete block of cell proliferation in cells lacking both Mdm2 and Mdm4. Together, these data argue for specific, nonoverlapping roles for Mdm2 and Mdm4 in inhibiting p53 function.
We next sought to confirm these observations in vivo. We focused on the nervous system for several reasons. The CNS has a well defined anatomy. Neuron-specific-Cre-transgenic and knock-in mouse lines were generated and extensively characterized, and Mdm2 and Mdm4 are expressed in the CNS (20, 26). We first crossed mice carrying the p53LSL mutation with the previously described mice deficient for mdm2 and mdm4 (14, 20). As expected, homozygous p53LSL/LSL mdm2 and/or mdm4-null animals were viable and fertile. These compound mice were next crossed with homozygous p53−/− mutants carrying different mdm2-null and mdm4-null alleles and the cre gene under the control of neuron-specific promoters. The control groups were composed of p53LSL/−, mdm2+/+ or mdm2+/− and/or mdm4+/+ or mdm4+/−, cre+, and all cre− genotypes. All of the control groups were phenotypically normal.
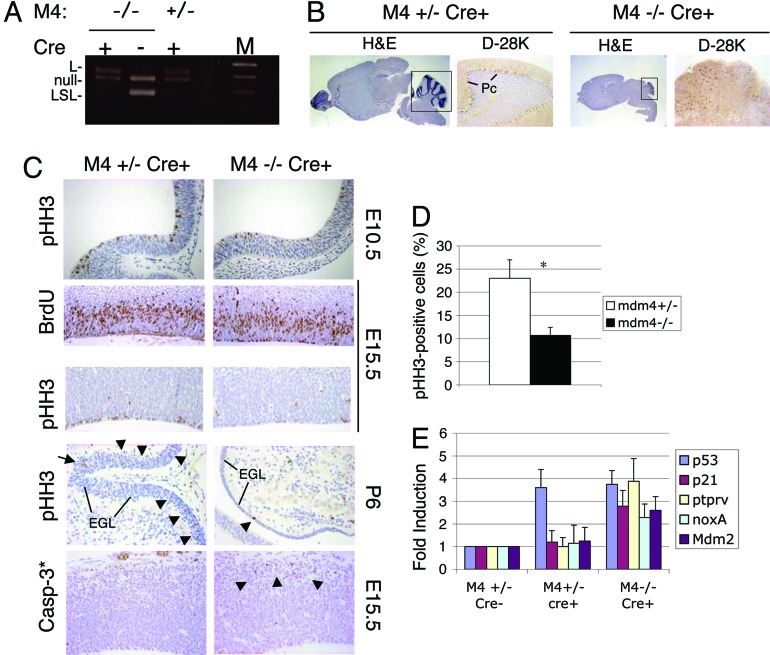
We used the Nes-Cre transgenic line in which the cre transgene was placed under the control of the rat nestin promoter (27). Its specific expression in mouse, as early as embryonic day 9.5, in neuronal progenitor cells and neural crest cells and their progeny has been verified (28). We first produced p53LSL/− mdm4−/− Nes-Cre mice in which both alleles of mdm2 remained wild-type. These mice were viable and, at birth, appeared indistinguishable from their littermates. We verified proper excision of the LSL element specifically in the CNS in vivo by PCR on postnatal day (P)21 brain genomic DNA from p53LSL/− mdm4−/− Nes-Cre mice and control mice (Fig. 2A). p53LSL/− mdm4−/− Nes-Cre mice exhibited growth retardation as early as P7 and reached only ≈50% of the size and weight of control mice at P21. All mutants displayed balance disorders, tremors, altered gait, and akinesis after P10. All these features are criteria of cerebellar ataxia. The brains from P6–P21 p53LSL/− mdm4−/− Nes-Cre mice showed microcephaly and greatly reduced cerebella (Fig. 2B). The mutant cerebella showed a marked agenesis of foliation associated with the disorganized granule cell layer and Purkinje cell layer. Immunostaining using a Purkinje cell-specific marker (calbindin-D28K) showed ectopic Purkinje cells lacking a characteristic lining pattern in mutant cerebella (Fig. 2B). To elucidate the cause of cortical hypoplasia, cerebellar developmental defects, and fate of the p53LSL/− mdm4−/− Nes-Cre progenitor cells, we examined cell proliferation and cell death. At embryonic day 15.5, the developing cortex in the lateral ventricle region of p53LSL/− mdm4−/− Nes-Cre embryos exhibited a statistically significant reduction (31%, P < 0.001) in the percentage of S-phase cells as determined by the extent of BrdUrd incorporation (Fig. 2C). To further examine the effects of Mdm4 loss on the cell cycle, sections were stained with antibody against phosphorylated histone H3 (pHH3), a marker for mitotic cells. Consistent with a reduction of the number of mitotic figures, very few pHH3-positive cells were found in the embryonic-day-15.5 lateral ventricular zone (Fig. 2C). A statistically significant reduction of pHH3-specific staining was also observed at embryonic day 10.5 (Fig. 2 C and D), in the external granular layer of the cerebellum primordium at embryonic day 18.5 (data not shown), and in the P6 cerebellum of p53LSL/− mdm4−/− Nes-Cre animals (Fig. 