Abstract
Gene expression patterns supply insight into complex biological networks that provide the organization in which viruses and host cells interact. Measles virus (MV) is an important human pathogen that induces transient immunosuppression followed by life-long immunity in infected individuals. Dendritic cells (DCs) are potent antigen-presenting cells that initiate the immune response to pathogens and are postulated to play a role in MV-induced immunosuppression. To better understand the interaction of MV with DCs, we examined the gene expression changes that occur over the first 24 h after infection and compared these changes to those induced by other viral, bacterial, and fungal pathogens. There were 1,553 significantly regulated genes with nearly 60% of them down-regulated. MV-infected DCs up-regulated a core of genes associated with maturation of antigen-presenting function and migration to lymph nodes but also included genes for IFN-regulatory factors 1 and 7, 2′5′ oligoadenylate synthetase, Mx, and TNF superfamily proteins 2, 7, 9, and 10 (TNF-related apoptosis-inducing ligand). MV induced genes for IFNs, ILs, chemokines, antiviral proteins, histones, and metallothioneins, many of which were also induced by influenza virus, whereas genes for protein synthesis and oxidative phosphorylation were down-regulated. Unique to MV were the induction of genes for a broad array of IFN-αs and the failure to up-regulate dsRNA-dependent protein kinase. These results provide a modular view of common and unique DC responses after infection and suggest mechanisms by which MV may modulate the immune response.
Keywords: immunosuppression, IFN, microarray, dsRNA, dependent protein kinase
Measles virus (MV) induces a generalized immune suppression that is responsible for most of the mortality associated with measles (1) and is concordant with an immune response that provides life-long protection. This paradoxical effect on immune responses is poorly understood and is likely to be influenced by early interactions of MV with cells of the immune system. Initial MV replication occurs in lung epithelial cells and then spreads to local lymphoid tissue and multiple other organs (2–4). It has been suggested that immature interstitial dendritic cells (DCs) capture and transport MV to regional lymph nodes, where the immune response is initiated and spread of infection is facilitated (5, 6). Immature DCs generated from monocytes can be infected with MV in vitro, and replication is facilitated by DC maturation and subsequent T cell activation (7–10). In vivo, monocyte-lineage cells are important sites of MV replication, although infection of DCs has not been documented (3, 4).
DCs are potent initiators of the immune response, but functions of cultured DCs are compromised by MV infection, a finding that is postulated to contribute to immunosuppression. For instance, in MV-infected DCs, CD40-CD40L signaling, IL-12 production, and stimulation of allogeneic CD4+ T cells are reduced (7, 8, 10, 11). MV may also induce Fas-mediated apoptosis in DCs and up-regulate TNF-related apoptosis-inducing ligand [TRAIL; also known as TNF superfamily protein (TNFSF)10] to induce apoptosis in T cells (10, 12–14). It is not clear which of these effects are unique to MV infection and which are common effects of DC infection with other viruses or other types of microbial pathogens that are not associated with immunosuppression.
To provide insight into the effects of MV infection on immature DCs we have used gene array analysis to provide a global view of changes in gene expression patterns in infected DCs. These data have been compared with those on the effects of other pathogens on DCs (15). Such comparisons have been hindered by the variety of array platforms, incomplete data sets, and differences between analysis techniques. We show that meaningful comparisons can be made between publicly available data sets to provide biological insights into unique and common pathogen–host cell interactions and that MV up-regulates and down-regulates a broad array of genes.
Results
Monocyte-derived DCs with the expected immature DC surface marker phenotype (CD14−, CD1a+, CD83−, CD80low, CD86low, and HLA-DRlow) were infected with MV or exposed to LPS. Flow cytometric analysis 24 h after infection showed that the MV-infected DCs had matured by down-regulating CD1a and up-regulating CD83, CD80, CD86, and HLA-DR similar to DCs stimulated with LPS (data not shown) and previously reported data for MV-infected DCs (8, 16). The mRNA expression changes for these molecules concurred with the flow cytometry results, except for HLA-DR, which is stored intracellularly in immature DCs for transport to the plasma membrane after maturation (17). Consistent with previous observations (10, 13), RT-PCR analysis for MV RNA showed that MV entered the DCs but replicated very little, if at all, during the first 24 h, although viral proteins may be synthesized. No new infectious virus was detected in supernatant fluids up to 72 h after infection (data not shown).
In MV-infected DCs there were 622 up-regulated genes and 931 down-regulated genes. Of these significantly regulated genes, 321 were unclassified and not analyzed further. To identify unique and common response pathways, we compared the publicly available gene array data of Huang et al. (15) and found 47 up-regulated genes in Candida albicans-infected DCs, 106 up-regulated genes in Escherichia coli-infected DCs, and 72 up-regulated genes in influenza virus-infected DCs. Of the C. albicans, E. coli, and influenza virus genes, 37 (79%), 86 (81%), and 60 (83%), respectively, overlapped those induced by MV. There were very few down-regulated genes detected in the C. albicans, E. coli, and influenza virus studies, so they were not analyzed further.
