Abstract
The bacterial outer-membrane vitamin B12 transporter, BtuB, undergoes a dramatic order-to-disorder transition in its N-terminal energy-coupling motif (Ton box) upon substrate binding. Here, site-directed spin labeling (SDSL) is used to show that a range of solutes prevents this conformational change when ligand is bound to BtuB, resulting in a more ordered Ton box structure. For each solute examined, the data indicate that solutes effectively block this conformational transition through an osmotic mechanism. The molecular weight dependence of this solute effect has been examined for a series of polyethylene glycols, and a sharp molecular weight cutoff is observed. This cutoff indicates that solutes are preferentially excluded from a cavity within the protein as well as the protein surface. Furthermore, the sensitivity of the conformational change to solution osmolality is consistent with a structural model predicted by SDSL. When the Ton box is unfolded by detergents or mutations (rather than by ligand binding), solutes, such as polyethylene glycols and salts, also induce a more structured compacted conformation. These results suggest that conformational changes in this class of outer membrane transporters, which involve modest energy differences and changes in hydration, may be modulated by a range of solutes, including solutes typically used in protein crystallization.
INTRODUCTION
The outer membrane of Gram-negative bacteria, such as Escherichia coli, contains a family of transport proteins that are designed to facilitate the uptake of rare nutrients from the external medium (1,2). These include proteins such as FhuA, FepA, and FecA, which transport various forms of iron, and BtuB, which transports vitamin B12. These transporters extract energy from the proton potential across the inner membrane by coupling with the transperiplasmic protein TonB. These TonB-dependent transporters are structurally homologous, consisting of a 22-stranded β-barrel, where an N-terminal domain or “hatch” region of the protein is folded within the interior of the barrel. A conserved feature in all these TonB-dependent transporters is a segment near their N-termini termed the “Ton box”, which is thought to play a role in coupling BtuB and other TonB-dependent transporters to TonB (see Fig. 1).
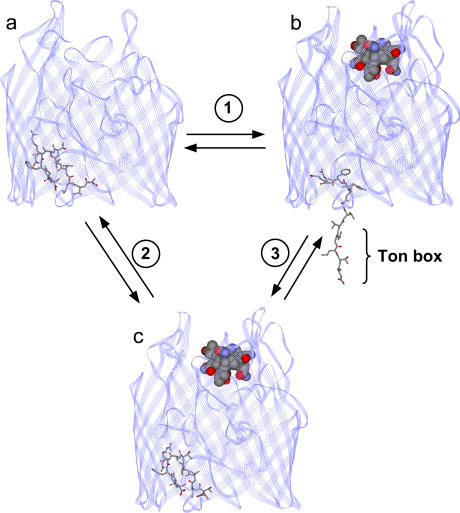
FIGURE 1.
Models for BtuB. (a) Crystal structure of BtuB in the absence of substrate (vitamin B12) (Protein Data Bank ID 1NQE), and (b) model for BtuB in the presence of substrate, where the configuration of the Ton box has been modified from that in the crystal structure to be consistent with the results of SDSL (3). In this case, SDSL indicates that the N-terminus, including the Ton box, unfolds to position 15 or 16. (c) Crystal structure of BtuB in the presence of substrate (Protein Data Bank ID 1NQH) (7). The Ton box includes residues 6–12. The portion of the structure highlighted in stick form represents residues 6–17. (1) A model for the conformational change that takes place under physiological conditions when substrate binds BtuB. (2) The conformational change that takes place under conditions of high osmolality upon the addition of substrate. (3) A conformational change that takes place upon the addition of osmolyte in the presence of substrate.
A number of experiments show that the Ton box in BtuB undergoes a substrate-dependent order-to-disorder transition. In particular, site-directed spin labeling (SDSL), chemical labeling, and cross-linking indicate that this segment is folded within the barrel of the transporter in the absence of substrate but unfolds and becomes unstructured in the presence of substrate (the undocked state) (Fig. 1 b) (3–5). The data indicate that these two conformational states exist in equilibrium, where the docked form is favored in the absence of substrate but the unfolded state is favored in the presence of substrate. This conformational transition may have functional significance, it may represent the first step in the transport process, and it appears to be the conformational trigger that initiates interactions between BtuB and TonB (6).
The order-to-disorder transition observed by SDSL and the increase in Ton box exposure indicated by chemical labeling and disulfide cross-linking are not revealed in the crystal structure of BtuB (7). Shown in Fig. 1 are crystal structures for BtuB in the absence (Fig. 1 a) and presence (Fig. 1 c) of substrate where the Ton box region is indicated. The crystal structure shows a much more subtle change in the structure of the Ton box upon substrate binding, and this segment remains folded within the hatch region of the protein. In comparison, Fig. 1 b shows a structure of BtuB where the Ton box has been placed in a configuration consistent with the results of our previous SDSL work (3). The spectroscopic result suggests that substrate binding unfolds the N-terminus of the protein and the Ton box so that it extends as much as 20–25 Å into the periplasmic space. We previously reported that the discrepancy between the crystallographic and spectroscopic models resulted from solutes in the buffer solutions in the sample preparations (8). In particular, solutes such as polyethylene glycol (PEG) 3350 were shown to prevent the Ton box from completely unfolding when BtuB is bound to ligand, a result consistent with the x-ray model.
