Abstract
Individuals with Down syndrome (DS) are predisposed to develop acute megakaryoblastic leukemia (AMKL), characterized by expression of truncated GATA1 transcription factor protein (GATA1s) due to somatic mutation. The treatment outcome for DS-AMKL is more favorable than for AMKL in non-DS patients. To gain insight into gene expression differences in AMKL, we compared 24 DS and 39 non-DS AMKL samples. We found that non-DS-AMKL samples cluster in two groups, characterized by differences in expression of HOX/TALE family members. Both of these groups are distinct from DS-AMKL, independent of chromosome 21 gene expression. To explore alterations of the GATA1 transcriptome, we used cross-species comparison with genes regulated by GATA1 expression in murine erythroid precursors. Genes repressed after GATA1 induction in the murine system, most notably GATA-2, MYC, and KIT, show increased expression in DS-AMKL, suggesting that GATA1s fail to repress this class of genes. Only a subset of genes that are up-regulated upon GATA1 induction in the murine system show increased expression in DS-AMKL, including GATA1 and BACH1, a probable negative regulator of megakaryocytic differentiation located on chromosome 21. Surprisingly, expression of the chromosome 21 gene RUNX1, a known regulator of megakaryopoiesis, was not elevated in DS-AMKL. Our results identify relevant signatures for distinct AMKL entities and provide insight into gene expression changes associated with these related leukemias.
Keywords: Down syndrome, GATA1
Down syndrome (DS) patients are highly predisposed to acute megakaryoblastic leukemia (AMKL). Treatment outcome is more favorable in DS-AMKL compared with non-DS individuals (1, 2). Discovery of molecular mechanisms involved in AMKL and identification of prognostic factors have been limited because of heterogeneity of the recurrent cytogenetic lesions involved (3). One defined entity is the infant form characterized by the recurrent translocation t(1;22) in non-DS patients (4, 5). The distinct biological features of DS-AMKL are reflected by differences in treatment response and the consistent presence of somatic mutations of the gene encoding transcription factor GATA1, leading to exclusive expression of a truncated form GATA1s. These mutations occur at early times in hematopoietic tissues during development. AMKL blasts express both erythroid and megakaryocytic markers, a finding that suggests transformation of an erythroid/megakaryocyte precursor of fetal origin (6, 7). In mice, loss of GATA1 causes maturation arrest and sustained proliferation of megakaryocyte precursors (8). Expression of GATA1s rescues the maturation deficit but fails to restrain proliferation of GATA1-null megakaryocytes (9). Furthermore, gene-targeted mice expressing GATA1s display sustained proliferation of a yolk sac/early fetal liver megakaryocyte progenitor (10). These effects of GATA1s are likely to reflect perturbed transcriptional control of a subset of genes normally regulated by GATA1.
Whether DS- and non-DS-AMKL represent a single disease entity, achieved through different molecular mechanisms, or should rather be understood as distinct subtypes of acute myeloid leukemia (AML) is a subject of debate. To gain insight into the molecular pathology of DS- and non-DS-AMKL, we established an international consortium to facilitate gene expression profiling of this rare disease. We report here molecular features that distinguish DS- and non-DS-AMKL and identify two subgroups within non-DS-AMKL. This analysis provides information regarding candidate genes for functional studies and genes patterns associated with marked differences in outcome. Cross-species comparison with genes that are modulated after induction of GATA1 expression in murine hematopoietic cells reveals patterns of altered GATA1 target gene expression in DS-AMKL, in particular the loss of repression mediated by GATA1s. Finally, we describe differences in expression levels of BACH1 and RUNX1, hematopoietic transcription factors previously implicated in megakaryocyte differentiation, in DS-AMKL with potential implications in the leukemogenic process.
Results
DS- and Non-DS-AMKL Have Distinct Gene Expression Profiles.
Seventy-two patient samples were studied (Table 1). We first compared our data set to a previously published pediatric AML data set (11) (see Supporting Text and Fig. 5, which are published as supporting information on the PNAS web site). We verified that differences in gene expression between AMKL and the myelomonocytic subtype of AML are highly concordant between the two data sets (enrichment score ≥ 0.95, nominal P value <0.001). This result indicates that the expression profiles obtained in independent data sets robustly reflect the same leukemia phenotype.
Table 1.
Patients included in this study
| DS-AMKL | non-DS-AMKL |
AML M4/5 | ||
|---|---|---|---|---|
| Children | Adults | |||
| Patients | 24 | 28* | 11† | 9 |
| Median age | 19 mo | 17 mo | 58 y | 5 y |
| Median blast, % | 60.0 | 70.0 | 56.0 | 74 |
| GATA1 mutants/n.d. | 18/6 | 2/14 | 0/6 | n.d. |
Blast % before Ficoll purification. Detailed information is in Table 4. n.d., not determined.
*Includes two cases with acquired trisomy 21.
†Includes one case with acquired trisomy 21.