2C). We next measured apoptosis by in situ end labeling (ISEL) to evaluate nuclear DNA fragmentation and by immunostaining using an antibody against cleaved caspase-3 (casp-3*). Loss of Mdm4 did not cause significant increase in the rate of apoptosis in the neuroepithelia of embryonic-day-10.5 and embryonic-day-12.5 embryos (data not shown). However, consistent with a readily detectable number of pyknotic nuclei in the nestin-negative layers of the enbryonic-day-15.5 cerebral cortex evident by hematoxylin and eosin (H&E) staining, an increased number of casp-3* cells and ISEL-positive cells were seen in p53LSL/− mdm4−/− Nes-Cre embryos (Fig. 2C). These observations indicate that expression of p53 in the absence of Mdm4 causes cell proliferation defects in the progenitor cells and apoptosis in postmitotic neurons. Note that these phenotypes are entirely p53-dependent because cell proliferation of the neural progenitor cells and levels of apoptosis observed in p53LSL/− mdm4−/− embryos were comparable with wild-type controls. Consistent with an increase in p53 activity, increase transcription of known p53 target genes was demonstrated by quantitative RT-PCR analysis on brain RNA extracts from p53LSL/− mdm4−/− Nes-Cre P21 animals (Fig. 2E). We next performed immunostaining and Western blotting analysis for p53. Interestingly, there was no significant increase in the level of p53 protein in embryonic-day-10.5 neuroepithelium, in embryonic-day-15.5 cerebral cortex of p53LSL/− mdm2+/+ mdm4−/− Nes-Cre embryos, in P6 cerebellum, or in P21 p53LSL/− mdm4−/− Nes-Cre brain extracts relative to controls (data not shown). Together, these results indicate that Mdm4 selectively regulates the specific activity of p53 in proliferating progenitors and postmitotic neurons, so that the function but not the stability of p53 is increased in p53LSL/− mdm4−/− Nes-Cre cells.
Fig. 2.
Mdm4 regulates proliferation of neural progenitors and prevents ataxia and cerebellar defects. Analyses are of Nes-Cre p53lsl/− mdm4+/− versus mdm4−/− animals (carrying wild-type mdm2 alleles). (A) PCR amplification of genomic DNA from p53LSL/− total-brain preparations allowing discrimination between the LSL, loxP (L), and null p53 alleles confirmed efficient excision of the LSL element in the brain of Cre-positive animals. (B) Histological analysis of P21 brains and cerebella (boxed area). H&E staining and calbindin-D28K (D-28K) staining. Pc, Purkinje cell. (Magnification: H&E, ×0.5; D-28K, ×10.) (C) In vivo BrdUrd (BrdU) labeling in the lateral ventricle region of the cerebral cortex at embryonic day 15.5 (E15.5) and immunohistological staining of pHH3 in the neuroepithelium at embryonic day 10.5 (E10.5), in the lateral ventricle region of the cerebral cortex at embryonic day 15.5, and in the cerebellum at P6. Staining for the cleaved form of casp-3* is shown in the lateral ventricle region of the cerebral cortex at embryonic day 15.5. (Magnification: pHH3–E10.5, ×30; BrdU–E15.5, ×20; pHH3–E15.5, ×20; pHH3–P6, ×20; Casp-3*–E15.5, ×20.) (D) Histogram of pHH3-positive cells as a percentage of cells present in the ventricular zone of the telencephalic vesicle of embryonic-day-10.5 embryos. ∗, P < 0.001. (E) Quantitative RT-PCR analysis shows induction of expression of p53 and various p53 target genes in P21 brain of mice with the indicated genotypes. The data were normalized to the level of expression in control, Cre-negative animals and represent the mean (± SD) of three independent experiments. M2, mdm2; M4, mdm4; M, marker.
