Abstract
The amplification of RNA viruses such as poliovirus is associated with high error rates, and the resulting diversity likely facilitates viral survival within an infected host. However, within individual tissues of infected hosts, there may be barriers to viral spread that limit genome sampling. We tested whether poliovirus population diversity was maintained during viral spread to the brain of poliovirus receptor-expressing mice. Each of four restriction enzyme site-tagged viruses was shown to be able to replicate in the mouse brain. However, when infection was initiated by i.m., i.v., or i.p. routes, only a subset of the members of the injected pool was detectable in the brain. This jackpot effect was the result of a bottleneck in viral transit from the inoculation site to the brain. The bottleneck was difficult to overcome, requiring a 107 increase in viral inoculum to allow representation of all or most members of the infecting pool. Therefore, the bottleneck is not likely to be a physical barrier but an antiviral state induced by a founder virus. We suggest that the innate immune response can limit viral pathogenicity by limiting the number and therefore the diversity of viruses during spread to vulnerable tissues.
Keywords: pathogenesis, virus transmission, population genetics, neurovirulence, innate immunity
Poliovirus causes paralytic poliomyelitis in humans. During poliovirus epidemics of the past, this very infectious enteric virus spread to nearly every nonimmune individual in the population but caused neurovirulent disease in only 1% of the people infected (1–3). The susceptibility of individuals to paralytic disease was correlated to varying extents with age, gender, fatigue, infecting viral strain, and the presence of recent or subsequent injuries (1–3). Why some individuals became paralyzed and some did not increased the uncertainty and terror associated with these epidemics.
Mice transgenic for the human poliovirus receptor CD155 (PVR mice) (4–6) develop symptoms similar to human poliomyelitis after i.m., i.v., or i.p. viral inoculations, although, unlike humans, they are not susceptible to enteric infection. Depending on the inoculation site, poliovirus traffics to the CNS by means of different routes (reviewed in ref. 7). When inoculated into the leg muscle (i.m.), poliovirus travels through neurons to the brain (3, 8, 9). When inoculated into the tail vein (i.v.), poliovirus travels through the blood, breaching the blood–brain barrier to gain access to the CNS (10). The routes by which i.p.-inoculated poliovirus reaches the CNS are not yet known.
RNA viruses display the highest replicative error rates in nature. With misincorporation frequencies of 10−3 to 10−5 per nucleotide per cycle, it is thought that each virus in the population differs from every other virus by at least one mutation (11–13). High replicative error may be important for the survival of the viral population (11, 14); a mutant virus with increased fidelity is attenuated in mice (15, 16).
Although high replicative error rates increase genetic diversity, other factors, such as selective pressure within the infected host, the dominance of defective genomes (17–21), and the presence of bottlenecks to viral spread, may serve to reduce quasispecies complexity. Bottlenecks restrict the numbers of genomes that pass through them, resulting in the enrichment of individual pool members in the replicative niches downstream of the bottleneck, and thus resulting in jackpot effects. Experimentally applied bottlenecks such as plaque-to-plaque passaging limit viral quasispecies diversity and viral fitness (22–29). Limitations of genetic diversity during microbial infection have been observed: e.g., for cucumber and tobacco mosaic viruses during spread within infected plants (30, 31) and for strains of Salmonella (32, 33) and Pneumocystis carnii (34) during spread in rodents. Jackpot or bottleneck effects must be common in bacterial pathogens, because they are often noted as an obstacle to population sampling after signature-tagged mutagenesis (35–38). Restricted, possibly stochastic spread of poliovirus has been observed in both humans and PVR mice by using mixtures of viruses whose relative fitnesses were either different or unknown (15, 39, 40). We report here the existence of a bottleneck during the spread of poliovirus in PVR mice by using an artificial quasispecies of four restriction site-tagged viruses of equivalent fitness. A bottleneck of surprising strength was identified that limits the genetic diversity of poliovirus transmitted to the murine brain.
Results
Assay for Quasispecies Maintenance.
In a previous study, we found an apparent barrier to poliovirus spread in the mouse between the injection site in the leg muscle and the brain (15). Upon competition between a restriction site-tagged mutant virus and WT virus after i.m. infection, only one of the two viruses was usually present in the brain, with WT virus “winning” most of the time. These results suggested that a host barrier that we term a bottleneck exists between the inoculation site and the brain. Careful examination of this bottleneck was limited, because there were only two detectable members of the quasispecies, and one of the members was attenuated (15).