We used supervised and unsupervised approaches to analyze the structure of the AMKL data set. DS and non-DS-AMKL samples exhibit distinct expression profiles by supervised analysis. By significance analysis of microarrays, 721 genes were expressed at higher levels in non-DS-AMKL and 332 in DS-AMKL with a false discovery ratio <0.01. We built a weighted voting model (12) to classify DS and non-DS samples by using features listed in Table 2. This predictor classified three non-DS samples as DS-AMKL. Cytogenetic data from leukemia and remission samples and cultured fibroblast indicated that blasts from these three patients had acquired trisomy 21 (nos. 39, 69, and 85; see Table 4, which is published as supporting information on the PNAS web site). We detected a GATA1 mutation in two infants (nos. 69 and 85) but not in the third adult sample with a complex karyotype (no. 39). These cases may, therefore, have a molecular phenotype similar to DS-AMKL, although mosaicism in the fetal liver hematopoiesis cannot be excluded in the infant cases. In agreement with immunophenotypic data (13), DS-AMKL samples express several erythroid cell markers, including glycophorin A, ankyrin 1, and transferrin receptor 2, together with higher levels of GATA1 (Table 5, which is published as supporting information on the PNAS web site). This signature is comparable with that recently described in ref. 14: Of 105 genes selected as DS-AMKL markers, 76 are also significant markers in our analysis. The mean expression value of GATA1 (as GATA1s) in our data set was increased 3.6-fold in DS- compared to non-DS-AMKL, an observation confirmed by quantitative PCR (2.7-fold; C.L., unpublished data).
Table 2.
DS-AMKL vs. non-DS-AMKL predictor by weighted voting
| Probe | HUGO | Chr | Fold |
|---|---|---|---|
| Higher in non-DS-AMKL | |||
| 218847_at | IMP-2 | 3q27.2 | 9.3 |
| 206414_s_at | DDEF2 | 2p25|2p24 | 3.2 |
| 201427_s_at | SEPP1 | 5q31 | 3.5 |
| 212071_s_at | SPTBN1 | 2p21 | 5.4 |
| 211555_s_at | GUCY1B3 | 4q31.3-q33 | 3.4 |
| 206310_at | SPINK2 | 4q12 | 5.0 |
| 204165_at | WASF1 | 6q21-q22 | 2.5 |
| 205609_at | ANGPT1 | 8q22.3-q23 | 3.1 |
| 214651_s_at | HOXA9 | 7p15-p14 | 4.1 |
| 217617_at | PBX1 | 1q23 | 3.8 |
| 203817_at | Unknown | Unknown | 3.0 |
| 211597_s_at | HOP | 4q11-q12 | 4.3 |
| 219161_s_at | CKLF | 16q22.1 | 2.7 |
| 212148_at | PBX1 | 1q23 | 4.0 |
| 208025_s_at | HMGA2 | 12q15 | 3.2 |
| 212063_at | CD44 | 11p13 | 4.2 |
| 205608_s_at | ANGPT1 | 8q22.3-q23 | 4.0 |
| 205612_at | MMRN1 | 4q22 | 2.8 |
| 200762_at | DPYSL2 | 8p22-p21 | 3.1 |
| 213056_at | FRMD4B | 3p14.1 | 2.1 |
| 201656_at | ITGA6 | 2q31.1 | 2.3 |
| 204069_at | MEIS1 | 2p14-p13 | 3.0 |
| 212614_at | ARID5B | 10q21.2 | 3.0 |
| 218223_s_at | CKIP-1 | 1q21.2 | 2.8 |
| 205253_at | PBX1 | 1q23 | 2.7 |
| 206761_at | CD96 | 3q13.13-q13.2 | 2.3 |
| 203408_s_at | SATB1 | 3p23 | 3.7 |
| 201596_x_at | KRT18 | 12q13 | 2.9 |
| 219789_at | NPR3 | 5p14-p13 | 2.7 |
| 209369_at | ANXA3 | 4q13-q22 | 2.5 |
| 208891_at | DUSP6 | 12q22-q23 | 2.4 |
| Higher in DS-AMKL | |||
| 216518_at | Unknown | Unknown | 2.2 |
| 212224_at | ALDH1A1 | 9q21.13 | 4.1 |
| 206023_at | NMU | 4q12 | 2.7 |
| 205159_at | CSF2RB | 22q13.1 | 4.3 |
| 217388_s_at | KYNU | 2q22.3 | 4.2 |
| 213515_x_at | HBG1 and 2 | 11p15.5 | 8.9 |
| 204419_x_at | HBG1 and 2 | 11p15.5 | 9.3 |
| 207883_s_at | TFR2 | 7q22 | 3.4 |
| 204848_x_at | HBG1 | 11p15.5 | 8.7 |
| 202283_at | SERPINF1 | 17p13.1 | 3.3 |
| 204637_at | CGA | 6q12-q21 | 3.8 |
| 203382_s_at | APOE | 113.2 | 3.2 |
| 210254_at | MS4A3 | 11q12 | 2.7 |
| 203917_at | CXADR | 21q21.1 | 3.6 |
| 205950_s_at | CA1 | 8q13-q22.1 | 6.2 |
| 204416_x_at | APOC1 | 19q13.2 | 4.5 |
| 211820_x_at | GYPA | 4q28.2-q31.1 | 3.6 |
| 208605_s_at | NTRK1 | 1q21-q22 | 2.9 |
| 209301_at | CA2 | 8q22 | 5.9 |
| 204561_x_at | APOC2 | 19q13.2 | 5.0 |
| 202411_at | IFI27 | 14q32 | 6.4 |
| 211821_x_at | GYPA | 4q28.2-q31.1 | 4.8 |
| 202007_at | NID | 1q43 | 3.1 |
| 206070_s_at | EPHA3 | 3p11.2 | 2.8 |
| 206488_s_at | CD36 | 7q11.2 | 6.3 |
| 209555_s_at | CD36 | 7q11.2 | 6.3 |
| 203381_s_at | APOE | 19q13.2 | 3.7 |
| 210215_at | TFR2 | 7q22 | 9.1 |
| 214433_s_at | SELENBP1 | 1q21-q22 | 7.1 |
| 213684_s_at | PDLIM5 | 4q22 | 4.4 |
| 211734_s_at | FCER1A | 1q23 | 20.0 |
| 207067_s_at | HDC | 15q21-q22 | 12.8 |
Sixty-three features selected by the algorithm listed by signal-to-noise ratio (see Supporting Text).