In this work, the effect of a number of different solutes that vary in size on the conformational equilibrium of the Ton box segment of BtuB is explored via SDSL. In addition to PEGs, we find that salts, sucrose, and trimethylamine N-oxide (TMAO) all alter the dynamics and structure of the Ton box through an osmotic mechanism. Both the effective excluded solvent volume and the effect of PEG molecular weight are consistent with a structural model where the Ton box unfolds upon binding of substrate to BtuB.
Additionally, previous work from our lab has shown that the Ton box can be unfolded (albeit by a different mechanism) in the absence of substrate by detergents such as octylglucoside (OG) (9) or by proline mutations within the Ton box (6,10). The results presented here demonstrate that a solution of high osmolality will induce a more ordered conformation of the Ton box even when its structure is disordered by detergents or mutations. These results demonstrate that osmotic stress generated by solutes promotes a more folded/compact conformation of the Ton box regardless of the mechanism by which it is unfolded/disordered.
METHODS
Materials
The sulfhydryl reactive methanethiosulfonate spin label (MTSL) [(1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl) methanethiosulfonate)] was purchased from Toronto Research Chemicals (North York, Ontario, Canada). Vitamin B12 (cyanocobalamin, CNCbl), polyethylene glycol 400 (PEG 400), sucrose (gradient grade), and Ficoll 400 (molecular weight (MWT) 400,000) were obtained from Sigma (St. Louis, MO). PEGs (PEG 1000, 2000, 3350, and 6000) and glycine betaine monohydrate were purchased from Fluka (St. Louis, MO). Lithium chloride, TMAO, HEPES, and sodium chloride were purchased from Fisher Scientific (Fair Lawn, NJ). OG (Anagrade) was purchased from Anatrace (Maumee, OH), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) was purchased from Avanti Polar Lipids (Alabaster, AL).
Expression, labeling with MTSL, purification, and reconstitution of BtuB
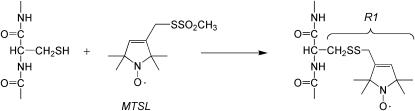
Expression, labeling with MTSL (Fig. 2), purification, and reconstitution of BtuB mutants were performed as previously described (11). Labeling with the MTSL generates the spin-labeled site chain R1 at a cysteine position, and single cysteine mutations that are derivatized with the MTSL are designated as R1. Reconstituted BtuB samples were prepared in a 10 mM HEPES, 130 mM NaCl, pH 6.5 buffer. The reservoir and soaking buffers used in protein crystallization were described previously (12).
FIGURE 2.
MTSL spin label is attached to a cysteine side chain to produce the spin-labeled side chain R1.
Measurement of solution osmolalities
Solution osmolalities were measured with a Wescor 5500 vapor pressure osmometer (Logan, UT). Sample sizes were 10 μl. Measurements were made in duplicate for each solution, and most solutions were prepared in duplicate to ensure reproducibility. The average values were used in all calculations and figures. Because electron paramagnetic resonance (EPR) sample volumes are typically <5 μl, separate solutions for osmolality measurements were prepared. These solutions mimicked those used for EPR except that there was no protein present in the samples used for osmometry measurements.
EPR spectroscopy
EPR spectroscopy was performed on a Varian E-line 102 X-band spectrometer equipped with a two-loop one-gap resonator (Medical Advances, Milwaukee, WI). LabView software, which was generously provided by Drs. Christian Altenbach and Wayne Hubbell (University of California, Los Angeles), was used for digital collection and analysis of data. All spectra for line shape analyses were recorded at 2.0 mW incident power with a modulation amplitude of 1.0 G, from samples in glass capillary tubes with a 0.6 mm i.d. (VitroCom, Mountain Lakes, NJ). Titration experiments were performed by preparation of individual protein samples where the appropriate amount of a stock solution of osmolyte was added. Osmolyte solutions were either added directly to BtuB reconstituted into POPC vesicles (with or without B12) or to samples of BtuB in POPC that were solubilized with OG (POPC/OG 1:17). For POPC samples where substrate or osmolyte are added, membranes are taken through five freeze-thaw cycles in liquid N2 to ensure that added solutes have access to both sides of vesicles. For incorporation into crystallization solutions, protein samples that were reconstituted into POPC were added to an equivalent volume of the crystallization solution that had been taken to dryness using a Savant Speed Vac concentrator (Holbrook, NY). In these samples, the detergent concentrations present in the crystallization buffers were sufficient to clarify the reconstituted membrane preparation, implying that a mixed micelle membrane mimetic system results.
Analysis of EPR spectra
In general, any protein conformation transition that involves a change in hydration will be sensitive to the addition of solutes. Consider the following simple equilibrium between two protein conformational states, where the protein in conformation 1, P1, undergoes a change in hydration to assume a second conformation, P2.
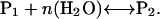 |
(1) |
In this equilibrium, the presence of solutes will lower the activity of water and may shift the equilibrium (Eq. 1) so that the protein assumes its less hydrated state, P1. Physical mechanisms have been discussed in detail to account for this behavior (13–15). In one mechanism, protein surfaces are envisioned to be differentially hydrated by solutes and solvent, so that the solute is excluded from a region near the protein surface. As a result of this negative interaction, work is done by the protein in a transition such as that in Eq. 1 to exclude solute from a region around the hydrated protein surface (15). In a second model, solutes are imagined to be inaccessible to hydrated cavities within the protein. To bring water into chemical equilibrium between the cavity and the bulk aqueous phase, an osmotic stress is generated across the protein that drives the protein toward its less hydrated conformation (P1) (14).