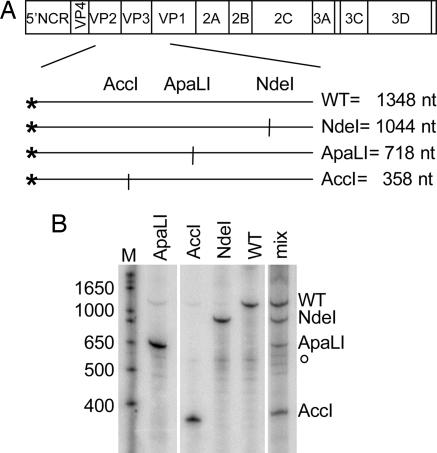
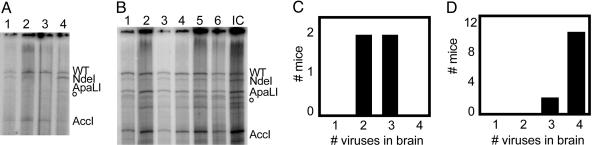
Here, monitored viral diversity within a population while minimizing fitness differences between members of the population (30). As shown in Fig. 1A, we used a mixture of four different polioviruses whose genomes could be distinguished after amplification of a particular region of the viral genome. Upon reverse transcription of the individual or pooled RNA genomes, a 1,348-bp DNA fragment was amplified by PCR and end-labeled by incorporating 32P into only one of the PCR primers. Then, digestion of the labeled DNA fragment yielded diagnostic products for the WT genome and the genomes that contained engineered NdeI, ApaLI, and AccI restriction sites. The restriction sites were introduced into the capsid coding region by means of silent mutagenesis to minimize potential fitness differences between the members of the pool, and, upon propagation of virus stocks, no growth differences were noted (see Materials and Methods). Additionally, replication kinetics of each of the viruses were indistinguishable in single-cycle growth curves (Fig. 6, which is published as supporting information on the PNAS web site). When HeLa cells were infected with single virus stocks, the expected labeled bands were observed upon electrophoresis of the NdeI-, ApaLI-, and AccI-digested, end-labeled products (Fig. 1B). When HeLa cells were infected with the quasispecies mixture, all four bands were observed and were well represented in the population (Fig. 1B). Therefore, neither a single cycle of growth in HeLa cells nor the manipulations used to amplify and quantify the resulting RNAs limited the observed quasispecies.
Fig. 1.
Assay for quasispecies integrity. (A) Genomic location of engineered restriction site polymorphisms used to generate artificial viral quasispecies. Restriction enzyme sites were created in the capsid-coding regions of the individually marked genomes by using silent mutagenesis. RT-PCR amplification of the identified sequences within the capsid region generated an end-labeled 1,348-bp DNA product; asterisks indicate the position of the radiolabeled primer. The sizes of the virus-specific labeled fragments generated upon digestion with NdeI, ApaLI, and AccI restriction enzymes are indicated. (B) Labeled products of RT-PCR restriction digestion from viral infections of HeLa cells. The four viruses, either singly or mixed, were used to infect HeLa cells at multiplicities of infection (mois) of 5 pfu per cell each; the total moi of the mixed infection was therefore 20 pfu/ml. At 4.5 h after infection, cell-associated RNA was harvested, amplified, and labeled by RT-PCR. Products of NdeI, ApaLI, and AccI digestion were displayed on a denaturing polyacrylamide gel. The sizes of selected bands in the marker lane (M) are indicated. o, location of a prevalent background band.
To assess quasispecies maintenance during murine infection, we inoculated 6- to 10-week-old mice by i.m. injection in the leg with a total of 2 × 107 plaque-forming units (pfu) of an equal mixture of all four viruses or with individual viruses. This dose of virus is 2- to 5-fold above the LD50 for these mice (ref. 4 and J.K.P., unpublished data). Animals were euthanized at the first sign of paralysis of the inoculated limb, usually 3 or 4 days after infection. All animal work was performed in compliance with protocols approved by the Stanford University Administrative Panel on Laboratory Animal Care. Total RNA from brain tissue was recovered, viral RNA was amplified by radioactive RT-PCR, and NdeI/ApaLI/AccI digestion products were displayed by gel electrophoresis. All four products were readily detected in the muscle tissue of the inoculated leg (Fig. 7, which is published as supporting information on the PNAS web site). Furthermore, when inoculated independently, all four viruses were shown to be capable of spreading to the brain, where the introduced mutation was maintained (data not shown).