The most up-regulated Swiss-Prot keyword classifications included IFN induction, antiviral, inflammatory response, cytokine, and chemotaxis (Table 1). The genes in these classification groups overlapped substantially and consisted of IFNs, chemokines, ILs, and IFN-induced proteins, including antiviral proteins. The metal-thiolate and chromosomal protein classification groups were composed of unique gene families.
Table 1.
Significantly up-regulated genes classified by Swiss-Prot keyword
| Swiss-Prot keyword | Measles |
C. albicans |
E. coli |
Influenza |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Up | Down | ES | Up | ES | Up | ES | Up | ES | |
| Interferon induction | 29 | 1 | 12.9 | 12 | 49.5 | 13 | 22.7 | 18 | 52.7 |
| Antiviral | 11 | 0 | 10.1 | 1 | 8.7 | 1 | 3.7 | 3 | 18.6 |
| Metal-thiolate cluster | 6 | 0 | 8.7 | 2 | 21.2 | 5 | 22.5 | 2 | 15.0 |
| Inflammatory response | 17 | 6 | 5.8 | 6 | 22.9 | 11 | 17.8 | 5 | 13.5 |
| Cytokine | 45 | 5 | 5.7 | 10 | 13.3 | 17 | 9.6 | 11 | 10.3 |
| Chemotaxis | 16 | 4 | 5.2 | 4 | 11.9 | 9 | 11.3 | 4 | 8.4 |
| Chromosomal protein | 13 | 3 | 3.6 | 0 | 0 | 1 | 1.0 | 1 | 1.6 |
ES, Enrichment score; Up, up-regulated; Down, down-regulated.
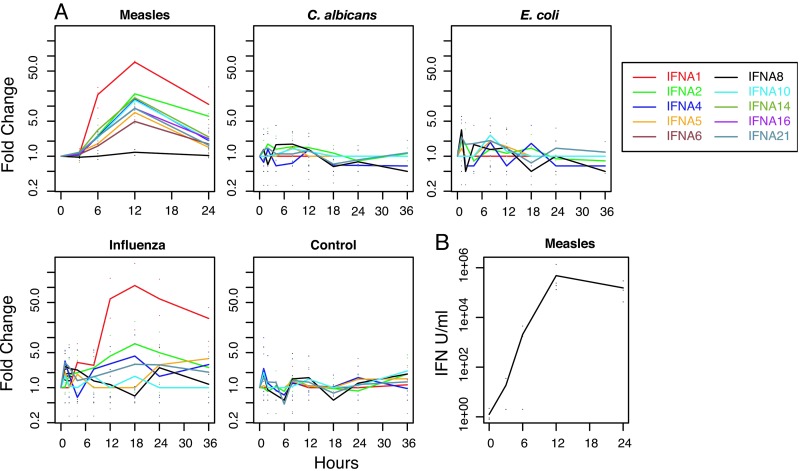
MV induced all of the IFN-α genes represented on the chip, except IFNA8, with peak mRNA levels at 12 h. Influenza virus induced IFNA1 and IFNA2, but somewhat later than MV (Fig. 1A). Data analysis by Huang et al. (15) also identified IFNA13, IFNA14, and IFNA16 induction by influenza virus. C. albicans and E. coli did not induce IFN-α genes. IFN-β mRNA was induced most quickly and was sustained in MV-infected DCs and influenza virus-infected DCs, whereas there was no induction by C. albicans, and induction by E. coli was transient (Fig. 2). MV induced IFN-ω more robustly and earlier than did influenza virus. Biologically active IFN from MV-infected DCs showed a rapid increase in production over the first 12 h, with sustained synthesis through 24 h (Fig. 1B).
Fig. 1.
Induction of IFN-α genes. (A) IFN-α gene expression. Time course of IFN-α gene expression for DCs exposed to MV, C. albicans, E. coli, influenza virus, or media. RNAs were collected at various time points, and expression levels were measured by microarray analysis. The lines show the mean fold changes normalized to the preexposure 0 time point, and the points show the actual fold changes. Data for C. albicans, E. coli, and influenza virus are from Huang et al. (15). (B) Production of biologically active IFN by MV-infected DCs. Points show the calculated IFN units per milliliter for the replicates and the line shows the means.
Fig. 2.
Induction of IFN-β, IFN-γ, and IFN-ω genes. Time courses of IFN-β, IFN-γ, and IFN-ω for DCs exposed to MV, C. albicans, E. coli, influenza virus, or media. RNA analysis, data sources, and plotting are as described for Fig. 1A.