The EPR spectra of the MTSL side chain (R1) reflect the local structure and dynamics at the labeled site (16,17), and EPR spectroscopy can be used to observe and measure conformational equilibria (18) such as those depicted in Eq. 1. In our analysis of the conformational states of substrate-bound BtuB, we will assume that the EPR spectra report two distinct conformation states of the Ton box that are in equilibrium: one that is folded and will be referred to as “trapped” and a second that is unfolded and will be referred to as “undocked”. These are the states pictured in Fig. 2, c and b, respectively. We can define a reaction quotient (Kobs) that describes the relative populations of these two states for the transformation
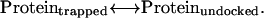 |
(2) |
Upon addition of solute, the equilibrium between trapped and undocked can be modulated, and in the osmotic pressure view, Kobs has the following dependence upon the solution osmotic pressure, π:
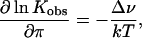 |
(3) |
where Δv is the change in hydration volume from which solute is excluded (19). It should be noted that Δν is not the hydration change associated with the conformation transition, because added solutes are unlikely to be completely excluded from the hydrated protein surface. Rather, if Δν has any physical meaning, it is likely to represent a minimum hydration change that takes place during this conformational transition (13).
Several assumptions are made in the analysis of our EPR spectra as a function of solution osmotic pressure. First, it is assumed that there are two conformational states of the Ton box in equilibrium. Second, we assume that the EPR spectra are a composite of these two states, such that changes in the EPR spectra upon solute addition reflect a shift in equilibrium of our two defined states. To determine the relative populations of these two states from the EPR spectra, one of two approaches was used. In the first approach, spectral subtraction was used to determine the relative populations and the equilibrium, Kobs, between the two states as described previously (20). This approach works well provided that the two line shapes of each individual state that comprise the composite line shape are known. In the second approach, we note that the normalized EPR amplitude, A, will be given by 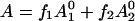 , where f1 and f2 represent the fractions of protein in states 1 and 2, respectively, and
, where f1 and f2 represent the fractions of protein in states 1 and 2, respectively, and 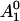 and
and 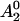 represent the normalized amplitudes of the EPR spectra that arise from protein solely in state 1 or 2, respectively. In this case a plot of A as a function of the osmotic pressure π is expected to have the following behavior:
represent the normalized amplitudes of the EPR spectra that arise from protein solely in state 1 or 2, respectively. In this case a plot of A as a function of the osmotic pressure π is expected to have the following behavior:
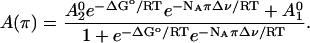 |
(4) |
Here, ΔGo is the standard free energy change for the process in Eq. 2 and is independent of π. NA is Avogadro's number, and R and T have their usual meanings. The term NAπΔν is an energy term that accounts for the work done to exclude solute from the volume Δν. From a fit of the data to Eq. 4, values of Kobs at each point may be determined, where Kobs is defined as 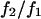 . Both approaches gave similar results for the data presented in Table 1, and were plotted as lnKobs versus π, according to Eq. 3.
. Both approaches gave similar results for the data presented in Table 1, and were plotted as lnKobs versus π, according to Eq. 3.
TABLE 1.
Effective solute-excluded hydration changes measured for a conformational transition in the Ton box at site V10R1 in BtuB
| Solute | Δv (Å3)* | No. waters |
|---|---|---|
| PEG 6000 | 9,200 | 307 |
| PEG 3350 | 7,800 | 260 |
| PEG 2000 | 7,300 | 240 |
| PEG 1000 | 5,600 | 190 |
| PEG 400 | 2,700 | 90 |
| Glycine Betaine | 780 | 26 |
| TMAO | 1,900 | 63 |
| LiCl | 4800 | 160 |
Values are the result of 1–4 measurements and have an estimated error of ±30% based upon error in the spectral subtraction or fit of the data to Eq. 4. Δv was divided by the volume of water molecule (∼30 Å3) to obtain number of waters.