Bottleneck Between Peripheral Tissues and the Brain upon Murine Infection by Various Routes.
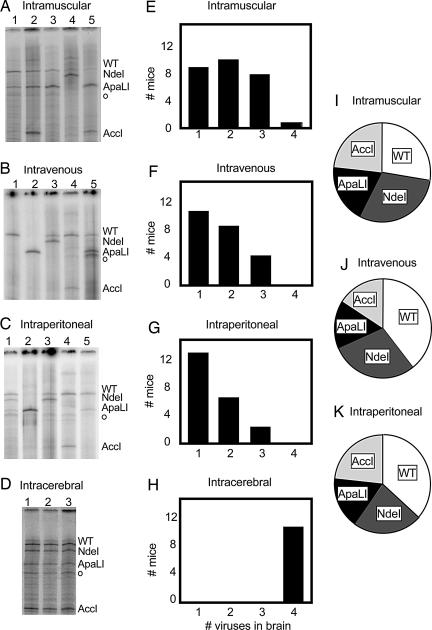
To determine whether the quasispecies complexity of the pooled viruses was preserved or altered during spread from the muscle to the brain, quasispecies in the brain were analyzed after i.m. inoculation. As shown in a representative gel in Fig. 2A, for any given mouse, only a subset of the members of the pool was usually detectable in the brain after i.m. inoculation. With approximately the same frequency, one (e.g., lane 4), two (e.g., lane 3), or three (e.g., lane 2) members of the quasispecies pool were detectable in any individual mouse (Fig. 2E). In only one of 27 mice, in fact, were all four viruses found in the brain at this virus dose. However, each of the four viruses in the artificial quasispecies had a roughly equivalent probability of being represented in the brain of an infected mouse.
Fig. 2.
Quasispecies diversity in the mouse brain after infection by various routes. (A–D) Gel electrophoretic patterns of labeled, digested RT-PCR products from poliovirus RNA genomes recovered from the brains of individual mice after i.m. (A), i.v. (B), i.p. (C), and intracerebral (D) injection of mixtures of viruses. Individual mice were infected with total inocula of 2 × 107 pfu for i.m. (n = 27), 8 × 107 pfu for i.v. (n = 22), 2 × 108 pfu for i.p. (n = 21), and 2 × 107 pfu for intracerebral (n = 10) infections. (E–H) For each route of infection, the numbers of mice that showed, after becoming ill, the presence of one, two, three, and four members of the quasispecies in the brain are indicated. (I–K) Pie graphs for the prevalence of each of the four viruses present in the brain samples for i.m., i.v., and i.p. inoculated animals, respectively.
Other inoculation sites were tested to determine whether this apparent bottleneck also existed for alternate routes of infection. Mice were injected i.v. with a total of 8 × 107 pfu of the virus mixture in the tail vein, an amount of virus 2- to 4-fold above the LD50 (4). Upon symptoms of disease (3–7 days), brain RNA was harvested, and the RT-PCR digestion assay was performed. As shown in Fig. 2 B, F, and J, a strong bottleneck was observed between the blood and the brain, although each of the viruses was capable of trafficking to the brain (Fig. 2J). Similarly, when brain samples were prepared from mice that were inoculated i.p. with 2 × 108 pfu of virus, 2-fold above the LD50 for this route (4), even more severe quasispecies restriction was observed (Fig. 2 C, G, and K). Although it is not yet known whether i.p.-inoculated virus traffics to the brain by means of a neuronal or blood route, animals injected in this way appear to display the strongest bottleneck. However, for all three routes of brain infection from peripheral tissues, all four viruses were capable of spreading to the brain, but the quasispecies restriction characteristic of a bottleneck was observed.