Chromosome 21 Gene Expression in DS-AMKL.
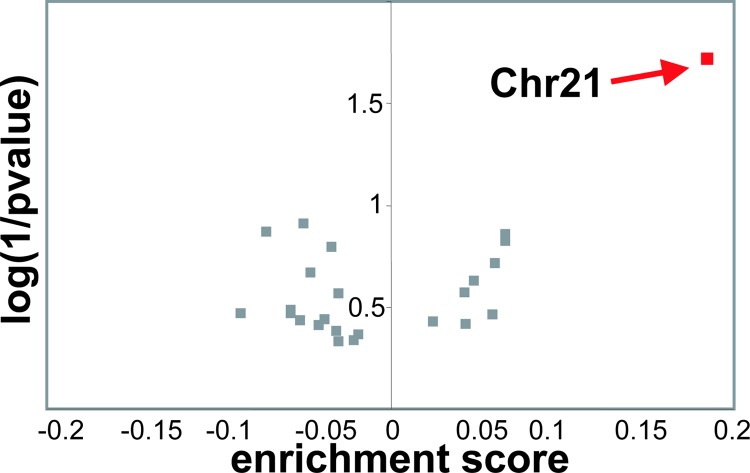
As anticipated, we detected an overall increase in expression of chromosome 21 genes in DS-AMKL, relative to non-DS-AMKL samples. Of the 225 genes predicted from the sequence of human chromosome 21, 155 (69%) are represented on the U133A array. By Gene Set Enrichment Analysis (gsea), chromosome 21 genes were enriched in DS-AMKL, in contrast to genes located on other chromosomes (Fig. 1). Forty-seven genes from chromosome 21 contribute most to the enrichment score, among them transcription factors BACH1, a transcriptional repressor expressed in the megakaryocyte lineage, and SON, a gene with homology to the proto-oncogene MYC family and MOS (Table 3). However, the distinction of DS- from non-DS-AMKL was not driven by the expression of chromosome 21 genes, because their subtraction did not influence the results of class prediction by weighted voting (WV) (data not shown). One of the genes selected by WV for this distinction is located on chromosome 21 (Table 2).
Fig. 1.
Chromosome 21 gene expression is increased in DS-AMKL. Genes were listed in sets according to their chromosomal location. Enrichment for chromosome sets was assessed by gsea for the comparison DS- vs. non-DS-AMKL. Positive enrichment score means higher expression in DS.
Table 3.
Transcription factors on chromosome 21 higher in DS-AMKL
| Gene | Description | Fold |
|---|---|---|
| BACH1 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 | 1.98 |
| SON | SON DNA-binding protein | 1.84 |
| C21orf66 | chromosome 21 open reading frame 66 | 1.64 |
| GABPA | GA binding protein transcription factor, α-subunit 60 kDa | 1.53 |
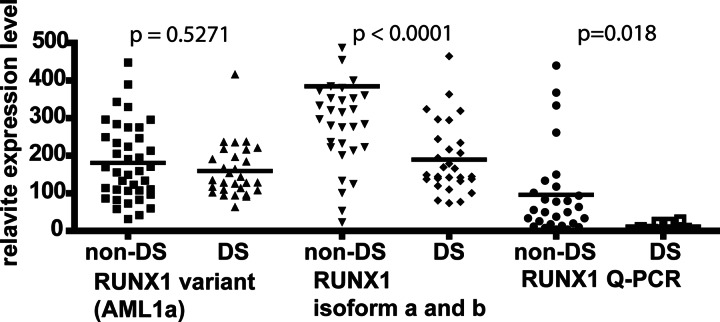
We observed that expression of the RUNX1 gene, which is located on chromosome 21 and essential for megakaryopoiesis, appears lower in DS-AMKL, as compared with non-DS-AMKL (Fig. 2). This difference was detected specifically with probes for the RUNX1 isoforms a and b present on the array and confirmed by quantitative PCR. Of note, the shorter RUNX1 variant AML1a, which is a minor transcript generated by alternative splicing, was expressed at comparable levels in the two subgroups, which suggests that differential expression of RUNX1 isoforms may be relevant for megakaryopoiesis. As a surrogate measure of RUNX1 target genes, we used comparative genomics (15) to search for the presence of conserved transcription factor binding sites in the upstream noncoding sequence of AMKL marker genes (see Supporting Text). Two gene sets defined by the presence of RUNX1 binding motifs were significantly enriched among genes with lower level of expression in DS-AMKL compared to non-DS-AMKL (P = 0.006, see Table 6, which is published as supporting information on the PNAS web site). Thus, both direct and indirect data suggest that RUNX1 gene expression is lower than anticipated in DS-AMKL.