RESULTS
Solutes modulate the equilibrium of the Ton box in BtuB
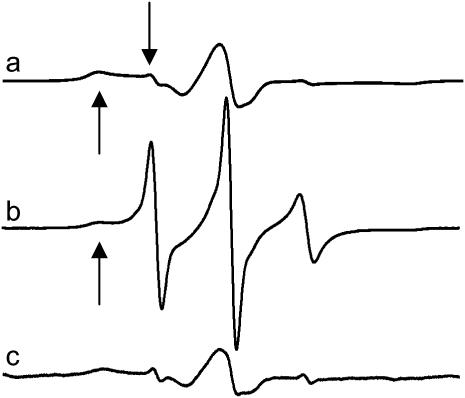
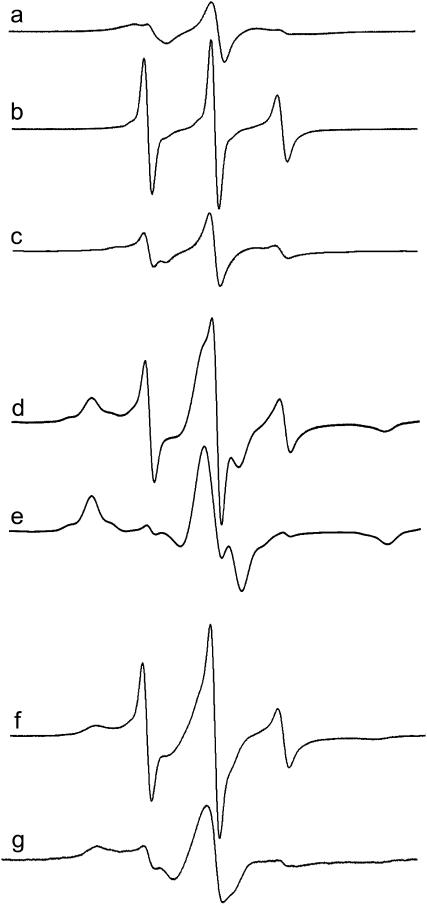
Shown in Fig. 3 are EPR spectra of the spin-labeled side chain at position 10 within the Ton box of BtuB (V10R1) reconstituted into POPC bilayers under three sets of conditions: a), in the absence and b), presence of substrate (vitamin B12), and c) in the presence of substrate, but in the solution used to produce protein crystals. As shown previously, the EPR line shapes in the absence of substrate (Fig. 3 a) arise from a nitroxide side chain that is in tertiary contact and buried within the protein interior (21). In addition, EPR spectra and the accessibilities of nitroxides along this segment are consistent with the Ton box being folded into the hatch region of the protein. In the presence of substrate, the Ton box undergoes a transition to a disordered state, and the spectrum in Fig. 3 b is obtained. We refer to this disordered state as an undocked or unfolded state because side-chain tertiary contacts are lost along the entire length of the Ton box and the aqueous exposures of the side chains are enhanced (3,21). Careful examination of the spectrum in Fig. 3 b indicates that it is a composite of two line shapes, which arise from an equilibrium between the docked and undocked states that are in slow exchange on the EPR timescale (8). The broad component in Fig. 3 is assigned to the docked conformation, whereas the narrower component (both indicated by arrows) arises from the unfolded state.
FIGURE 3.
Normalized X-band EPR spectra of purified reconstituted BtuB labeled in the Ton box at position 10 (V10R1): (a) in POPC without substrate, and (b) in POPC with substrate (vitamin B12) added. The addition of substrate induces an unfolding in the Ton box. (c) In the soaking solution used for protein crystallization with substrate added. Each spectrum is a composite arising from a highly mobile R1 population (downward-facing arrow), corresponding to the unfolded state and a motionally restricted R1 population, corresponding to the folded or docked state (upward-facing arrow). Spectra are 100 gauss scans.
As shown previously (8), if the BtuB samples are prepared using the soaking buffer used to produce crystals, this transition is effectively blocked and the EPR spectrum shown in Fig. 3 c is obtained. Similar spectra for V10R1 are obtained when the sample solution contains high concentrations of PEG 3350, a major solute in the crystallization buffer (8). The EPR line shape with solute addition does not result from changes in solution viscosity, as a 15% Ficoll 400 solution (a 400,000 MWT polymer) produces no significant change in the EPR spectrum of V10R1. (Note osmotic pressure is a colligative property. With Ficoll it is possible to achieve the same viscosity of a sucrose, glycerol, or PEG solution, albeit at a lower molality.)
The EPR spectrum in Fig. 3 c closely resembles that obtained for the unliganded (apo) state (Fig. 3 a), indicating that the Ton box is now folded; however, the spectrum in Fig. 3 c has a slightly different line shape from that of the apo form (as do other labels along the Ton box), indicative of a different conformational state. This result is reasonable because the x-ray structures show the Ton box in different, although still folded, conformations in the absence and presence of substrate. The folded state that is obtained in the presence of ligand and high solute concentrations will be referred to as a trapped conformational state, and the EPR data in Fig. 3, a–c, are consistent with the structures shown in Fig. 1, a–c, respectively.
Osmotic stress alters the equilibrium of the N-terminal Ton box
Solutes, such as PEGs, sucrose, and ions and natural osmolytes, such as TMAO and glycine betaine are all found to modulate the conformational equilibrium of the BtuB Ton box. Each of these solutes appears to modulate the Ton box equilibrium by altering the activity of water or the osmotic pressure of the solution.