To ensure that the observed quasispecies restriction did not result from the method of tissue processing or the inability of all four viruses to replicate in the brain at the same time, we inoculated 2-week-old mice intracerebrally with 2 × 107 pfu of virus and processed the samples after symptoms appeared. In this case, products for all four viruses were readily detectable in the brain (Fig. 2 D and H), demonstrating that all four viruses can replicate simultaneously and be detected in the brain when inoculated directly.
Analysis of the Bottleneck in Mice with Altered Quasispecies Pools.
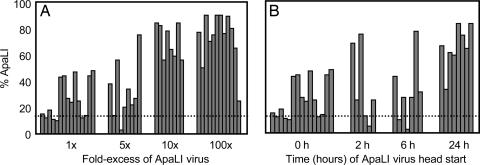
To explore the nature of the observed bottleneck, we attempted to define inoculation conditions that would overcome the quasispecies restriction. First, we inquired whether increasing the proportion of a single member of the quasispecies would increase its probability of spreading to the brain. Equivalent total inocula (2 × 107 pfu per mouse) of different virus mixtures that contained either an equivalent amount of the ApaLI variant or that contained five-fold, 10-fold, or 100-fold excesses of the ApaLI virus were injected i.m. Brain tissues were processed, the labeled RT-PCR digestion assay was performed, and the percentage of the ApaLI product signal in each lane was determined. As shown in Fig. 3A, mice inoculated with an equal mixture of the four viruses (1×) showed, as in Fig. 2A, sporadic recovery of the ApaLI virus, with only 8 of the 15 mice tested displaying an ApaLI genomic signal over the background of the assay. However, when the inocula contained an increasing proportion of the ApaLI virus, an increase in the representation of the ApaLI product in infected brains was observed. Interestingly, even at 100-fold excess of ApaLI virus, the ApaLI product was not always predominant: 1 of the 12 inoculated mice showed very little ApaLI product in its infected brain (Fig. 3A). This finding is more consistent with a mechanism that restricts the initial access of viruses to the brain than one in which the quasispecies is restricted after many viruses have gained access to brain tissue.
Fig. 3.
Analysis of the bottleneck in mice with altered quasispecies pools. (A) The effect of increasing the representation of one member of the inoculated quasispecies (ApaLI) on transmission to the brain. Mice were injected i.m. with a total of 2 × 107 pfu containing various fold-excess (5×, 10×, and 100×) of the ApaLI virus. Mice from 1× samples received equal amounts of all four viruses. Brain samples were processed, labeled RT-PCR products were digested, and the products were displayed by gel electrophoresis. The percentage of total product represented in the ApaLI-specific band is plotted for each individual mouse; the detection limit is indicated by a dotted line. (B) The effect of preinoculation with ApaLI virus. Mice were injected i.m. either with the pooled quasispecies (0 h) or first with the ApaLI virus and then with the remaining three viruses 2, 6, or 24 h later. A total of 2 × 107 pfu virus, 5 × 106 pfu for each pool member, was inoculated into each animal. Samples were processed as in A. The dotted line indicates the limit of detection in this assay.
We also tested whether the bottleneck for an individual virus could be overcome by giving that virus a temporal “head start.” First, either 2 × 107 pfu per mouse of the quasispecies or 5 × 106 pfu per mouse of the ApaLI virus alone was delivered by i.m. injection. Then, at various later times (2, 6, and 24 h), the remaining WT, NdeI, and AccI viruses were injected into the same tissue. As shown in Fig. 3B, the representation of the ApaLI product remained sporadic even when the ApaLI virus was given a head start of up to 6 h. However, when the ApaLI virus was given a 24-h head start, the ApaLI product predominated, even though other viruses could also be represented. Therefore, although the bottleneck for an individual virus to reach the brain can be overcome by increasing its prevalence in the population, either by dose (Fig. 3A) or 24-h head start (Fig. 3B), these benefits are not always sufficient to ensure that the advantaged virus the sole winner. This finding is consistent with a bottleneck that allows passage of small numbers of viruses that are randomly sampled from the population.
The Bottleneck Can Be Overcome by High Dose in Young Mice.