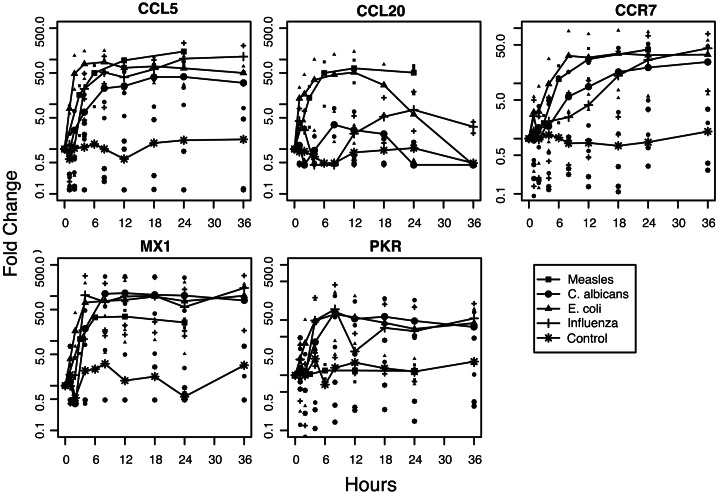
Consistent with the increase in IFN production, mRNAs for many IFN-induced proteins were up-regulated in MV and influenza virus-infected DCs but also often in E. coli-infected DCs and C. albicans-infected DCs. mRNAs encoding proteins with known antiviral activity that were universally up-regulated included 2′5′-oligoadenylate synthetase, MxA, guanylate-binding protein, ISG-12, ISG-15, and APOBEC3B (Fig. 3). Interestingly, the IFN-induced antiviral gene dsRNA-dependent protein kinase (PKR) was not induced by MV infection but was induced by all other stimuli (Fig. 3). The failure of MV to up-regulate PKR mRNA was confirmed by RT-PCR with LPS-treated DCs as a positive control (data not shown).
Fig. 3.
Induction of representative chemokine and antiviral protein genes. Time courses for CCL5, CCL20, CCR7, Mx1, and PKR gene expression for DCs exposed to MV, C. albicans, E. coli, influenza virus, or media. RNA analysis, data sources, and plotting are as described for Fig. 1A.
Maturation-associated genes coded for chemokines and IFN-regulatory factors (IRFs) (examples in Fig. 3). The chemokine mRNAs induced by all pathogens included CCL5, CCL8, CCL19, CXCL2, CXCL8, CXCL9, CXCL10, and CXCL11. The chemokines CCL1, CCL2, CCL7, CCL15, CCL20, CXCL3, and CXCL9 were differentially regulated. For instance, MV and E. coli promptly induced high levels of CCL20 (MIP-3α) mRNA, whereas C. albicans and influenza virus induction was slower and lower (Fig. 3). The gene for CCR7, a chemokine receptor important for targeting DCs to secondary lymphoid tissue (18–20), was strongly induced by all pathogens, as were CXCR4 (19) and CCRL2, an orphan chemokine receptor (21) (Fig. 3). Up-regulated IRFs included IRF1 and IRF7.
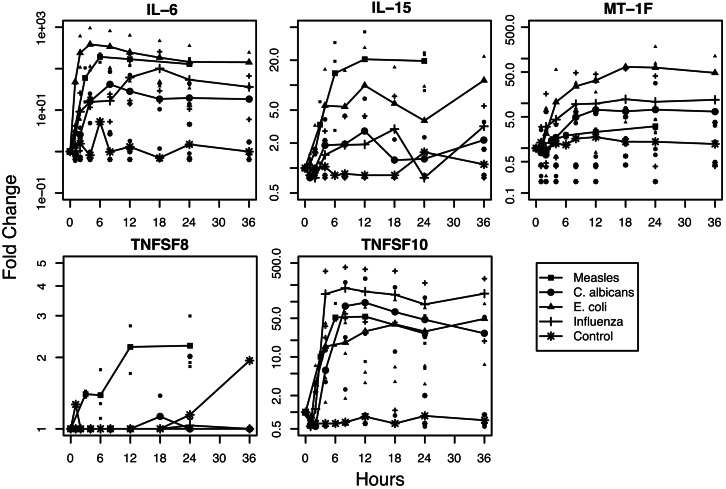
MV up-regulated a variety of interleukins, including IL-1β, 6, 11, 12A, 12B and 15 and TNFSF genes 1, 2, 4, 7, 8, 9 and 10. IL-6 and TNFSF 2 (TNF), 7 (CD27L), 9 (4IBBL), and 10 (TRAIL) were up-regulated by all pathogens, whereas TNFSF8 (CD30L) was only induced by MV (examples in Fig. 4). Five of the seven metallothionein-1 genes were significantly regulated in MV-infected DCs, and MT-1F and MT-1H were induced in DCs exposed to all of the pathogens (examples in Fig. 4). The chromosomal protein group was composed mostly of histone 1, H2b family members.
Fig. 4.
Induction of representative IL, metallothionein, and TNFSF member genes. Time courses for IL-6, IL-15, MT-1F, TNFSF8, and TNFSF10 for DCs exposed to MV, C. albicans, E. coli, influenza virus, or media. RNA analysis, data sources, and plotting are as described for Fig. 1A.