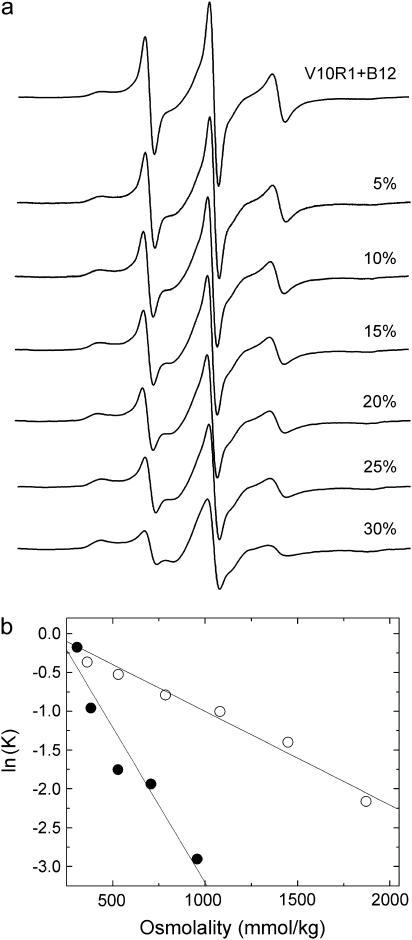
Shown in Fig. 4 a are plots of the EPR spectra of V10R1 with the addition of PEG 400 in the presence of substrate. The decrease in amplitude and increase in EPR line width upon addition of this solute results from an equilibrium shift from the unfolded to trapped form. Each spectrum can be analyzed in terms of two spectra resulting from the fraction of trapped (folded) versus unfolded Ton box, and the equilibrium constant Kobs is then calculated from the ratio of these two populations at each solute concentration. Fig. 4 b shows plots for the response of equilibrium populations as a function of osmotic pressure for two different osmolytes, PEG 400 and PEG 3350. As indicated above (Methods), we define the observed equilibrium constant as Kobs = [unfolded]/[trapped]. The different slopes between these two curves are not unexpected and arise from differences in the manner in which the two solutes are excluded from the protein surface in the two conformational states of the Ton box. For PEGs and all the solutes explored here, a roughly linear dependence for free energy of this conformational transition upon π (osmotic pressure) is observed.
FIGURE 4.
Series of EPR spectra from V10R1 in the presence of substrate with the addition of 0% to 30% w/v PEG 400 to a buffer solution of 10 mM HEPES, 130 mM NaCl, pH 6.5. The decrease in amplitude reflects the conversion of the Ton box from an undocked to a docked (or trapped state). Also shown are plots of ln(K) versus osmolality obtained for V10R1 when either PEG 3350 (•) or PEG 400 (○) is used as the solute. The different slopes reflect different thermodynamic hydration volume changes probed by these solutes (see Table 1).
Table 1 summarizes the results for several different solutes and lists the effective hydration volume changes from which solutes are excluded, Δv, during the unfolding of the Ton box. As indicated above, the values of Δv cannot be interpreted as the hydration changes associated with the conformational transition but represent the effective volume change occupied by water from which each particular solute is excluded during the conformational transition. Glycine betaine was examined because it has been reported to be highly excluded from protein surfaces and to provide the best estimate for the minimum hydration volume of a protein surface (13). However, when TMAO is compared to glycine betaine, a much larger effective volume change is measured for TMAO. Based upon a comparison of solvent exclusion of TMAO versus glycine betaine seen in BSA (13), we would have expected a much larger effect for the glycine betaine. We do not fully understand this result; however, it could reflect differences in the proteins being studied and different interactions that are made with the protein surfaces by these solutes.
The changes in hydration volume are in approximate agreement with a structural model
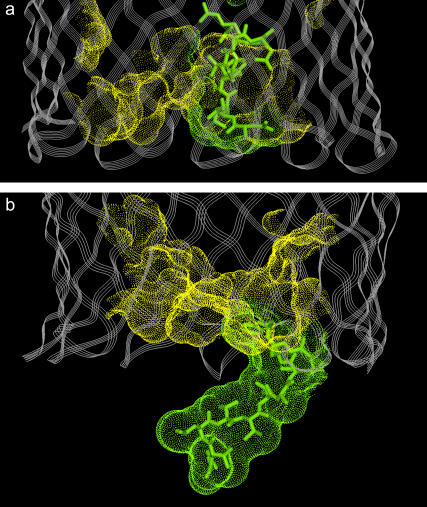
As discussed above, SDSL suggests that the Ton box of BtuB is unstructured and may extend toward the periplasmic space upon the addition of substrate (see Fig. 2 b) (3,21). This conformational transition should change the periplasmic surface of BtuB in two ways. First, the Ton box should now be extended into the solution and be fully accessible to solvent. Second, this structural change should leave behind a water-filled cavity or hydrated surface within the barrel. As discussed above, both these structural features may make this Ton box structural transition sensitive to solution osmolality. Shown in Fig. 5 are solvent-exposed surfaces on the periplasmic side of BtuB for the ligand-bound crystal structure (Fig. 5 a) and a model predicted based upon SDSL (Fig. 5 b). In Fig. 5 a, the Ton box is folded into the BtuB barrel but projects outward toward the periplasm in Fig. 5 b.
FIGURE 5.
Renderings of the solvent-exposed surface (Connolly surface) on the periplasmic end of BtuB for the hatch region (residues 6–135) of BtuB. Surface area for residues 6–15 is shown in green; surface area for residues 16–135 is shown in yellow. (a) The BtuB crystal structure in the Ca2+- and B12-bound state (Protein Data Bank ID 1NQH), and (b) a model of BtuB in the Ca2+- and substrate-bound state where the conformation of the Ton box is consistent with the results of SDSL (see Fig. 1 b) and unfolded from residues 6–15.
Our hydration/cavity model was tested in two ways. First, we examined the effect of a number of different sized PEGs on the Ton box equilibrium. In previous work, the gating of ion channels or activity of enzymes was shown to be modulated differently by different sized PEGs (19,22,23). In these cases, the dependence of Δv upon PEG molecular weight could be interpreted in terms of the size of a channel pore or cavity in the protein. As shown in Table 1, PEGs of 2000 MWT and above have large and roughly consistent values of Δv. PEGs that are 1000 MWT and smaller exhibit smaller values of Δv, and PEG 400 has an excluded hydrated volume that is 2–3 times smaller than that of the larger PEGs. This is consistent with the idea that solute exclusion is due in part to steric exclusion of the larger polymers from a hydrated protein surface (22), perhaps a cavity or channel in the core of the barrel.