Fig. 2.
RUNX1 expression in DS-AMKL. Runx 1 levels were decreased in DS- compared with non-DS-AMKL as detected with the probe set 209360_s_at [specific for RUNX1 isoforms a and b, National Center for Biotechnology Information (NCBI) NM_001001890] and by quantitative PCR (Q-PCR) with a probe amplifying RUNX1 isoforms a and b but not the shorter variant AML1a, NCBI D43967 (probe Hs00231079_m1, Applied Biosystems). Variant AML1a detected by probe set 210365_at. Statistical analysis: two-tailed nonparametric Mann–Whitney test (GraphPad prism 4.0, San Diego).
Cross-Species Gene Expression Analysis Identifies GATA1s Signature.
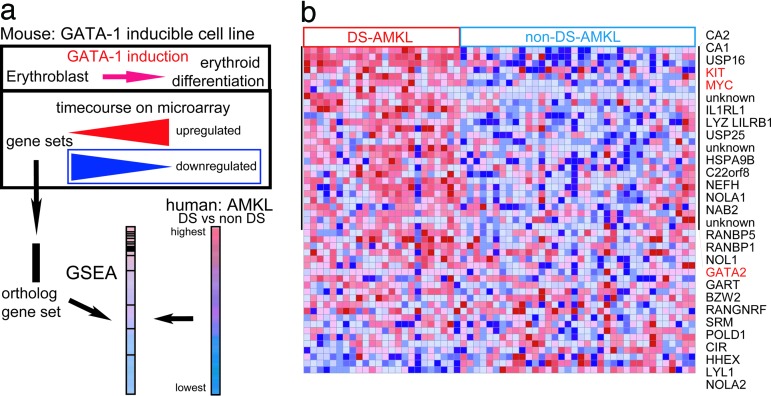
To gain insight into patterns associated with GATA1s expression, we took advantage of prior transcriptome analysis of G1-ER4 cells, GATA1-null cells that undergo erythroid maturation upon restoration of GATA1 expression (16). We generated human ortholog sets for genes that are up- or down-regulated after GATA1 activation (Table 7, which is published as supporting information on the PNAS web site). With gsea, the genes in the leukemia data set were first rank-ordered from highest to lowest by degree of differential expression when comparing DS-AMKL to non-DS-AMKL. The relative positions of the genes that are up- or down-regulated after GATA1 induction in G1-ER4 cells were scored by their degree of enrichment to the top or bottom of the ranked ordered list of genes in the AMKL data set (Fig. 3a). As anticipated, erythroid genes and candidate GATA1 targets, including BACH1 and KLF1, are enriched among genes with the highest expression in DS-AMKL (P = 0.044, Table 7). Genes that are normally down-regulated in G1E-ER4 after GATA1 activation, notably KIT, MYC, and GATA-2, are enriched among markers that are present at relatively higher levels in DS-AMKL (P = 0.029, Fig. 3b and Table 7). These observations indicate that in the context of GATA1s expression in leukemic cells, subsets of GATA1 target genes may be aberrantly regulated and, in particular, that loss of repression mediated by GATA1s may contribute to the pathogenesis of DS-AMKL.
Fig. 3.
Cross-species analysis with a murine GATA1 inducible system. (a) Principle of GSEA. Ortholog genes correspond to genes induced or repressed by GATA1 in the mouse system (see Table 7) are listed in sets. The human genes are rank-ordered based on differential expression for DS-AMKL vs. non-DS-AMKL. gsea scores the relative position of the each gene in the mouse set in the leukemia signature. (b) gsea results. Genes down-modulated by GATA1 in the mouse experiment are listed as they appear in the rank-ordered list of markers for DS vs. non-DS-AMKL, from top-highest to bottom-lowest relative level of expression. Enrichment score = 0.69, P = 0.029. Twenty-nine genes with significant differences between DS and non-DS are listed on the right.
Unsupervised Analysis Reveals Two Distinct Entities in Non-DS-AMKL.