Genes down-regulated by MV fell into three general groups (Table 2). One group contained the overlapping ubiquinone, hydrogen ion transport, inner membrane, NAD, mitochondrion, transit peptide, and oxidoreductase groups that encode the proteins that form the five complexes of the oxidative phosphorylation machinery: NADH dehydrogenase, succinate-Q reductase, ubiquinol-cytochrome c reductase, cytochrome c oxidase, and ATP synthase. Overall, mRNAs for 31 of 59 oxidative phosphorylation protein genes on the chip were down-regulated. The second group included the ribosomal protein, initiation factor, and protein biosynthesis classifications that code for nuclear and mitochondrial ribosomal proteins and eukaryotic translation initiation factors. The third chromatin regulator group was composed of histone deacetylase and chromobox homolog genes.
Table 2.
Significantly down-regulated genes classified by Swiss-Prot keyword
| Swiss-Prot keyword | Measles |
||
|---|---|---|---|
| Up | Down | ES | |
| Ubiquinone | 0 | 13 | 6.4 |
| Ribosomal protein | 0 | 36 | 4.3 |
| Hydrogen ion transport | 0 | 13 | 3.5 |
| Inner membrane | 0 | 15 | 3.5 |
| Chromatin regulator | 2 | 12 | 3.4 |
| Initiation factor | 1 | 12 | 3.3 |
| NAD | 3 | 32 | 3.1 |
| Mitochondrion | 11 | 82 | 2.7 |
| Transit peptide | 6 | 58 | 2.7 |
| Protein biosynthesis | 3 | 17 | 2.5 |
| Oxidoreductase | 14 | 66 | 2.3 |
ES, enrichment score; Up, up-regulated; Down, down-regulated.
Discussion
We have analyzed the gene expression changes of immature, monocyte-derived DCs infected with MV, and we compared the results to publicly available data from DCs infected with a bacterium (E. coli), a fungus (C. albicans), and another virus (influenza virus). After infection with MV, 1,553 genes changed by 2-fold or more within 24 h. Included within these genes were those previously identified as part of the core response of DCs to a pathogen (15). A high percentage of up-regulated genes were shared in common with all pathogens studied, with the highest percentage shared with influenza virus. However, the transcriptional response of DCs to MV was unique in the broad array of IFN-α genes induced and the failure to induce PKR. The MV-induced response differed from influenza virus in induction of CCL1 (I-309), IL-12B, and IL-15.
The few previous studies of the cellular antiviral response to MV have described the induction of IFNs, 2′5′-oligoadenylate synthetase, IRF, and Mx genes in various cells (22–27). Microarrays were used to study MV infection of PBMCs at 48 h (28). Of the 10 induced genes identified, 6 were on the U95Av2 chip and 5 of those, NF-κB p52, IRF7, 2′5′-oligoadenylate synthetase, IFN-α, and IFN-β, were significantly up-regulated in our study.
The induction of IFN was a predominant part of the response of DCs to MV and has been reported to be the mechanism by which MV induces DC maturation (16). Increases in IFN have not been identified during natural infection, perhaps reflecting a lack of analysis of samples early during infection (29). MV induces production of IFN-α/β by a lung epithelial cell line (30), but there is limited synthesis of IFN by MV-infected PBMCs (31). Plasmacytoid DCs are generally considered to be responsible for early IFN production in response to infection (32), but MV inhibits IFN production by these cells (33). An important early source of IFN in MV infection may be myeloid DCs.
Not all genes known to be IFN-inducible were increased in MV-infected DCs, most notably PKR. An interesting aspect of MV biology is the robust induction of IFN but selective inhibition of IFN-responsive genes. The role of IFN in control of MV replication is unclear. Inhibition by MxA is cell-type-specific (24, 25, 34), and IFN has a limited ability to interfere with MV replication in PBMCs (31, 35). Little infectious MV is produced during the first 1–3 d of DC infection, but, with maturation and T cell activation syncytia-formation, virus production and cell death are reported (10, 13, 36).
DCs affect the induction of the adaptive immune response. Measles is associated with an early CD4 T helper (Th)1 and CD8 T cell response and later with a predominant Th2 CD4 response (37, 38). CXCL9 (mig), CXCL10 (IP-10), and CXCL11 (I-TAC), ligands for CXCR3, a chemokine receptor expressed on activated Th1 cells, were increased by all pathogens. Through the course of an immune response, DC priming conditions can change from Th1 to Th2 (39) and genes, such as TNFSF8 (CD30L), implicated in Th2 CD4T cell maturation (40) and in generation of memory CD8 T cells (41) were also induced by MV. This network may promote effector cell function for virus clearance, the establishment of memory T cells, and the long-term immunity characteristic of measles.
DCs may play a central role in MV-associated immunosuppression. One suggested mechanism is TRAIL–mediated apoptosis of T cells (42). However, TRAIL was similarly induced by all pathogens and therefore is not a unique mechanism for immunosuppression in measles. How immunosuppression and the induction of a robust immune response to MV coexist is unclear, but one possibility is that apoptotic MV-infected DCs facilitate cross-presentation of MV antigens to lymphocytes by uninfected DCs (43).