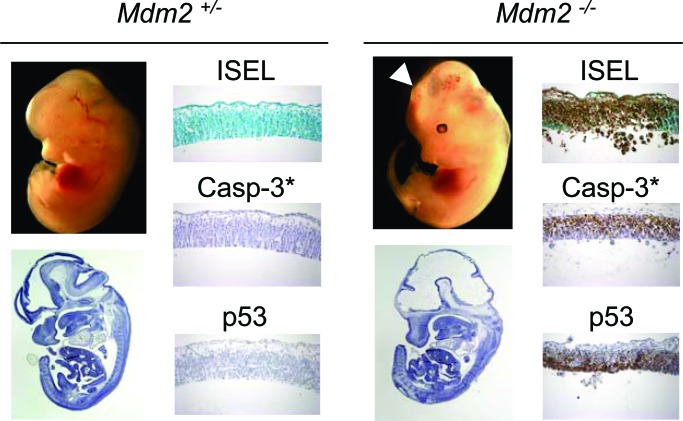
In contrast to p53LSL/− mdm4−/− Nes-Cre, p53LSL/− mdm2−/− Nes-Cre mice were not viable. Macroscopic examination of p53LSL/− mdm2−/− Nes-Cre embryos revealed a protruding head phenotype as early as embryonic day 12.5 (Fig. 3). At this stage, the neuroepithelia in lateral ventricle, spinal cord, and spinal ganglia were dramatically reduced in thickness, and many pyknotic nuclei were readily detectable from the H&E staining in all those regions. As anticipated, loss of Mdm2 increases the rate of apoptosis throughout the CNS (Fig. 3). Therefore, in contrast to Mdm4, Mdm2 does regulate cell survival of the proliferating neuronal progenitors and, thus, in a p53-dependent manner because no apoptosis could be detected in p53LSL/− mdm2−/− embryos. To determine whether loss of Mdm2 allowed the level of p53 protein to increase, we performed immunohistochemistry (IHC) for p53. A dramatic increase in the level of p53 was observed in the nuclei of p53LSL/− mdm2−/− Nes-Cre embryos (Fig. 3). Thus, Mdm2 regulates both p53 apoptotic function and p53 protein levels in vivo.
Fig. 3.
Mdm2 is essential for neuronal progenitors survival. Analyses are of Nes-Cre p53lsl/− mdm2+/− versus mdm2−/− embryos (carrying wild-type mdm4 alleles). External views and H&E staining are of embryonic-day-12.5 embryos. (Magnification: ×0.5.) Also shown is ISEL staining and IHC for p53 and casp-3* in the neuroepithelium of the forebrain region of embryonic-day-12.5 embryos. (Magnification: ×20.) The arrowhead indicates obvious head developmental defect in the forebrain area.
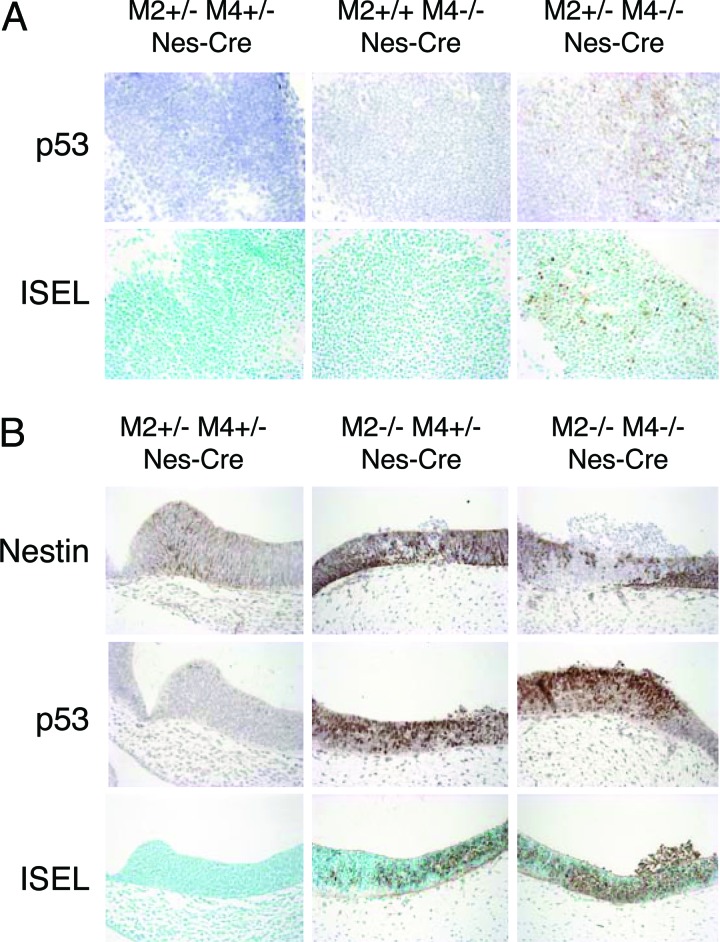
Interestingly, whereas no detectable increase in p53 levels could be detected in p53LSL/− mdm2+/+ mdm4−/− Nes-Cre embryos, a slight but reproducible increase was observed in p53LSL/− mdm2+/− mdm4−/− Nes-Cre embryos (Fig. 4A). Consequently, apoptosis was detectable in the neuroepithelium of p53LSL/− mdm2+/− mdm4−/− Nes-Cre embryos at embryonic day 10.5 (Fig. 4A). These data suggest that Mdm2 and Mdm4 have a genetic dosage effect on inhibiting p53 function in vivo.