In a second test of this model, the change in the hydrated surface and hydrated volume that takes place during the substrate-induced unfolding of the Ton box was estimated by comparing the models in Fig. 5, a and b (Fig. 1, c and b). An estimate of the change in hydrated surface area was made by measuring the change in surface that is accessible to a sphere of radius 1.4 Å (approximating the size of a water molecule) between the two models. This yielded an area change of ∼1,500 Å2. We then made a rough estimate of the volume change associated with solute exclusion by assuming solute exclusion from a 4 Å layer around this surface (24). This yielded a value of 6,000 Å3. Although there are many factors that may influence the size of Δv, this number is roughly the same magnitude as the changes reported in Table 1, suggesting that size of Δv is consistent with the structural change depicted in Fig. 2.
The Ton box is unfolded by certain mutations but may be folded by solutes
Certain mutations within the Ton box result in a disordered unfolded conformation of the Ton box in the unliganded state (3,6). Here, we tested the ability of solutions of high osmolality to induce a more ordered conformation of the Ton box when its “structure” is perturbed by proline mutations. Shown in Fig. 6 a is an EPR spectrum of T7R1 reconstituted into POPC. This spectrum results from an R1 side chain that is in tertiary contact and is consistent with the position of residue T7 in the crystal structure of BtuB. As shown previously, mutations in the Ton box that produce a transport-defective phenotype, such as V10P, appear to disorder the Ton box (6,10), and previous spin-label scanning along the Ton box in the presence of this mutation indicated that there was a loss of tertiary contacts along this segment, suggesting that the Ton box was unfolded even in the absence of substrate. The spectrum shown in Fig. 6 b arises from a double mutant that includes T7R1 where the mutation V10P has been included in the Ton box. When this same sample is placed within the crystallization soaking buffer, the spectrum in Fig. 6 c is observed. This spectrum resembles that obtained for T7R1 alone and indicates that solutes used in the crystallization solution will produce a more ordered and compact Ton box structure in the presence of the V10P mutant.
FIGURE 6.
When the Ton box is disturbed, it may be refolded by solutes. EPR spectrum of (a) T7R1 in POPC, (b) T7R1/V10P in POPC, and (c) T7R1/V10P in the crystallization soaking buffer. EPR spectra for N13R1 in (d) POPC and (e) the crystallization soaking buffer. EPR spectra of V10R1 in (f) POPC/OG (1:17) mixed micelles and (g) POPC/OG mixed micelles plus 30% PEG 3350. Spectra are 100 gauss scans.
We observed previously that substitution of the R1 side chain at certain positions destabilizes the folded conformation of the Ton box, thus increasing the fraction of the unfolded form. For example, the substitution of phenylalanine or R1 for threonine at position 11 unfolds the Ton box (3). In fact, the spectra for sites 13 and 14 appear to be a composite of mobile and immobilized spectra corresponding to the unfolded and folded conformations. This behavior is expected. The nitroxide-labeled side chain R1 is readily accommodated into protein structures and is no more perturbing than many single amino acid substitutions; however, it may destabilize a protein fold when placed within the core of a protein by as much as 3–5 kcal/mole (16). Because the folded state of the Ton box is unlikely to be stabilized by an energy that exceeds 3–5 kcal/mole (20), substitutions that are buried with the interior of the hatch of BtuB may destabilize the folded state.
Shown in Fig. 6 d is an EPR spectrum from N13R1. This spectrum arises from two motional components of the R1 label that appear to arise from a folded state and an unfolded state of the N-terminus. The line shape for the narrower component matches the line shape that is obtained at position 13 in the presence of substrate (3). The fraction of unfolded protein in the unliganded spectrum is not large and represents ∼13% of the total protein in the sample. We speculate that the increased fraction of unfolded protein in the absence of substrate is due to the unfavorable energetics of the N13R1 substitution. Fig. 6 e shows the effect that the crystallization soaking solution has upon the EPR spectrum of N13R1. This solution eliminates the narrower spectral component that is assigned to the unfolded form of the Ton box. Thus, it would appear that when the Ton box equilibrium is perturbed by placing R1 at position 13, the equilibrium can be shifted back to the folded form by solutions of high osmolality.
The Ton box is unfolded by detergents but refolded by high solute concentrations
In addition to being unfolded by mutations and substitutions, the Ton box in BtuB can be unfolded without substrate in mixed micelles of OG (9) as well as a number of other nonionic and ionic detergents (D. Ho and D. Cafiso, unpublished results). Fig. 6 f shows an EPR spectrum of V10R1 when reconstituted into mixed micelles of POPC/OG (1:17), and it resembles the spectrum seen for V10R1 when BtuB is bound to substrate (Fig. 3 b). When solutes, such as PEG 3350, are added back to this micellar system, the EPR spectrum (Fig. 6 g) broadens and resembles the spectrum seen in the docked or trapped state. The data indicate that solutes that are used in the crystallization of BtuB and other TonB-dependent transporters have the capacity to refold the Ton box when it is unfolded by detergents.