The mechanism by which MV induces transcriptional changes in DCs is not known. The H glycoprotein is a determinant of MV infection of DCs (36) and can bind to Toll-like receptor-2 on macrophages (44) and to the MV receptors CD46 and SLAM (45). DCs express CD46 and increasing amounts of SLAM with differentiation (45), and the Chicago-1 strain of MV can use both receptors. In addition, the MV nucleocapsid can directly activate the IRF3 pathway intracellularly (46). Therefore, triggering could be directly through signaling at the cell surface or through cytoplasmic detection of MV infection. The pattern of early (by 3h) production of transcripts for IFN-β, IRF7, CCL5, and TRAIL are consistent with IRF3 signaling and with a subsequent IRF7-mediated increase in IFN-α mRNAs at 6 h.
It has long been observed that host cell transcription and translation are altered during viral infection. Gene array studies often find more down-regulated genes than up-regulated genes after viral infection (47). Two large gene groups were down-regulated after MV infection: oxidative phosphorylation and protein biosynthesis. Whether shutting down oxidative phosphorylation is an antiviral response or part of the apoptotic process or whether it has some other role in the virus–host relationship is unclear. Down-regulating the mRNA for ribosomal proteins could help block viral protein production.
Currently, the various array platforms, analysis algorithms, and annotation schemes make it difficult for laboratories to compare microarray results. The use of standard experimental designs and statistical procedures will increase the resolution of microarray data comparisons, which will yield new insights into virus–cell interactions.
Materials and Methods
Virus and Cell Culture.
The Chicago-1 strain of MV (48) was grown in Vero cells, concentrated by centrifugation at 80,000 × g, and assayed by plaque formation. Stocks had concentrations of LPS between 0.01 and 0.04 ng/ml (49).
PBMCs acquired from anonymous donors by the Johns Hopkins Hemapheresis Center were isolated over a Ficoll–Hypaque gradient. Monocyte-derived DCs were produced by standard methods (50). Briefly, 107 cells per ml cultured overnight in RPMI medium 1640 with 30% autologous serum were washed with PBS (pH 7.2), and adherent cells were removed by gentle scraping. Lymphocytes were removed with magnetic beads (Miltenyi Biotec, Auburn, CA) resulting in 86.3 ± 7.3% CD14+ cells that were cultured at 5 × 105 cells per ml in RPMI medium 1640 with 10% FBS, 2 mM l-glutamine, 2 mM Hepes, 100 μM nonessential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin (GIBCO), 1,000 units/ml IL-4, and 50 ng/ml granulocyte–macrophage colony-stimulating factor (BD Biosciences). On day 6, the cells were treated with 200 ng/ml LPS (Sigma-Aldrich) or infected with MV at a multiplicity of infection of 5, incubated for 1 h at 37°C, and washed. Fresh medium with cytokines was then added. Virus replication was assessed by plaque formation and by quantitative RT-PCR.
IFN Assay.
Serially diluted supernatant fluids from MV-infected DCs were incubated overnight with Hep-2 cells and challenged with 100 tissue culture 50% infective doses of vesicular stomatitis virus. The cytopathic effect was assessed after 24 h. IFN concentration (in units per milliliter) for each sample was calculated from an IFN-α standard curve (R & D Systems).
Microarray Analysis.
Total RNA from four separate experiments was isolated from control cells, and infected cells at 3, 6, 12, and 24 h after infection using an RNeasy kit (Qiagen, Valencia, CA). RNA (15 μg) was used to synthesize cDNA in two steps using the Superscript Choice System (GIBCO/BRL) and the reverse-transcription primer T7-(dt)24, [5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG(T)24] (GENSET, San Diego). The cDNA product was purified by using phase-lock gels (Brinkmann Instruments) and used to synthesize biotin-labeled cRNA with the BioArray high-yield RNA transcript labeling kit (Enzo Diagnostics). The cRNA was precipitated, washed twice with 80% ethanol, dried, and resuspended in RNase-free water. RNA (20 μg) were fragmented by metal-induced hydrolysis and products verified by gel electrophoresis.
Fragmented cRNA was hybridized to a human U95Av2 GeneChip array along with 50 pM control oligo B2 (5′-bioGTCAAGATGCTACCGTTCAG-3′), a control RNA of 150 pM Bio B, 500 pM Bio C, 2.5 nM Bio D, and 10 mM Cre X (Affymetrix), and 0.1 mg/ml herring sperm DNA. Hybridization was at 42°C and 60 rpm for 16 h. The chips were washed with Affymetrix fluidics protocol EukWSH-4 and stained with streptavidin-phycoerythrin (Molecular Probes) at 10 μg/ml in 6× standard saline phosphate/EDTA buffer (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA) with 1 mg/ml acetylated BSA (Sigma–Aldrich). The chips were washed twice and scanned with an HP GeneArray scanner (Hewlett–Packard).