If the observed bottleneck were a barrier that restricted viral passage, it is likely that it could be overcome by large increases in the inoculum. When the inoculum of mixed viruses delivered i.m. into 6- to 8-week-old mice was increased to 1 × 109 pfu per mouse, a dose ≈1,000-fold above the LD50, an average of three viruses of the pool was present in the brain, suggesting that the bottleneck was not completely overcome (data not shown).
To increase the concentration range of inoculum that could be tested, we infected 2-week-old mice with the artificial quasispecies mixture. Two-week-old mice are extremely susceptible to poliovirus, displaying an LD50 of ≈100 pfu per mouse (ref. 4 and J.K.P., unpublished data). When very young mice were infected with 1 × 106 pfu per mouse, an amount 10,000-fold above the LD50, quasispecies restriction of viruses obtained from the brains was observed (Fig. 4A and C). A similar result was obtained with an input of 1 × 103 pfu per mouse (data not shown). However, when very young mice were injected with 1 × 109 pfu per mouse, the bottleneck was largely overcome, with all four viruses being represented in the brains of most of the animals (Fig. 4 B and D). These results argue that the bottleneck is difficult to overcome, requiring a dose of virus >10,000-fold above the LD50 in very young mice.
Fig. 4.
Analysis of the bottleneck in young mice inoculated with high doses of virus. (A and B) Two-week-old mice were injected i.m. with a total of 1 × 106 pfu (A) or 1 × 109 pfu (B) of the four quasispecies viruses. Brain samples were processed, and the digested, labeled RT-PCR products were displayed by gel electrophoresis. The o marks a background band. (C and D) The results from all mice are summarized, as in Fig. 2 E–H, in C (1 × 106 pfu) and D (1 × 109 pfu).
Discussion
Here, we describe a bottleneck to viral spread in mice, documented by using an artificial quasispecies. We found that only a subset of peripherally inoculated viruses was detectable in the brain, even though each of the viruses was capable of spreading and replicating in the brain (Fig. 2). Increasing the prevalence of a particular member of the quasispecies in the population, or giving a member a temporal head start, increased its probability of being sampled through the bottleneck but did not ensure its representation (Fig. 3). These results suggest that breaching the bottleneck, either through the sciatic nerve from infected muscle or through the blood–brain barrier from the bloodstream, is a stochastic event. This finding is consistent with previously observed quasispecies restriction within populations of polioviruses of varying fitness during passage in both mice and humans (39, 40). The poliovirus bottleneck is strong and could only be overcome by using hypersusceptible young mice at extremely high doses of virus.
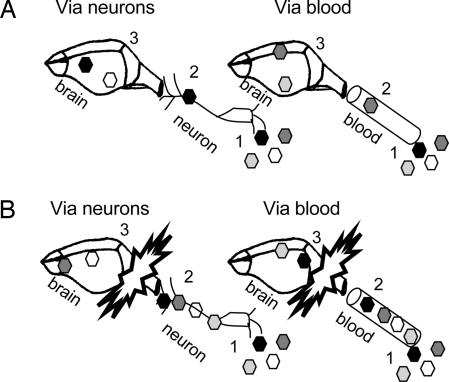
Two classes of mechanism (Fig. 5) could explain the observed bottleneck from infected peripheral tissues to the brain. A “tough-transit model” (Fig. 5A) proposes that trafficking within the infected animal is difficult and dangerous for viruses, with each particle having a very low probability of successfully trafficking through the sciatic nerve to the CNS and brain, in the case of i.m. inoculation, or past the blood–brain barrier, in the case of i.v. inoculation. Once in the CNS, the winning virus or viruses act as founders, reestablishing a population with initially limited diversity. In this model, an increase in virus in the periphery of 100-fold, for example, should have a noticeable impact on the number of founder viruses. A “burned-bridge” model (Fig. 5B) proposes that it is not that difficult physically for the first viruses to reach the gateway to the brain but that the first few viruses to enter very quickly trigger an antiviral state that limits the successful entry and spread of later viruses. Given that the bottleneck is observed within hours of infection and is found even in 2-week-old mice, it is much more likely to be part of the host innate immune response than any aspect of acquired immunity. We are currently testing whether the absence of the IFN α/β receptor from the PVR mice (43) will eliminate the bottleneck.
Fig. 5.