Fig. 4.
p53 levels and apoptosis in neuronal progenitors lacking both Mdm2 and Mdm4. (A) ISEL staining and IHC for p53 in the spinal cord of embryonic-day-10.5 embryos. (Magnification: ×30.) (B) ISEL staining and IHC for Nestin and p53 in the neuroepithelium of the telencephalic vesicle of embryonic-day-10.5 embryos. (Magnification: ×20.) All embryos analyzed are p53LSL/− and Nes-Cre. M2, mdm2; M4, mdm4.
Strikingly, the extent of apoptosis was significantly greater in the neuroepithelium of p53LSL/− mdm2−/− mdm4−/− Nes-Cre embryos than in p53LSL/− mdm2−/− mdm4+/− Nes-Cre embryos (Fig. 4B; see also Fig. 7A, which is published as supporting information on the PNAS web site). This observation indicates that p53 activity is more pronounced in cells lacking both Mdm2 and Mdm4 than in cells lacking Mdm2 alone and therefore confirms that Mdm4 controls p53 activity in vivo independent of Mdm2.
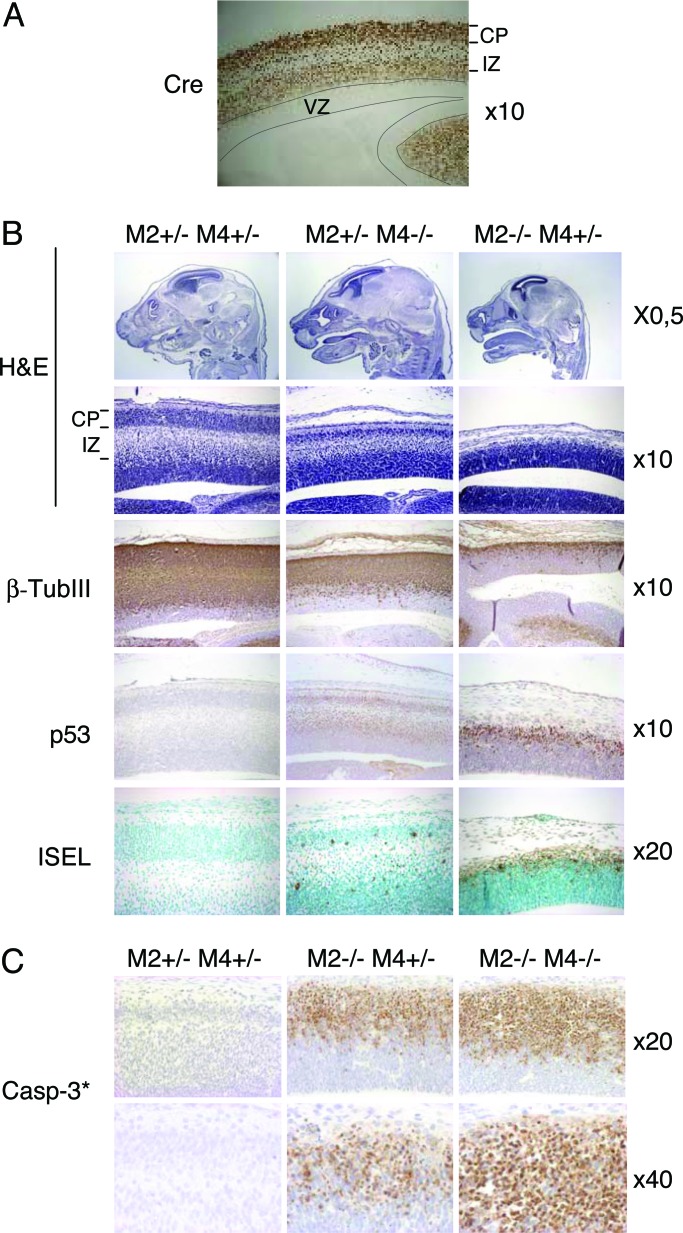
The relevance of Mdm2 and Mdm4 in quiescent cells in vivo is unknown. The above data suggest that Mdm4 is essential for the survival of postmitotic neurons (Figs. 2C and 4A). To further confirm this observation and assess the role of Mdm2 in these cells, we set up crosses using the NEX-Cre knock-in mice. The NEX-Cre mice express cre specifically in differentiated neurons (29). Cre expression and activity was monitored by IHC and by using the highly sensitive β-actin-lacZ reporter mouse strain (S. Goebbels, M. H. Swab and K.-A. Nave, unpublished data). Expression was evident at embryonic day 16.5 in postmitotic cells of the cerebral cortex but was absent from the ventricular zone containing the Nestin-positive cells (Fig. 5A). Histological examination of the cerebral cortex in the lateral ventricle region of embryonic-day-16.5 p53LSL/− mdm2+/− mdm4−/− NEX-Cre embryos revealed hypocellularity in the intermediate zone and cortical plate (Fig. 5B). Immunostaining confirmed that the number of cells positive for β-tubulin III (βTubIII), an early neuronal marker, was significantly reduced (Fig. 5B). Moreover, a more pronounced decrease in βTubIII-positive cells was observed in p53LSL/− mdm2−/− mdm4+/− NEX-Cre embryos (Fig. 5B). To elucidate the cause of reduced neuron numbers, we