The December 7, 2004 Affymetrix U95Av2 release was used to annotate the data. The affy package v1.5.8 for r v2.0.1 was used to analyze the data (51). Exploratory data analysis verified the quality of the hybridizations. Background correction, normalization, and summarization used quantile normalization and robust multiarray analysis. The data set is under accession no. GSE980 in the Gene Expression Omnibus database (available at www.ncbi.nlm.nih.gov/geo). Significance was determined by using sam 1.21 (Significance Analysis of Microarrays) (52). Four controls and three samples from the 6-h and 24-h time points were compared with a two-class, unpaired algorithm. Significantly regulated genes identified at each time point were combined. Only genes with a fold change of 2.0 or more were considered for significance. A false discovery rate of <10% at the 90th percentile was chosen. A higher threshold with a false discovery rate of <1% at the 90th percentile yielded similar conclusions.
Comparisons were made with the data of Huang et al. (15), who studied changes in transcription over 36 h in monocyte-derived DCs exposed to C. albicans, mannose, E. coli, LPS, influenza virus, and dsRNA using the Affymetrix HuGeneFL chip (15). The results from this study were handled in the same way as the MV data; however, the raw data were not available, so the background subtraction, normalization, and summarization procedures were different. Data from the C. albicans, E. coli, and influenza virus-infected DCs, available in triplicate, were used for comparison with data from MV-infected DCs. The significantly regulated genes were annotated using DRAGON (Database Referencing of Array Genes Online) to assign Swiss-Prot keywords (53, 54). None of the classifications were exclusive. An enrichment score was calculated for the Swiss-Prot keyword annotations: [(significantly regulated genes with keyword)/(total genes with keyword)]/[(total significantly regulated genes)/(total genes on chip)] (55).
For graphing, replicate measurements at each time point were averaged and divided by the time 0 (control) mean to calculate the fold change. Duplicated genes were averaged. When probes from the HuGeneFL chip overlapped two or more probes on the U95Av2 chip, the U95Av2 probe with the highest percent similarity to the probe on the HuGeneFL chip was graphed. The graphs show the average fold change. Individual points represent the actual gene expression measurements. The DC controls and duplicate genes had up to 15 measurements at each time point, but only three randomly chosen measurements are displayed.
Acknowledgments
We thank Anne Jedlicka and Margaret V. Mintz for invaluable help. Affymetrix GeneChip experimentation and analyses were provided by the Johns Hopkins Malaria Research Institute Gene Array Core Facility. This work was supported by National Institutes of Health Research Grant R01 AI023047 and Training Grant T32 ES07141 and by the Bill and Melinda Gates Foundation.
Abbreviations
- MV
measles virus
- DC
dendritic cell
- TNFSF
TNF superfamily
- PKR
dsRNA-dependent protein kinase
- TRAIL
TNF-related apoptosis-inducing ligand
- IRF
IFN-regulatory factors
- Th
T helper.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The time-course infection data have been deposited in the Gene Expression Omnibus database, www.ncbi.nlm.nih.gov/geo (accession no. GSE980).
References
- 1.Stein C. E., Birmingham M., Kurian M., Duclos P., Strebel P. J. Infect. Dis. 2003;187(Suppl. 1):S8–S14. doi: 10.1086/368114. [DOI] [PubMed] [Google Scholar]
- 2.Griffin D. E. Curr. Top. Microbiol. Immunol. 1995;191:117–134. doi: 10.1007/978-3-642-78621-1_8. [DOI] [PubMed] [Google Scholar]
- 3.Esolen L. M., Ward B. J., Moench T. R., Griffin D. E. J. Infect. Dis. 1993;168:47–52. doi: 10.1093/infdis/168.1.47. [DOI] [PubMed] [Google Scholar]
- 4.Moench T. R., Griffin D. E., Obriecht C. R., Vaisberg A. J., Johnson R. T. J. Infect. Dis. 1988;158:433–442. doi: 10.1093/infdis/158.2.433. [DOI] [PubMed] [Google Scholar]
- 5.Schneider-Schaulies S., Bieback K., Avota E., Klagge I., ter Meulen V. J. Mol. Med. 2002;80:73–85. doi: 10.1007/s00109-001-0299-x. [DOI] [PubMed] [Google Scholar]
- 6.Kaiserlian D., Dubois B. Semin. Immunol. 2001;13:303–310. doi: 10.1006/smim.2001.0326. [DOI] [PubMed] [Google Scholar]