DISCUSSION
In this work, we show that a range of solutes alter the equilibrium between a folded (trapped) conformation of the Ton box and an unfolded, undocked form. Solutes drive the Ton box to this folded (less hydrated) state by lowering the activity of water; as a result, the energy difference between these two protein conformations is directly related to the solution osmolality. We show that for a series of PEGs, the effect of solution osmolality drops dramatically below a molecular weight of ∼2000, indicating that a water-filled cavity or pore is formed within the barrel when the Ton box unfolds. In addition, we show that when the folded form of the Ton box is destabilized by mutations or detergents, it may be stabilized or refolded by the addition of solutes such as PEGs. It should be noted that the ordering induced by solutes when the Ton box is disordered by detergents or mutations may not represent the same folded state. Indeed previous work from our lab has shown that the disordered state in the Ton box induced by detergents represents a different conformation than that produced by substrate addition (9).
The results presented here are consistent with a large body of work indicating that solutes can affect the folding, stability, and conformational equilibrium of proteins. For example, solutes such as sugars and polyols have been shown to modulate conformational transitions in ion channels, enzymes, and hemoglobin (19,22,23,25). Naturally occurring osmolytes such as TMAO have been observed to increase the thermal denaturation temperature of proteins, force the folding of unstructured proteins, and refold proteins that have been unfolded by destabilizing mutations (26–30). In this work, we demonstrate that the equilibrium of a specific conformational transition can be monitored in an active transport protein by SDSL and that this conformational equilibrium is modified by a wide range of solutes through an osmotic mechanism. Furthermore, the magnitude of the osmotic effect is consistent with a structural model that has been proposed for this transition.
In general, water activity will alter the energy of a protein because protein surfaces are differentially solvated by solute and solvent. This may occur because solute is excluded from a hydrated cavity, such as an open ion channel, or because solvent is excluded from an exposed hydrated protein surface (14,15). In the case of BtuB, both mechanisms are likely to be important. The unfolding of the Ton box leads to an increase in solvent-exposed protein surface due to the hydration of the Ton box but also due to the creation of a solvated surface or cavity within the protein β-barrel (see Fig. 5). The data shown in Table 1 are consistent with this conclusion. Smaller molecular weight solutes such as TMAO, glycine betaine, and PEG 400 all give rise to smaller values of Δv. Larger PEGs produce the largest values of Δv, revealing the presence of a cavity that selectively excludes polymers of larger size. The complete unfolding of the Ton box in the liganded form of BtuB is blocked in the presence of high solute concentrations because the unfolded, more hydrated state is no longer favored in the presence of high osmolyte concentrations.
At this time, we do not know how general or specific the effects of solutes are on membrane proteins; however, it is reasonable to assume that any conformational change that involves modest energy differences and significant changes in hydration will be sensitive to the effects of solute exclusion. We have observed that solutes induce an ordering and compaction of the extracellular binding loops of other outer membrane transporters (M. Kim, Q. Xu, and D. Cafiso, unpublished), and a general prediction that can be made from these results is that membrane proteins will likely be in a more compact and less dynamic state in high solute concentrations.
In summary, the work presented here indicates that a range of solutes modulate the structure of the outer membrane transport protein BtuB; in particular, solutes have the capacity to inhibit a conformational transition that takes place upon substrate binding. The data are consistent with a structural model for the unfolding of the Ton box proposed based upon SDSL, where solutes are excluded from the protein surface and from a cavity within the protein. When the structure of the Ton box is destabilized by detergents or by mutations, it may be refolded by the addition of solutes. In general, this work suggests that conformational transitions in membrane proteins that involve modest energy differences and significant changes in hydration may be blocked or modified by high solute concentrations.
Acknowledgments
This work was supported by National Institutes of Health grant GM35215 (to D.S.C.).
References
- 1.Faraldo-Gómez, J. D., and M. S. P. Sansom. 2003. Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 4:105–116. [DOI] [PubMed] [Google Scholar]
- 2.Postle, K., and R. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869–882. [DOI] [PubMed] [Google Scholar]
- 3.Fanucci, G. E., K. A. Coggshall, N. Cadieux, M. Kim, R. J. Kadner, and D. S. Cafiso. 2003. Substrate-induced conformational changes of the periplasmic N-terminus of an outer-membrane transporter by site-directed spin labeling. Biochemistry. 42:1391–1400. [DOI] [PubMed] [Google Scholar]