assayed for apoptosis. A substantial number of ISEL-positive and casp3*-positive cells were found in the p53LSL/− mdm2+/− mdm4−/− NEX-Cre embryos. The frequency of spontaneous apoptosis was increased in p53LSL/− mdm2−/− mdm4+/− embryos and, importantly, even more dramatic in p53LSL/− mdm2−/− mdm4−/− NEX-Cre embryos (Figs. 5 B and C and 7B). Induction of apoptosis in mdm2-null and mdm4-null cells is entirely p53-dependent because no effect was observed in cells that do not express the cre transgene. Together, these data indicate that Mdm2 and Mdm4 are critical and synergize for restraining p53 activity in terminally differentiated postmitotic neurons. Finally, although no p53-positive cells could be detected in Cre+ controls, low levels of p53 were observed in p53LSL/− mdm2+/− mdm4−/− NEX-Cre embryos. Strikingly, most of the remaining βTubIII-positive cells found in p53LSL/− mdm2−/− NEX-Cre embryos were strongly p53-positive. Together, these data indicate that Mdm2 contributes to the regulation of p53 stability in vivo, regardless of the differentiation or proliferation status of the cells.
Fig. 5.
Role of Mdm2 and Mdm4 in terminally differentiated neurons. Shown are H&E staining and IHC for Cre, p53, βTubIII, and cleaved casp-3* and ISEL staining in the lateral ventricle region of the cerebral cortex of embryonic-day-16.5 (A and B) and embryonic-day-14.5 embryos (C). (A) Immunodetection of Cre in a Nex-Cre embryo using a Cre-specific antibody. (B and C) All embryos analyzed are p53LSL/− and NEX-Cre. Magnifications are indicated on the right. VZ, ventricular zone; CP, cortical plate; IZ, intermediate zone; M2, mdm2; M4, mdm4.
Discussion
We show herein that Mdm2 is required to maintain p53 at low levels both in proliferating progenitors and terminally differentiated cells. Our data therefore support a model in which constitutive degradation of p53 in vivo occurs in a strict Mdm2-dependent manner. Surprisingly, our data fail to support a role for Mdm2 in the regulation of p53 transcriptional activity. This observation is consistent with independent observations recently made using another mouse model evaluating p53 structure–function relationships (F. Toledo and G. Wahl, personal communication).