Possible mechanisms for bottleneck. Different members of viral quasispecies are depicted as different shades of hexagons. In the tough- transit model (A), a physical barrier prevents viral trafficking, either through neurons or through the blood–brain barrier for i.m. and i.v. routes of inoculation, respectively. In each case, one or a few viruses breach the physical barrier and act as founders in the brain. In the burned-bridge model (B), the first virus or viruses to reach the brain, by means of neurons or the bloodstream, trigger a rapid antiviral response that prevents the entry or amplification of later viruses.
During natural infections with poliovirus, a predominantly enteric virus, the efficiency of viral growth and fecal-oral spread is likely to be dictated by enteric growth, not growth in the CNS or brain. Therefore, a bottleneck between peripheral tissues and the brain should have little genetic consequence for viral transmission. However, random sampling of only small numbers of viruses by the brain and CNS could drastically reduce the frequency of neurovirulent disease that results even from robust enteric infection and could serve to explain the stochastic nature of poliomyelitis in poliovirus-infected populations. The ability of induced antimicrobial states to limit quasispecies diversity by means of bottlenecks is likely to add a stochastic element to microbial virulence in individuals.
Materials and Methods
Plasmid Construction.
The restriction site-tagged virus plasmids, called NdeI, ApaLI, and AccI, were made by silent site-directed mutagenesis of the viral cDNA clone (41). Sense primers for NdeI, ApaLI, and AccI were AAACTGTTGGTGTCATATGCGCCTCCTGGAG, CCAGTCACCGTGTGCACTGCCTGAATTTGATG, and AGGTTCTGCCCGGTCGACTACCTCCTTGGAAAT, respectively (nucleotide changes are shown in italics). For each plasmid, the entire PCR-generated region was confirmed by sequencing (Sequetech, Mountain View, CA). WT poliovirus was the fourth pool member.
Cells, Mice, and Viruses.
HeLa cells were propagated in DMEM supplemented with 10% calf serum. PVR mice expressing the human poliovirus receptor (CD155) from the β-actin promoter (4) were a kind gift from R. Andino (University of California, San Francisco). Mahoney serotype 1 poliovirus stocks were grown from single plaques generated by transfection of the viral cDNA clone (41). Care was taken to ensure that specific infectivities of all viruses were similar. High-titer stocks were obtained by infecting HeLa cells with individual plaque stocks at 37°C, and stocks were titered as described in ref. 42.
Tissue Culture Infections.
Each of the WT, NdeI, ApaLI, and AccI virus stocks alone or in combination was used to infect 1 × 106 HeLa cells at a multiplicity of infection of 5 pfu per cell for each virus (Fig. 1). At 4.5 h after infection, RNA was harvested with TRIzol (Invitrogen) according to the manufacturer’s instructions.
Mouse Infections and Tissue Processing.
For i.m. injections, 50 μl of virus stock was injected into the right hind leg of 2- to 10-week-old PVR mice with a 28-gauge needle. For i.p. injections, 50–100 μl of virus stock was injected into the peritoneal cavity. For i.v. injections, the tail was warmed with a heat pack and swabbed with ethanol, and 50–100 μl of virus stock was inoculated into the tail vein. For intracerebral inoculations, 2-week-old animals were anesthetized (15), and 20 μl of virus stock was injected into the cerebrum through the right temple. In all cases, mice were observed for symptoms of disease twice daily and were euthanized at the first sign of disease. Brain tissue was stored at −80°C until being ground into a fine powder under liquid nitrogen with a mortar and pestle. RNA was extracted from 200–400 mg of total tissue by using TRIzol.
RT-PCR Digestion Assay.
Superscript II (Invitrogen) reverse transcription reactions, performed according to the supplied protocol, contained 5–10 μg of RNA and reverse primer GAATTCTAACCCCTGTGCTAGCGCTT for tissue culture experiments or a nested primer, ATGCTTTCAAGCATCTGACCTAACC, for mouse experiments. In all cases, 20% of the reverse transcription reaction was PCR-amplified by using Taq (Qiagen, Valencia, CA) with sense primer GAATTCCCAGACGTCGCTGCATGC and antisense primer GAATTCTAACCCCTGTGCTAGCGCTT. The sense primer was end-labeled by using T4 polynucleotide kinase (NEB, Beverly, MA) and was used at a 1:5 ratio with the unlabeled sense primer (15). PCR products were digested with NdeI, ApaLI, and AccI (NEB) at 37°C, ethanol-precipitated, and run on denaturing polyacrylamide gels as described in ref. 15.