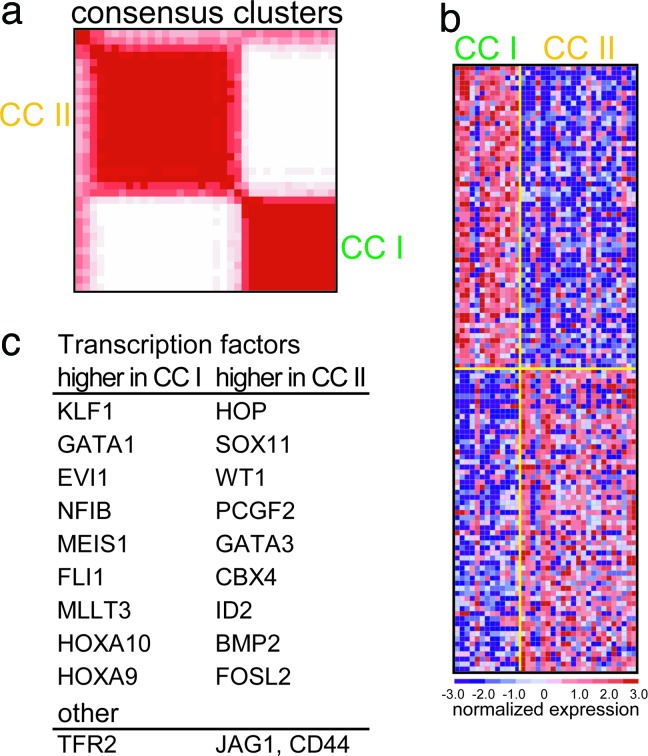
By unsupervised consensus clustering, an algorithm used for class discovery in lymphoma (17), non-DS-AMKL samples cluster in two subgroups (Fig. 4a). Marker selection for these clusters (Fig. 4b and Table 8) reveals features that distinguish these subgroups from each other and from the DS-AMKL subgroup. Cluster I (n = 13) shares some features with DS-AMKL, including expression of KLF, GATA1, and TRF2, but is remarkable for expression of members of the HOX/TALE family, MEIS1, HOXA9, and HOXA10. MLLT3, the most frequent fusion partner of MLL in t(9;11), is also represented in the top 50 markers of Cluster I; yet t(9;11) was not identified by cytogenetics in Cluster I. Of note, the majority of adult AMKL cases are found in Cluster I (6 of 10). The markers of Cluster II (n = 23) include HOP, a small homeodomain-only protein and a cofactor of the serum response factor (SRF). Interestingly, the five t(1;22) positive samples, in which the translocation results in a fusion product involving MKL1, another cofactor of serum response factor, are all included in Cluster II. A selection of the most significant markers of Cluster I and II is presented in Fig. 4c.
Fig. 4.
Identification of consensus cluster in non-DS-AMKL. (a) Consensus matrix produced by hierarchical clustering (K = 2). The samples are listed in the same order on the x and y axis. The intensity of the red color of the square for each sample combination corresponds to the frequency the samples cluster together in the iterations of dataset perturbation. Cluster I (CC I), 13 samples; Cluster II (CC II), 23 samples. (b) Expression profiles of the two AMKL clusters. The top 60 genes associated with each AMKL clusters are shown. Color scale at bottom indicates relative expression to the median. Red, high-level expression; blue, low-level expression. (c) List of selected genes (complete data in Table 8, which is published as supporting information on the PNAS web site).
Discussion
GATA1, a principal regulator of erythroid/megakaryocytic differentiation, is expressed as the mutant variant GATA1s in DS-AMKL. Apart from the translocation t(1;22), which is unique to AMKL in infants, somatic events associated with non-DS-AMKL are largely undefined. Through a consortium, we assembled sufficient samples representing these infrequent disease entities to discriminate previously undescribed AMKL subgroups based on gene expression profiling. First, we show that DS- and non-DS-AMKL constitute distinct molecular phenotypes. Second, we identify chromosome 21 genes with increased expression in DS-AMKL with respect to non-DS-AMKL, notably BACH1, a repressor of normal megakaryopoiesis and possibly a target of GATA1, and SON, a gene with homology to the MYC family. Moreover, we show that RUNX1, a chromosome 21 gene essential for normal megakaryopoisis and a candidate gene for a dosage effect in DS-AMKL, is paradoxically expressed at lower levels in DS-AMKL, implicating a mechanism that may contribute to a block in differentiation in AMKL. Third, we identify a signature for DS-AMKL that highlights a relatively increased expression of GATA1 transcripts (as GATA1s) and failure to down-regulate proliferation-promoting genes that are normally repressed by GATA1. Finally, we describe two distinct molecular phenotypes in non-DS-AMKL, which provide candidate genes for further study.
Chromosome 21 Gene Expression in DS-AMKL.
Trisomy 21 is presumed to be critical for in vivo selection of cells expressing GATA1s, but its involvement in maintenance of a leukemic clone is uncertain. A dosage effect of candidate oncogenic genes from chromosome 21 has been invoked as a contributor to DS-AMKL pathogenesis (7, 18, 19).
We detected BACH1 among the chromosome 21 genes whose expression is highly represented in DS-AMKL. BACH1 heterodimerizes with Maf and represses targets of NFE2, a factor required for platelet production. Transgenic mice that overexpress BACH1 exhibit reduced proplatelet formation and impaired endomitosis (20). Our data suggest that BACH1, which is up-regulated upon GATA1 activation in erythroid cells (16), is aberrantly regulated by GATA1s in DS-AMKL. Given the potential role of BACH1 in megakaryopoiesis, it may be of interest to examine the effects of increased BACH1 expression in the setting of GATA1s in mice.
Another chromosome 21-encoded transcription factor expressed at higher dosage in DS-AMKL is SON, which bears homology to MYC and MOS. Its increased expression might contribute to maintain proliferation. In contrast, proto-oncogenes ETS2 and ERG, which have been proposed as candidates for gene dosage-sensitive effects in DS-AMKL (21), were not differentially expressed in DS vs. non-DS-AMKL, a finding confirmed by quantitative PCR (C.L., unpublished data).