Mdm4 also appears essential for keeping p53 activity in check in a nonredundant manner in proliferating and postmitotic neuronal cells. Therefore, Mdm2 and Mdm4 function to inhibit p53 activity in the same cell types and confirm the notion that Mdm2 cannot compensate for Mdm4 loss in vivo. In addition, we provide a genetic demonstration that Mdm4 inhibits p53 transcriptional activity independent of Mdm2. Our data suggest that Mdm2 and Mdm4 cooperate in vivo to limit p53 activity, irrespective of the proliferation/differentiation status of the cells. Data from ref. 30 and from F. Toledo and G. Wahl (personal communication) further support this view.
The precise mechanism by which Mdm4 regulates p53 activity needs further investigation. Chromatin immunoprecipitation assays suggest that Mdm4 does not modulate the DNA-binding activity of p53 (data not shown). As proposed earlier for Mdm2, Mdm4 might inhibit the recruitment of essential cofactors such as p300/CBP and/or inhibit acetylation of p53. Alternatively, it is possible that Mdm4 mediates or promotes other specific posttranslational modifications that inactivate p53, such as neddylation.
As opposed to Mdm2, loss of Mdm4 consistently leads to only a moderate increase in p53 activity in vivo. This difference can be explained, at least in part, by the fact that mdm2, in contrast to mdm4, is a direct p53 target gene (7, 8). Thus, in absence of Mdm4, p53 transcriptional activity is enhanced, leading to the stimulation of the p53–mdm2 negative feedback loop. In agreement, Mdm4 loss leads to a moderate increase in Mdm2 protein levels in vitro (Fig. 1B) and an increase in mdm2 transcription in vivo (Fig. 2E). This stimulation complicates the interpretation of the results from Mdm4 deficiency. In particular, it has been difficult to evaluate the contribution of Mdm4 to the regulation of p53 stability. In this study, we provide a genetic demonstration that Mdm4 does not participate in the regulation of p53 stability independent of Mdm2. However, whether it does so in a Mdm2-dependent manner remains unclear. We show that loss of Mdm4 does not lead to detectable increase in p53 levels by Western blotting and IHC, both in fibroblasts and in tissues in vivo. However, two possibilities remain: (i) Mdm4 does not participate in the regulation of p53 levels. In this scenario, Mdm4 loss would lead to increased p53 turnover as a result of the stimulation of the p53–Mdm2 negative feedback loop. (ii) Mdm4 stimulates Mdm2-mediated degradation of p53, as proposed in ref. 31 and, therefore, contributes to the regulation of p53 degradation in an Mdm2-dependent manner. In this case, overall p53 levels would be the result of a decrease in Mdm2 activity but in the same time an increase in Mdm2 expression. Our data favor the second possibility because the overall p53 level does not decrease in mdm4-null MEFs despite the stimulation of the p53–mdm2 negative feedback loop, and a slight increase in p53 immunostaining was observed in tissues from p53LSL/− mdm2+/− mdm4−/− NES-Cre embryos compared with p53LSL/− mdm2+/− mdm4+/− NES-Cre embryos. However, to formally resolve this issue, one needs to carefully measure p53 half-life in the various genetic backgrounds. Regardless, because no significant increase in p53 levels is observed in the absence of Mdm4 alone, it appears that Mdm2 can induce efficient degradation of p53 in vivo in the absence of Mdm4.
High levels of Cre in cultured fibroblasts cause activation of a DNA damage response, most likely because of the recognition and Cre-mediated recombination between cryptic loxP sites (32). Accordingly, phosphorylation of H2AX and p53 on Ser-18 (human Ser-15), an ATM-mediated activating phosphorylation, was detected in Ad-Cre-infected p53LSL/− MEFs. Importantly, however, IHC and Western blotting analysis failed to detect increased basal level of DNA damage and Ser-18 phosphorylation in tissues from Nes-Cre and Nex-Cre p53LSL/− mdm4+/− embryos and from mdm2+/− embryos (data not shown). Consistently, Cre-dependent genotoxic effects were reported in only one of >70 published Cre-transgenic mouse strains (33). Our data therefore support the notion that p53 can execute its biological functions in vivo in the absence of detectable genotoxic lesions and/or activating posttranslational modifications.
Materials and Methods
Mice and PCR Genotyping.
Mice carrying an LSL element in the p53 gene were generated by D. Tuveson and T. Jacks (unpublished data). Genotyping of the p53LSL allele was done by PCR using three primers. Two primers are in the first intron (primers a and b), and one primer is in the stop element (primer c) (Fig. 1A). Primer sequences are available upon request. Genotyping of the p53-null, mdm2-null, and mdm4-null alleles were done as described in refs. 14, 20, and 22.
Histology, IHC, ISEL Staining, and Western Blotting Analysis.