- 7.Grosjean I., Caux C., Bella C., Berger I., Wild F., Banchereau J., Kaiserlian D. J. Exp. Med. 1997;186:801–812. doi: 10.1084/jem.186.6.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Servet-Delprat C., Vidalain P. O., Bausinger H., Manie S., Le Deist F., Azocar O., Hanau D., Fischer A., Rabourdin-Combe C. J. Immunol. 2000;164:1753–1760. doi: 10.4049/jimmunol.164.4.1753. [DOI] [PubMed] [Google Scholar]
- 9.Helin E., Salmi A. A., Vanharanta R., Vainionpaa R. Virology. 1999;253:35–42. doi: 10.1006/viro.1998.9460. [DOI] [PubMed] [Google Scholar]
- 10.Fugier-Vivier I., Servet-Delprat C., Rivailler P., Rissoan M.-C., Liu Y. J., Rabourdin-Combe C. J. Exp. Med. 1997;186:813–823. doi: 10.1084/jem.186.6.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnorr J.-J., Xanthakos S., Keikavoussi P., Kampgen E., ter Meulen V., Schneider-Schaulies S. Proc. Natl. Acad. Sci. USA. 1997;94:5326–5331. doi: 10.1073/pnas.94.10.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh M., Billeter M. J. Gen. Virol. 1999;80:101–106. doi: 10.1099/0022-1317-80-1-101. [DOI] [PubMed] [Google Scholar]
- 13.Servet-Delprat C., Vidalain P., Azocar O., LeDeist F., Fischer A., Rabourdin-Combe C. J. Virol. 2000;74:4387–4393. doi: 10.1128/jvi.74.9.4387-4393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vidalain P., Azocar O., Lamouille B., Astier A., Rabourdin-Combe C., Servet-Delprat C. J. Virol. 2000;74:556–559. doi: 10.1128/jvi.74.1.556-559.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Q., Liu D., Majewski P., Schulte L. C., Korn J. M., Young R. A., Lander E. S., Hacohen N. Science. 2001;294:870–875. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- 16.Klagge I. M., ter Meulen V., Schneider-Schaulies S. Eur. J. Immunol. 2000;30:2741–2750. doi: 10.1002/1521-4141(200010)30:10<2741::AID-IMMU2741>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 17.Kleijmeer M., Ramm G., Schuurhuis D., Griffith J., Rescigno M., Ricciardi-Castagnoli P., Rudensky A. Y., Ossendorp F., Melief C. J., Stoorvogel W., et al. J. Cell Biol. 2001;155:53–63. doi: 10.1083/jcb.200103071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Randolph G. J., Angeli V., Swartz M. A. Nat. Rev. Immunol. 2005;5:617–628. doi: 10.1038/nri1670. [DOI] [PubMed] [Google Scholar]
- 19.Sallusto F., Schaerli P., Loetscher P., Schaniel C., Lenig D., Mackay C. R., Qin S., Lanzavecchia A. Eur. J. Immunol. 1998;28:2760–2769. doi: 10.1002/(SICI)1521-4141(199809)28:09<2760::AID-IMMU2760>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 20.Sallusto F., Palermo B., Lenig D., Miettinen M., Matikainen S., Julkunen I., Forster R., Burgstahler R., Lipp M., Lanzavecchia A. Eur. J. Immunol. 1999;29:1617–1625. doi: 10.1002/(SICI)1521-4141(199905)29:05<1617::AID-IMMU1617>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Shimada T., Matsumoto M., Tatsumi Y., Kanamaru A., Akira S. FEBS Lett. 1998;425:490–494. doi: 10.1016/s0014-5793(98)00299-3. [DOI] [PubMed] [Google Scholar]
- 22.Tilles J. G., Balkwill F., Davilla J. Proc. Soc. Exp. Biol. Med; 1987. pp. 70–74. [DOI] [PubMed] [Google Scholar]
- 23.Volckaert-Vervliet G., Billiau A. J. Gen. Virol. 1977;37:199–203. doi: 10.1099/0022-1317-37-1-199. [DOI] [PubMed] [Google Scholar]
- 24.Schneider-Schaulies S., Schneider-Schaulies J., Schuster A., Bayer M., Pavlovic J., ter Meulen V. J. Virol. 1994;68:6910–6917. doi: 10.1128/jvi.68.11.6910-6917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnorr J.-J., Schneider-Schaulies S., Simon-Jodicke A., Pavlovic J., Horisberger M. A., ter Meulen V. J. Virol. 1993;67:4760–4768. doi: 10.1128/jvi.67.8.4760-4768.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chelbi-Alix M. K., Quignon F., Pelicano L., Koken M. H. M., de Thé H. J. Virol. 1998;72:1043–1051. doi: 10.1128/jvi.72.2.1043-1051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marie I., Durbin J. E., Levy D. E. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolt G., Berg K., Blixenkrone-Moller M. J. Gen. Virol. 2002;83:1157–1165. doi: 10.1099/0022-1317-83-5-1157. [DOI] [PubMed] [Google Scholar]
- 29.Griffin D. E., Ward B. J., Jauregui E., Johnson R. T., Vaisberg A. Clin. Exp. Immunol. 1990;81:218–224. doi: 10.1111/j.1365-2249.1990.tb03321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helin E., Vainionpaa R., Hyypia T., Julkunen I., Matikainen S. Virology. 2001;290:1–10. doi: 10.1006/viro.2001.1174. [DOI] [PubMed] [Google Scholar]