- 4.Cadieux, N., P. G. Phan, D. S. Cafiso, and R. J. Kadner. 2003. Differential substrate-induced signaling through the TonB-dependent transporter BtuB. Proc. Natl. Acad. Sci. USA. 100:10688–10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadieux, N., and R. J. Kadner. 1999. Site-directed disulfide bonding reveals an interaction site between energy coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. USA. 96:10673–10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coggshall, K. A., N. Cadieux, C. Piedmont, R. Kadner, and D. S. Cafiso. 2001. Transport-defective mutations alter the conformation of the energy-coupling motif of an outer membrane transporter. Biochemistry. 40:13946–13971. [DOI] [PubMed] [Google Scholar]
- 7.Chimento, D. P., A. K. Mohanty, R. J. Kadner, and M. C. Wiener. 2003. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Biol. 10:394–401. [DOI] [PubMed] [Google Scholar]
- 8.Fanucci, G. E., J. Y. Lee, and D. S. Cafiso. 2003. Spectroscopic evidence that osmolytes used in crystallization buffers inhibit a conformation change in a membrane protein. Biochemistry. 42:13106–13112. [DOI] [PubMed] [Google Scholar]
- 9.Fanucci, G. E., J. Y. Lee, and D. S. Cafiso. 2003. Membrane mimetic environments alter the conformation of the outer membrane protein BtuB. J. Am. Chem. Soc. 125:13932–13933. [DOI] [PubMed] [Google Scholar]
- 10.Cadieux, N., C. Bradbeer, and R. Kadner. 2000. Sequence changes in the Ton box region of BtuB affect its transport activities and the interaction with TonB protein. J. Bacteriol. 182:5954–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fanucci, G. E., N. Cadieux, C. A. Piedmont, R. J. Kadner, and D. S. Cafiso. 2002. Structure and dynamics of the β-barrel of the membrane transporter BtuB by site-directed spin labeling. Biochemistry. 41:11543–11551. [DOI] [PubMed] [Google Scholar]
- 12.Chimento, D. P., A. K. Mohanty, R. J. Kadner, and M. C. Wiener. 2003. Crystallization and initial x-ray diffraction of BtuB, the integral membrane cobalamin transporter of Escherichia coli. Acta Crystallogr. D59:509–511. [DOI] [PubMed] [Google Scholar]
- 13.Courtenay, E. S., M. W. Capp, C. F. Anderson, and M. T. Record Jr. 2000. Vapor pressure osmometry studies of osmolyte-protein interactions: implications for the action of osmoprotectants in vivo and for the interpretation of “osmotic stress” experiments in vitro. Biochemistry. 39:4455–4471. [DOI] [PubMed] [Google Scholar]
- 14.Parsegian, V. A., R. P. Rand, and D. C. Rau. 2000. Osmotic stress, crowding, preferential hydration, and binding. A comparison of perspectives. Proc. Natl. Acad. Sci. USA. 97:3987–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timasheff, S. N. 2002. Protein hydration, thermodynamic binding, and preferential hydration. Biochemistry. 41:13473–13482. [DOI] [PubMed] [Google Scholar]
- 16.Mchaourab, H. S., M. A. Lietzow, K. Hideg, and W. L. Hubbell. 1996. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 35:7692–7704. [DOI] [PubMed] [Google Scholar]
- 17.Columbus, L., and W. L. Hubbell. 2004. Mapping backbone dynamics in solution with site-directed spin labeling: GCN4–58 bZip free and bound to DNA. Biochemistry. 43:7273–7287. [DOI] [PubMed] [Google Scholar]
- 18.Hubbell, W. L., D. S. Cafiso, and C. A. Altenbach. 2000. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 7:735–739. [DOI] [PubMed] [Google Scholar]
- 19.Vodyanoy, I., S. M. Bezrukov, and V. A. Parsegian. 1993. Probing alamethicin channels with water-soluble polymers. Size-modulated osmotic action. Biophys. J. 65:2097–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fanucci, G. E., N. Cadieux, R. Kadner, and D. S. Cafiso. 2003. Competing ligands stabilize alternate conformations of the energy coupling motif of a TonB-dependent outer membrane transporter. Proc. Natl. Acad. Sci. USA. 100:11382–11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merianos, H. J., N. Cadieux, C. H. Lin, R. Kadner, and D. S. Cafiso. 2000. Substrate-induced exposure of an energy-coupling motif of a membrane transporter. Nat. Struct. Biol. 7:205–209. [DOI] [PubMed] [Google Scholar]
- 22.Zimmerberg, J., F. Bezanilla, and V. A. Parsegian. 1990. Solute inaccessible aqueous volume changes during opening of the potassium channel of the squid giant axon. Biophys. J. 57:1049–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reid, C., and R. P. Rand. 1997. Probing protein hydration and conformational states in solution. Biophys. J. 72:1022–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhat, R., and S. N. Timasheff. 1992. Steric exclusion is the principal source of the preferential hydration of proteins in the presence of polyethylene glycols. Protein Sci. 1:1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colombo, M. F., D. C. Rau, and V. A. Parsegian. 1992. Protein solvation in allosteric regulation: a water effect on hemoglobin. Science. 256:1335–1336. [DOI] [PubMed] [Google Scholar]
- 26.Bolen, D. W. 2004. Effects of naturally occurring osmolytes on protein stability and solubility: issues important in protein crystallization. Methods. 34:312–322. [DOI] [PubMed] [Google Scholar]
- 27.Mello, C. C., and D. Barrick. 2003. Measuring the stability of partly folded proteins using TMAO. Protein Sci. 12:1522–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baskakov, I., A. Wang, and D. W. Bolen. 1998. Trimethylamine-N-oxide counteracts urea effects on rabbit muscle lactate dehydrogenase function: a test of the counteraction hypothesis. Biophys. J. 74:2666–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baskakov, I., and D. W. Bolen. 1998. Forcing thermodynamically unfolded proteins to fold. J. Biol. Chem. 273:4831–4834. [DOI] [PubMed] [Google Scholar]
- 30.Qu, Y., C. L. Bolen, and D. W. Bolen. 1998. Osmolyte-driven contraction of a random coil protein. Proc. Natl. Acad. Sci. USA. 95:9268–9273. [DOI] [PMC free article] [PubMed] [Google Scholar]