Embryos were fixed overnight in 4% paraformaldehyde/1× PBS, paraffin-embedded, sectioned (6–7 μm), and stained in H&E. For BrdUrd experiments, pregnant females were injected i.p. with 100 μg of BrdUrd per gram of body weight and killed 2 h later.
The following antisera were used for IHC: anti-βTubIII (TUJ1; Covance Research Products, Denver, PA), anti-calbindin-D28K (clone CB955; Sigma), anti-Nestin (clone rat 401; Becton Dickinson), anti-p53 (CM5, NovoCastra, Newcastle, U.K.), anti-cre (Babco, Richmond, CA), anti-BrdUrd (BU33; Sigma), anti-cleaved caspase-3 (Cell Signaling Technology, Beverly, MA), and anti-pHH3 (Calbiochem). All antibodies were used on paraffin-embedded sections. Detection was performed with the Envision Kit (DAKO) according to the manufacturer’s instructions. Sections were counterstained with hematoxylin.
ISEL staining was performed with the FragEL kit (Calbiochem) according to the manufacturer’s directions. Sections were counterstained with methyl green.
The antibodies used for Western blot analysis were anti-p53 sheep polyclonal (Ab-7, 1:300; Oncogene Science) and anti-Mdm2 mouse monoclonal 2A10, and anti-Vinculin mouse monoclonal (clone hVIN-1) was used for normalization.
MEF Preparation, Adenoviral Infections, and BrdUrd Incorporation Assays.
MEFs were prepared from embryonic-day-13.5 embryos and maintained as described in ref. 20. We used an empty adenovirus, adeno-GFP, or adeno-Cre-GFP (Ad5 CMV-based vectors; Vector Development Lab, Baylor College of Medicine) to infect MEFs with an approximate multiplicity of infection of 100. MEFs were incubated in medium containing adenovirus for 12 h to allow for efficient infection and LSL excision at the p53 locus.
For the BrdUrd incorporation assays, 2 × 104 cells were seeded on chamber slides for 24 h before treatment to prevent contact inhibition. The cultures were pulse-labeled with 10 μM BrdUrd (Sigma) for 1 h, and slides were fixed 10 min in 4% paraformaldehyde/1× PBS. The BrdUrd-positive cells were identified by indirect immunofluorescence as described in ref. 20.
TaqMan Real-Time Quantitative RT-PCR Assays.
Total RNA was prepared from cell pellets by using an RNeasy minikit (Qiagen, Valencia, CA) according to manufacturer’s protocol. One microgram of total RNA was reverse-transcribed with a SuperScript kit (Invitrogen). These assays were performed by following the manufacturer’s specifications (Applied Biosystems). Primer pairs and TaqMan probes were designed by Applied Biosystems (Assays-on-Demand). Each sample was analyzed in triplicate.
Supplementary Material
Acknowledgments
We thank Dieter Defever and Ines Bonk for excellent technical help; Caroline Vandenbroeke for critical histopathological evaluations; Tyler Jacks (Massachusetts Institute of Technology, Cambridge) for providing the p53LSL/LSL mice; Klaus-Armin Nave (Max Planck Institute of Experimental Medicine, Goettinger, Germany) for providing the NEX-Cre knock-in mice; Guillermina Lozano (University of Texas M. D. Anderson Cancer Center, Houston) for providing the mdm2-deficient mice; and Aart Jochemsen, Stéphane Schurmans, and Pierre Vanderhaeghen for comments on the manuscript. S.F. and P.F. were supported by grants from Fonds National de la Recherche Scientifique, Télévie and Fondation pour la Recherche Médicale, respectively. This work was supported in part by grants from the Association for International Cancer Research and the Belgium Federation Against Cancer and by European Commission Research and Technological Development Framework Programme Grant EC FP6.