- 31.Naniche D., Yeh A., Eto D. S., Manchester M., Friedman R., Oldstone B. A. J. Virol. 2000;74:7478–7484. doi: 10.1128/jvi.74.16.7478-7484.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna K., Beignon A. S., Bhardwaj N. J. Virol. 2005;79:17–27. doi: 10.1128/JVI.79.1.17-27.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlender J., Hornung V., Finke S., Gunthner-Biller M., Marozin S., Brzozka K., Moghim S., Endres S., Hartmann G., Conzelmann K. K. J. Virol. 2005;79:5507–5515. doi: 10.1128/JVI.79.9.5507-5515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haller O., Kochs G. Traffic. 2002;3:710–717. doi: 10.1034/j.1600-0854.2002.31003.x. [DOI] [PubMed] [Google Scholar]
- 35.Leopardi R., Hyypia T., Vainionpaa R. Acta Pathol. Microbiol. Immunol. Scan. 1992;100:125–131. doi: 10.1111/j.1699-0463.1992.tb00850.x. [DOI] [PubMed] [Google Scholar]
- 36.Ohgimoto S., Ohgimoto K., Niewiesk S., Klagge I. M., Pfeuffer J., Johnston I. C., Schneider-Schaulies J., Weidmann A., ter Meulen V., Schneider-Schaulies S. J. Gen. Virol. 2001;82:1835–1844. doi: 10.1099/0022-1317-82-8-1835. [DOI] [PubMed] [Google Scholar]
- 37.Griffin D. E., Ward B. J. J. Infect. Dis. 1993;168:275–281. doi: 10.1093/infdis/168.2.275. [DOI] [PubMed] [Google Scholar]
- 38.Moss W. J., Ryon J. J., Monze M., Griffin D. E. J. Infect. Dis. 2002;186:879–887. doi: 10.1086/344230. [DOI] [PubMed] [Google Scholar]
- 39.Langenkamp A., Messi M., Lanzavecchia A., Sallusto F. Nat. Immunol. 2000;1:311–316. doi: 10.1038/79758. [DOI] [PubMed] [Google Scholar]
- 40.Rossi F. M., Degan M., Mazzocut-Zecchin L., Di Francia R., Aldinucci D., Pinto A., Gattei V. FEBS Lett. 2001;508:418–422. doi: 10.1016/s0014-5793(01)03076-9. [DOI] [PubMed] [Google Scholar]
- 41.Nishimura H., Yajima T., Muta H., Podack E. R., Tani K., Yoshikai Y. J. Immunol. 2005;175:4627–4634. doi: 10.4049/jimmunol.175.7.4627. [DOI] [PubMed] [Google Scholar]
- 42.Servet C., Zitvogel L., Hosmalin A. Curr. Mol. Med. 2002;2:739–756. doi: 10.2174/1566524023361907. [DOI] [PubMed] [Google Scholar]
- 43.Bhardwaj N. J. Exp. Med. 1997;186:795–799. doi: 10.1084/jem.186.6.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bieback K., Lien E., Klagge I. M., Avota E., Schneider-Schaulies J., Duprex W. P., Wagner H., Kirschning C. J., ter Meulen V., Schneider-Schaulies S. J. Virol. 2002;76:8729–8736. doi: 10.1128/JVI.76.17.8729-8736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murabayashi N., Kurita-Taniguchi M., Ayata M., Matsumoto M., Ogura H., Seya T. Microbes Infect. 2002;4:785–794. doi: 10.1016/s1286-4579(02)01598-8. [DOI] [PubMed] [Google Scholar]
- 46.Tenoever B. R., Servant M. J., Grandvaux N., Lin R., Hiscott J. J. Virol. 2002;76:3659–3669. doi: 10.1128/JVI.76.8.3659-3669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geiss G., Jin G., Guo J., Bumgarner R., Katze M. G., Sen G. C. J. Biol. Chem. 2001;276:30178–30182. doi: 10.1074/jbc.c100137200. [DOI] [PubMed] [Google Scholar]
- 48.Rota J. S., Hummel K. B., Rota P. A., Bellini W. J. Virology. 1992;188:135–142. doi: 10.1016/0042-6822(92)90742-8. [DOI] [PubMed] [Google Scholar]
- 49.Garcia M., Yu X. F., Griffin D. E., Moss W. J. J. Virol. 2005;79:9197–9205. doi: 10.1128/JVI.79.14.9197-9205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cella M., Salio M., Sakakibara Y., Langen H., Julkunen I., Lanzavecchia A. J. Exp. Med. 1999;189:821–829. doi: 10.1084/jem.189.5.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Irizarry R. A., Gautier L., Cope L. M. In: The Analysis of Gene Expression Data: Methods and Software. Parmigiani G., Garrett E. S., Irizarry R. A., Zeger S. I., editors. New York: Springer; 2003. [Google Scholar]
- 52.Tusher V. G., Tibshirani R., Chu G. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouton C. M., Pevsner J. Bioinformatics. 2000;16:1038–1039. doi: 10.1093/bioinformatics/16.11.1038. [DOI] [PubMed] [Google Scholar]
- 54.Bairoch A., Boeckmann B. Nucleic Acids Res. 1994;22:3578–3580. [PMC free article] [PubMed] [Google Scholar]
- 55.Zeeberg B. R., Feng W., Wang G., Wang M. D., Fojo A. T., Sunshine M., Narasimhan S., Kane D. W., Reinhold W. C., Lababidi S., et al. Genome Biol. 2003;4:R28.1–R28.8. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]