RUNX1 Expression Is Reduced in DS-AMKL.
Increased RUNX1 gene dosage due to trisomy has been proposed as a potential mechanism favoring AMKL in DS (18). However, to date, leukemogenic effects of RUNX1 have been associated with loss of RUNX1 function. Heterozygosity for RUNX1 predisposes patients to AML (22). Acquired loss of function is common in AML, and found in AML with acquired trisomy 21, suggesting that this event is relevant in these cases (18). The recurrent translocation t(8;21), which involves RUNX1, results in dominant repression of RUNX1 target genes in AML (18). Conditional inactivation of RUNX1 in adult mice leads to impaired megakaryopoiesis reminiscent of that seen in the absence of GATA1 (23). We found that levels of the RUNX1 major isoforms were relatively decreased in DS-AMKL and identified RUNX1 binding sites by comparative genomics upstream of genes that were down-regulated in DS-AMKL. We propose that reduced RUNX1 gene dosage in the context of GATA1s may contribute to the pathogenesis of DS-TMD and AMKL, for example, by favoring self-renewal properties at the expense of differentiation, by a mechanism to be determined because RUNX1 mutations are exceedingly rare in AMKL (19). Alterated expression of upstream regulators of RUNX1 could account for lower RUNX1 expression despite its increase genomic dosage. Conversely, the relative lower expression RUNX1 in DS-AMKL might reflect enhanced expression in non-DS-AMKL, although this possibility seems less likely based on available evidence.
GATA1s Are Associated with a Loss of Repression of a Subset of GATA-1 Target Genes.
Transcription repression mediated by GATA1 is likely to be relevant for the observed leukemogenic effect of GATA1s. GATA1 modulates repression of pro-proliferative genes such as MYC, MYB, KIT, and NAB2 (16, 24) and of GATA-2 (25), a factor itself important for hematopoiesis (9). Our data indicate that GATA-2, MYC, and KIT are inadequately repressed in the context of GATA1s expression in DS-AMKL. This observation is consistent with gene expression profiling of megakaryocytes derived from fetal livers of GATA1s-expressing mice (10). In this model, GATA1s-expressing megakaryocyte progenitors display a proliferative advantage, which is correlated with increased expression of GATA2, MYC, and KIT. The notion that GATA1s may have a predominant effect on cell proliferation is supported by recent experiments in which GATA1s rescued the differentiation block but not the hyperproliferative phenotype of a GATA1 “low” mouse megakaryocytes (9).
AMKL Can Be Subdivided in Two Molecular Phenotypes in Non-DS Individuals.
Differences in outcome between DS- and non-DS-AMKL with current management likely reflect distinctive biological features (1). We identified one cluster of AMKL samples (Cluster I) characterized by the expression of HOX/TALE transcription factors that have been implicated in the pathogenesis of AML in prior studies. HOX genes, in particular HOXA9, were found to be overexpressed in human AML and involved in recurrent chromosomal translocations (26). Evidence from mouse models suggest that HOXA9 and MEIS1 function as dominant cooperating oncogenes (27). This HOX expression pattern is also reminiscent of MLL signatures identified in AML (11). Rearrangements involving the MLL gene have been described for non-DS-AMKL in ref. 28 but were not identified by conventional cytogenetics in patients from Cluster I. The second AMKL cluster (Cluster II) included all samples with the translocation t(1:22), which involves the cofactor of serum response factor named MKL1 (4, 5) and samples that overexpress HOP, another cofactor of serum response factor essential for heart development (29). Cluster II is also characterized by increased expression of GATA3, which is essential for T cell development. Of note, coexpression of T cell markers is frequent in AMKL.
In conclusion, global transcriptional profiling provides previously undescribed insights into genetic heterogeneity of AMKL and suggests testable hypotheses for subsequent studies. With respect to DS-AMKL, we have identified a pattern of impaired repression of “pro-proliferative” genes that are also likely targets (either direct or indirect) of GATA1. Similar findings have been obtained in GATA1s-expressing mice in the absence of trisomy (10). Patient data are highly suggestive that trisomy 21 is required for the in vivo selection of cells in which somatic mutation to GATA1s has occurred spontaneously. Whether the trisomic state is relevant for the cellular phenotype at a later stage in AMKL pathogenesis is uncertain. Data in humans and mice underscore the link between GATA1s and increased cellular proliferation. We speculate that perturbed expression of BACH1, and perhaps RUNX1, may influence cellular differentiation in DS-AMKL. Indeed, BACH1 is postulated to function as a negative regulator of megakaryopoiesis. As a candidate GATA1 target gene, BACH1 expression might be sustained in the context of GATA1s expression. In addition, persistent GATA-2 expression might perturb expression of genes normally regulated by GATA1 in megakaryopoiesis and, thereby, modify the balance between proliferation and differentiation. Finally, the finding of unexpectedly low levels of RUNX1 in DS-AMKL is provocative. Impaired gene activation by RUNX1 activation could also contribute to the leukemic phenotype. Our results are consistent with a model in which seemingly small changes in the expression of critical hematopoietic regulators in context of GATA1s ultimately contributes to the phenotype of DS-AMKL.
Materials and Methods
Leukemia Samples.