Abbreviations
- βTubIII
β-tubulin III
- casp-3*
cleaved caspase-3
- H&E
hematoxylin and eosin
- IHC
immunohistochemistry
- ISEL
in situ end labeling
- LSL
loxP-stop-loxP
- MEF
mouse embryonic fibroblast
- pHH3
phosphorylated histone H3
- Pn
postnatal day n
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Vousden K. H., Lu X. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 2.Michalak E., Villunger A., Erlacher M., Strasser A. Biochem. Biophys. Res. Commun. 2005;331:786–798. doi: 10.1016/j.bbrc.2005.03.183. [DOI] [PubMed] [Google Scholar]
- 3.el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 4.Doumont G., Martoriati A., Beekman C., Bogaerts S., Mee P. J., Bureau F., Colombo M., Alcalay M., Bellefroid E., Marchesi F., et al. EMBO J. 2005;24:3093–3103. doi: 10.1038/sj.emboj.7600769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 6.Chen J., Marechal V., Levine A. J. Mol. Cell. Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barak Y., Juven T., Haffner R., Oren M. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X., Bayle J. H., Olson D., Levine A. J. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 9.Haupt Y., Maya R., Kazaz A., Oren M. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 10.Honda R., Tanaka H., Yasuda H. FEBS. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 11.Kubbutat M. H., Jones S. N., Vousden K. H. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 12.Honda R., Yasuda H. Oncogene. 2000;19:1473–1476. doi: 10.1038/sj.onc.1203464. [DOI] [PubMed] [Google Scholar]
- 13.Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., Gu W. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 14.Montes de Oca Luna R., Wagner D. S., Lozano G. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 15.Jones S. N., Roe A. E., Donehower L. A., Bradley A. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 16.Mendrysa S. M., McElwee M. K., Michalowski J., O’Leary K. A., Young K. M., Perry M. E. Mol. Cell. Biol. 2003;23:462–472. doi: 10.1128/MCB.23.2.462-473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shvarts A., Steegenga W. T., Riteco N., Van Laar T., Dekker P., Bazuine M., Van Ham R. C., Van der Houven, Van Oordt W., Hateboer G., et al. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 18.Sharp D. A., Kratowicz S. A., Sank M. J., George D. L. J. Biol. Chem. 1999;274:38189–38196. doi: 10.1074/jbc.274.53.38189. [DOI] [PubMed] [Google Scholar]
- 19.Parant J., Chavez-Reyes A., Little N. A., Yan W., Reinke V., Jochemsen A. G., Lozano G. Nat. Genet. 2001;29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 20.Migliorini D., Lazzerini-Denchi E., Danovi D., Jochemsen A., Capillo M., Gobbi A., Helin K., Pelicci P. G., Marine J. C. Mol. Cell. Biol. 2002;22:5527–5538. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marine J. C., Jochemsen A. G. Biochem. Biophys. Res. Commun. 2005;331:750–760. doi: 10.1016/j.bbrc.2005.03.151. [DOI] [PubMed] [Google Scholar]
- 22.Jacks T., Remington L., Williams B. O., Schmitt E. M., Halachmi S., Bronson R. T., Weinberg R. A. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 23.Grier J. D., Yan W., Lozano G. Genesis. 2002;32:145–147. doi: 10.1002/gene.10066. [DOI] [PubMed] [Google Scholar]
- 24.Grier J. D., Xiong S., Elizondo-Fraire A. C., Parant J. M., Lozano G. Mol. Cell. Biol. 2006;26:192–198. doi: 10.1128/MCB.26.1.192-198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe S. W., Ruley H. E. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 26.Leveillard T., Gorry P., Niederreither K., Wasylyk B. Mech. Dev. 1998;74:189–193. doi: 10.1016/s0925-4773(98)00074-4. [DOI] [PubMed] [Google Scholar]
- 27.Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P. C., Bock R., Klein R., Schutz G. Nat. Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 28.Graus-Porta D., Blaess S., Senften M., Littlewood-Evans A., Damsky C., Huang Z., Orban P., Klein R., Schittny J. C., Muller U. Neuron. 2001;31:367–379. doi: 10.1016/s0896-6273(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 29.Schwab M. H., Bartholomae A., Heimrich B., Feldmeyer D., Druffel-Augustin S., Goebbels S., Naya F. J., Zhao S., Frotscher M., Tsai M. J., Nave K. A. J. Neurosci. 2000;20:3714–3724. doi: 10.1523/JNEUROSCI.20-10-03714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong S., Van Pelt C. S., Elizondo-Fraire A. C., Liu G., Lozano G. Proc. Natl. Acad. Sci. USA. 2006;103:3226–3231. doi: 10.1073/pnas.0508500103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linares L. K., Hengstermann A., Ciechanover A., Muller S., Scheffner M. Proc. Natl. Acad. Sci. USA. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loonstra A., Vooijs M., Beverloo H. B., Al Allak B., van Drunen E., Kanaar R., Berns A., Jonkers J. Proc. Natl. Acad. Sci. USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt E. E., Taylor D. S., Prigge J. R., Barnett S., Capecchi M. R. Proc. Natl. Acad. Sci. USA. 2000;97:13702–13707. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.