Supplementary Material
Acknowledgments
We thank Peter Sarnow for helpful comments on the manuscript and Shane Crotty and Raul Andino for the provision of PVR mice. This work was supported by National Institutes of Health Grant AI-48756. J.K.P. is a Rebecca Ridley Kry Fellow of the Damon Runyon Cancer Research Foundation.
Abbreviation
- pfu
plaque-forming unit.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Modlin J. F. In: Human Enterovirus Infections. Rotbart H. A., editor. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 195–220. [Google Scholar]
- 2.Pallansch M. A., Roos R. P. In: Virology. Fields B. N., Knipe D. M., Howley P. M., Chanock R. M., Monath T. P., Melnick J. L., Roizman B., Straus S. E., editors. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 723–775. [Google Scholar]
- 3.Gromeier M., Wimmer E. J. Virol. 1998;72:5056–5060. doi: 10.1128/jvi.72.6.5056-5060.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crotty S., Hix L., Sigal L. J., Andino R. J. Gen. Virol. 2002;83:1707–1720. doi: 10.1099/0022-1317-83-7-1707. [DOI] [PubMed] [Google Scholar]
- 5.Ren R. B., Costantini F., Gorgacz E. J., Lee J. J., Racaniello V. R. Cell. 1990;63:353–362. doi: 10.1016/0092-8674(90)90168-e. [DOI] [PubMed] [Google Scholar]
- 6.Koike S., Taya C., Aoki J., Matsuda Y., Ise I., Takeda H., Matsuzaki T., Amanuma H., Yonekawa H., Nomoto A. Arch. Virol. 1994;139:351–363. doi: 10.1007/BF01310797. [DOI] [PubMed] [Google Scholar]
- 7.Ohka S., Nomoto A. Trends Microbiol. 2001;9:501–506. doi: 10.1016/s0966-842x(01)02200-4. [DOI] [PubMed] [Google Scholar]
- 8.Ren R., Racaniello V. R. J. Infect. Dis. 1992;166:747–752. doi: 10.1093/infdis/166.4.747. [DOI] [PubMed] [Google Scholar]
- 9.Ohka S., Yang W. X., Terada E., Iwasaki K., Nomoto A. Virology. 1998;250:67–75. doi: 10.1006/viro.1998.9360. [DOI] [PubMed] [Google Scholar]
- 10.Yang W. X., Terasaki T., Shiroki K., Ohka S., Aoki J., Tanabe S., Nomura T., Terada E., Sugiyama Y., Nomoto A. Virology. 1997;229:421–428. doi: 10.1006/viro.1997.8450. [DOI] [PubMed] [Google Scholar]
- 11.Domingo E., Holland J. J. Annu. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 12.Drake J. W., Charlesworth B., Charlesworth D., Crow J. F. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crotty S., Maag D., Arnold J. J., Zhong W., Lau J. Y., Hong Z., Andino R., Cameron C. E. Nat. Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 14.Domingo E., Menendez-Arias L., Holland J. J. Rev. Med. Virol. 1997;7:87–96. doi: 10.1002/(sici)1099-1654(199707)7:2<87::aid-rmv188>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 15.Pfeiffer J. K., Kirkegaard K. PLoS Pathog. 2005;1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vignuzzi M., Stone J. K., Arnold J. J., Cameron C. E., Andino R. Nature. 2005;439:344–347. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitaker-Dowling P., Youngner J. S. Microbiol. Rev. 1987;51:179–191. doi: 10.1128/mr.51.2.179-191.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charpentier N., Davila M., Domingo E., Escarmis C. Virology. 1996;223:10–18. doi: 10.1006/viro.1996.0450. [DOI] [PubMed] [Google Scholar]