Blast percentages were determined from diagnostic smears and may be higher in the cryopreserved samples after Ficoll purification than reported in Table 1. We did not observe an influence of blast percentage on our results (see Supporting Text). In this study, we have labeled most DS leukemia cases as AMKL, keeping the differences in the expression of megakaryocytic and erythroid markers between DS- and non-DS-AMKL in mind, as described recently in ref. 13. We sequenced exon 2 of GATA1 from similar numbers of TMD-AMKL/DS-AMKL and non-DS-AMKL samples (Table 4). Characteristic mutations in GATA1 that predict expression of GATA1s were detected in 19 DS-AMKL, 5 TMD samples, and in 2 non-DS infants with acquired trisomy 21 (see note in Table 4). No mutations were observed in non-DS-AMKL. Material from six DS samples was inadequate for analysis.
Sample Preparation.
Samples were lysed in TRIzol (GIBCO/BRL), and RNA was purified on a Promega SV column. RNA quality was assessed by size fractionation by a microfluidics instrument (Agilent Technologies, Palo Alto, CA). cRNA preparation and hybridization on Affymetrix U133A was performed as described in ref. 12. Only samples with >10% presence call and a 3′/5′ signal ratio <3.0 for the HUMGAPH probe were selected for further analysis.
Data Set Normalization.
Affymetrix MicroArray Suite version 5.0 was used to scan and quantify the GeneChips by using default scan settings. For normalization of gene expression data we used Robust MultiArray (RMA). RMA files were generated with the rmaexpress software v.0.3 (B. Bolstad, University of California, Berkeley) by using default settings (background adjustment and quantile normalization). The data set has been deposited at Gene Expression Omnibus.
Supervised Analysis.
After data set preprocessing by log transformation, we used significance analysis of microarrays for marker selection. For class prediction, we used the weighted voting algorithm in genecluster 2 to build a 50-feature model that we evaluated by using the leave-one-out crossvalidation as described in Supporting Text.
gsea.
gsea was performed as described in ref. 30. For cross-species comparison, conversion for ortholog probe sets for gsea was made from mouse to human with tools available from the Affymetrix web site by using the NetAffx annotation database.
Unsupervised Analysis.
We used agglomerative hierarchical clustering (Pearson correlation and complete linkage) to explore the data set for biologically meaningful subclasses and the consensus clustering algorithm (17) to establish the number of clusters and clusters boundaries. Consensus clustering provides a method to represent the consensus across multiple runs of a clustering algorithm as a robust statistical measure of the number and stability of clusters in the data. For each iteration, data set perturbation was obtained by randomly selecting a subset of 36 non-DS-AMKL samples. Consensus matrices were built and evaluated for structures including two to five clusters to represent the frequency at which samples cluster together in this iterative process (see Supporting Text).
Supplementary Material
Acknowledgments
We thank Karen Niss and Maria Martinez for technical assistance; Charles Roberts for critical review of the manuscript; and J.-M. Cazuela (Hôpital St. Louis), F. Valensi (Hôpital Necker Enfants-Malades), B. Quilichini (Laboratoire de Cytogenetique Oncologique, Hôpital la Timone, Marseilles, France), F. Niggli and D. Betts (Universitäts-Kinderklinik Zurich), and M. Dworzak (St.-Anna Kinderspital and Children’s Cancer Research Institute, Vienna) for providing patient samples. J.-P.B. was supported by grants from the Leukemia Research Foundation and the Lady Tata Foundation. T.R.G. and S.H.O. are Investigators of the Howard Hughes Medical Institute. This work was supported in part by a grant from the National Institutes of Health (to S.H.O.). The French network to study AMKL is funded by Groupement d’Intérêt Scientifique-Maladies Rares.
Abbreviations
- AMKL
acute megakaryoblastic leukemia
- AML
acute myeloid leukemia
- DS
Down syndrome
- gsea
Gene Set Enrichment Analysis.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The array data reported in this paper have been deposited in the Gene Expression Omnibus database, www.ncbi.nlm.nih.gov/geo (GEO accession no. GSE4119).
References
- 1.Athale U. H., Razzouk B. I., Raimondi S. C., Tong X., Behm F. G., Head D. R., Srivastava D. K., Rubnitz J. E., Bowman L., Pui C. H., Ribeiro R. C. Blood. 2001;97:3727–3732. doi: 10.1182/blood.v97.12.3727. [DOI] [PubMed] [Google Scholar]
- 2.Creutzig U., Reinhardt D., Diekamp S., Dworzak M., Stary J., Zimmermann M. Leukemia. 2005;19:1355–1360. doi: 10.1038/sj.leu.2403814. [DOI] [PubMed] [Google Scholar]
- 3.Dastugue N., Lafage-Pochitaloff M., Pages M. P., Radford I., Bastard C., Talmant P., Mozziconacci M. J., Leonard C., Bilhou-Nabera C., Cabrol C., et al. Blood. 2002;100:618–626. doi: 10.1182/blood-2001-12-0241. [DOI] [PubMed] [Google Scholar]
- 4.Ma Z., Morris S. W., Valentine V., Li M., Herbrick J. A., Cui X., Bouman D., Li Y., Mehta P. K., Nizetic D., Kaneko Y., et al. Nat. Genet. 2001;28:220–221. doi: 10.1038/90054. [DOI] [PubMed] [Google Scholar]
- 5.Mercher T., Coniat M. B., Monni R., Mauchauffe M., Khac F. N., Gressin L., Mugneret F., Leblanc T., Dastugue N., Berger R., Bernard O. A. Proc. Natl. Acad. Sci. USA. 2001;98:5776–5779. doi: 10.1073/pnas.101001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hitzler J. K., Zipursky A. Nat. Rev. Cancer. 2005;5:11–20. doi: 10.1038/nrc1525. [DOI] [PubMed] [Google Scholar]
- 7.Gurbuxani S., Vyas P., Crispino J. D. Blood. 2004;103:399–406. doi: 10.1182/blood-2003-05-1556. [DOI] [PubMed] [Google Scholar]
- 8.Shivdasani R. A., Fujiwara Y., McDevitt M. A., Orkin S. H. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muntean A. G., Crispino J. D. Blood. 2005;106:1223–1231. doi: 10.1182/blood-2005-02-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z., Godinho F. J., Klusmann J. H., Garriga-Canut M., Yu C., Orkin S. H. Nat. Genet. 2005;37:613–619. doi: 10.1038/ng1566. [DOI] [PubMed] [Google Scholar]
- 11.Ross M. E., Mahfouz R., Onciu M., Liu H. C., Zhou X., Song G., Shurtleff S. A., Pounds S., Cheng C., Ma J., et al. Blood. 2004;104:3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 12.Golub T. R., Slonim D. K., Tamayo P., Huard C., Gaasenbeek M., Mesirov J. P., Coller H., Loh M. L., Downing J. R., Caligiuri M. A., et al. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 13.Langebrake C., Creutzig U., Reinhardt D. Klin. Padiatr. 2005;217:126–134. doi: 10.1055/s-2005-836510. [DOI] [PubMed] [Google Scholar]
- 14.Ge Y., Dombkowski A. A., Lafiura K. M., Tatman D., Yedidi R. S., Stout M. L., Buck S. A., Massey G., Becton D. L., Weinstein H. J., et al. Blood. 2006;107:1570–1581. doi: 10.1182/blood-2005-06-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie X., Lu J., Kulbokas E. J., Golub T. R., Mootha V., Lindblad-Toh K., Lander E. S., Kellis M. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welch J. J., Watts J. A., Vakoc C. R., Yao Y., Wang H., Hardison R. C., Blobel G. A., Chodosh L. A., Weiss M. J. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- 17.Monti S., Savage K. J., Kutok J. L., Feuerhake F., Kurtin P., Mihm M., Wu B., Pasqualucci L., Neuberg D., Aguiar R. C., et al. Blood. 2005;105:1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 18.Speck N. A., Gilliland D. G. Nat. Rev. Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 19.Izraeli S. Hematol. Oncol. 2005. Nov 4, 10.1002/hon.758.
- 20.Toki T., Katsuoka F., Kanezaki R., Xu G., Kurotaki H., Sun J., Kamio T., Watanabe S., Tandai S., Terui K., et al. Blood. 2005;105:3100–3108. doi: 10.1182/blood-2004-07-2826. [DOI] [PubMed] [Google Scholar]
- 21.Rainis L., Toki T., Pimanda J. E., Rosenthal E., Machol K., Strehl S., Gottgens B., Ito E., Izraeli S. Cancer Res. 2005;65:7596–7602. doi: 10.1158/0008-5472.CAN-05-0147. [DOI] [PubMed] [Google Scholar]
- 22.Song W. J., Sullivan M. G., Legare R. D., Hutchings S., Tan X., Kufrin D., Ratajczak J., Resende I. C., Haworth C., Hock R., et al. Nat. Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 23.Ichikawa M., Asai T., Saito T., Yamamoto G., Seo S., Yamazaki I., Yamagata T., Mitani K., Chiba S., Hirai H., et al. Nat. Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 24.Rylski M., Welch J. J., Chen Y. Y., Letting D. L., Diehl J. A., Chodosh L. A., Blobel G. A., Weiss M. J. Mol. Cell. Biol. 2003;23:5031–5042. doi: 10.1128/MCB.23.14.5031-5042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letting D. L., Chen Y.-Y., Rakowski C., Reedy S., Blobel G. A. Proc. Natl. Acad. Sci. USA. 2004;101:476–481. doi: 10.1073/pnas.0306315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drabkin H. A., Parsy C., Ferguson K., Guilhot F., Lacotte L., Roy L., Zeng C., Baron A., Hunger S. P., Varella-Garcia M., et al. Leukemia. 2002;16:186–195. doi: 10.1038/sj.leu.2402354. [DOI] [PubMed] [Google Scholar]
- 27.Wang G. G., Pasillas M. P., Kamps M. P. Blood. 2005;106:254–264. doi: 10.1182/blood-2004-12-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morerio C., Rapella A., Tassano E., Rosanda C., Panarello C. Leuk. Res. 2005;29:1223–1226. doi: 10.1016/j.leukres.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Kook H., Epstein J. A. Trends Cardiovasc. Med. 2003;13:261–264. doi: 10.1016/s1050-1738(03)00107-5. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.