- 19.Crowder S., Kirkegaard K. Nat. Genet. 2005;37:701–709. doi: 10.1038/ng1583. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez-Lopez C., Arias A., Pariente N., Gomez-Mariano G., Domingo E. J. Virol. 2004;78:3319–3324. doi: 10.1128/JVI.78.7.3319-3324.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grande-Perez A., Lazaro E., Lowenstein P., Domingo E., Manrubia S. C. Proc. Natl. Acad. Sci. USA. 2005;102:4448–4452. doi: 10.1073/pnas.0408871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novella I. S., Quer J., Domingo E., Holland J. J. J. Virol. 1999;73:1668–1671. doi: 10.1128/jvi.73.2.1668-1671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manrubia S. C., Escarmis C., Domingo E., Lazaro E. Gene. 2005;347:273–282. doi: 10.1016/j.gene.2004.12.033. [DOI] [PubMed] [Google Scholar]
- 24.Bergstrom C. T., McElhany P., Real L. A. Proc. Natl. Acad. Sci. USA. 1999;96:5095–5100. doi: 10.1073/pnas.96.9.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke D. K., Duarte E. A., Moya A., Elena S. F., Domingo E., Holland J. J. Virol. 1993;67:222–228. doi: 10.1128/jvi.67.1.222-228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazaro E., Escarmis C., Perez-Mercader J., Manrubia S. C., Domingo E. Proc. Natl. Acad. Sci. USA. 2003;100:10830–10835. doi: 10.1073/pnas.1332668100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duarte E., Clarke D., Moya A., Domingo E., Holland J. Proc. Natl. Acad. Sci. USA. 1992;89:6015–6019. doi: 10.1073/pnas.89.13.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duarte E. A., Clarke D. K., Moya A., Elena S. F., Domingo E., Holland J. J. Virol. 1993;67:3620–3623. doi: 10.1128/jvi.67.6.3620-3623.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novella I. S., Ebendick-Corpus B. E. J. Mol. Biol. 2004;342:1423–1430. doi: 10.1016/j.jmb.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Li H., Roossinck M. J. J. Virol. 2004;78:10582–10587. doi: 10.1128/JVI.78.19.10582-10587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider W. L., Roossinck M. J. J. Virol. 2001;75:6566–6571. doi: 10.1128/JVI.75.14.6566-6571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meynell G. G., Stocker B. A. J. Gen. Microbiol. 1957;16:38–58. doi: 10.1099/00221287-16-1-38. [DOI] [PubMed] [Google Scholar]
- 33.Meynell G. G. J. Gen. Microbiol. 1957;16:396–404. doi: 10.1099/00221287-16-2-396. [DOI] [PubMed] [Google Scholar]
- 34.Keely S. P., Cushion M. T., Stringer J. R. Infect. Immun. 2003;71:47–60. doi: 10.1128/IAI.71.1.47-60.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mecsas J., Bilis I., Falkow S. Infect. Immun. 2001;69:2779–2787. doi: 10.1128/IAI.67.5.2779-2787.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiang S. L., Mekalanos J. J. Mol. Microbiol. 1998;27:797–805. doi: 10.1046/j.1365-2958.1998.00726.x. [DOI] [PubMed] [Google Scholar]
- 37.Hava D. L., Camilli A. Mol. Microbiol. 2002;45:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- 38.Darwin A. J., Miller V. L. Mol. Microbiol. 1999;32:51–62. doi: 10.1046/j.1365-2958.1999.01324.x. [DOI] [PubMed] [Google Scholar]
- 39.Georgescu M. M., Balanant J., Ozden S., Crainic R. J. Gen. Virol. 1997;78:1819–1828. doi: 10.1099/0022-1317-78-8-1819. Part 8. [DOI] [PubMed] [Google Scholar]
- 40.Hovi T., Lindholm N., Savolainen C., Stenvik M., Burns C. J. Gen. Virol. 2004;85:369–377. doi: 10.1099/vir.0.19518-0. [DOI] [PubMed] [Google Scholar]
- 41.Racaniello V. R., Baltimore D. Proc. Natl. Acad. Sci. USA. 1981;78:4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfeiffer J. K., Kirkegaard K. Proc. Natl. Acad. Sci. USA. 2003;100:7289–7294. doi: 10.1073/pnas.1232294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ida-Hosonuma M., Iwasaki T., Yoshikawa T., Nagata N., Sato Y., Sata T., Yoneyama M., Fujita T., Taya C., Yonekawa H., Koike S. J. Virol. 2005;79:4460–4469. doi: 10.1128/JVI.79.7.4460-4469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.