Abstract
Ubiquitin-mediated proteolysis plays a key role in many pathways inside the cell and is particularly important in regulating cell cycle transitions. SCF (Skp1/Cul1/F-box protein) complexes are modular ubiquitin ligases whose specificity is determined by a substrate-binding F-box protein. Dia2 is a Saccharomyces cerevisiae F-box protein previously described to play a role in invasive growth and pheromone response pathways. We find that deletion of DIA2 renders cells cold-sensitive and subject to defects in cell cycle progression, including premature S-phase entry. Consistent with a role in regulating DNA replication, the Dia2 protein binds replication origins. Furthermore, the dia2 mutant accumulates DNA damage in both S and G2/M phases of the cell cycle. These defects are likely a result of the absence of SCFDia2 activity, as a Dia2 ΔF-box mutant shows similar phenotypes. Interestingly, prolonging G1-phase in dia2 cells prevents the accumulation of DNA damage in S-phase. We propose that Dia2 is an origin-binding protein that plays a role in regulating DNA replication.
INTRODUCTION
The duplication of a single cell requires multiple complex regulatory networks controlled by cyclin/CDK (cyclin-dependent kinase) complexes responsible for cell cycle transitions. It is clear that the ubiquitin-proteasome system is also important for the regulation of cell cycle progression, most notably in regulating the degradation of cyclins and cyclin-dependent kinase inhibitors (reviewed in Deshaies, 1999; Koepp et al., 1999). In the ubiquitin-proteasome pathway, substrate proteins are covalently attached to the small polypeptide ubiquitin in an enzymatic cascade. The addition of long chains of ubiquitin polymers, linked using lysine 48 of each ubiquitin monomer, to substrate proteins leads to destruction of the substrate protein via the 26S proteasome (reviewed in Pickart, 1997). The primary means of specificity in the ubiquitination reaction is accomplished by the E3 ubiquitin ligase, as this enzyme is responsible for recruiting substrate proteins (Ciechanover et al., 1982; Hershko et al., 1983).
Members of the highly conserved SCF (Skp1/Cdc53/F-box protein) ubiquitin ligase family have been shown to control cell proliferation by regulating the ubiquitin-mediated proteolysis of key cell cycle regulators (Mathias et al., 1996; Willems et al., 1996; Feldman et al., 1997; Skowyra et al., 1997, 1999; Carrano et al., 1999; Tsvetkov et al., 1999; Strohmaier et al., 2001; Koepp et al., 2001). SCF complexes are modular ubiquitin ligases whose specificity is determined by a substrate-binding F-box protein (Feldman et al., 1997; Skowyra et al., 1997). The 40–50 amino acid F-box motif is required for binding to Skp1. Thus, mutation of the F-box motif renders F-box proteins incapable of targeting their substrate proteins for ubiquitination (Bai et al., 1996). Many F-box proteins have been identified in both humans and model eukaryotic systems, suggesting that SCF pathways reflect a highly conserved mechanism for controlling protein function. In Saccharomyces cerevisiae, at least 17 F-box proteins have been identified, but many of these proteins have not been well characterized (Patton et al., 1998; Deshaies, 1999).
The F-box protein Dia2 was initially described as a mutant that exhibits a switch from apical to bipolar budding pattern and is hyperinvasive (Palecek et al., 2000). Dia2 has recently been proposed to target a transcription factor in the pheromone-response pathway for ubiquitination (Bao et al., 2004), although this conclusion is controversial (Chou et al., 2004). Other evidence suggests that Dia2 plays a role in cell cycle progression. The dia2 deletion mutant accumulates large-budded cells, indicative of a block in mitotic progression, and has been found to be synthetically lethal with a pds1 mutant, which exhibits sister chromatid segregation defects at anaphase (Sarin et al., 2004). In addition, Dia2 is related to the Schizosaccharomyces pombe F-box protein Pof3, which has been shown to be important for maintaining genomic stability (Katayama et al., 2002).
To maintain genomic integrity, the timing of S-phase initiation is linked with growth control mechanisms in the G1-phase of the cell cycle. In budding yeast, the S-phase cyclins Clb5 and Clb6 are required for proper entry into S-phase from G1 (Schwob and Nasmyth, 1993). The control of the G1 to S phase transition is particularly important, as genes involved in this pathway are often mutated in tumor cells. DNA replication initiates at replication origins, called autonomously replicating sequences (ARSs) in S. cerevisiae. Activation, or firing, of origins is precisely controlled to ensure that each chromosome is only replicated once per cell cycle (reviewed in Bell and Dutta, 2002). Here we present evidence that Dia2 has a role in cell cycle regulation by demonstrating that Dia2 binds replication origins and is involved in regulating the timing of the G1 to S-phase transition of the cell cycle.
MATERIALS AND METHODS
Media and Cell Culture
Yeast were maintained and cultured according to standard methods (Rose et al., 1990). Yeast strains used in this study are listed in Table 1.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| Y80 (wild type) | MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Bai et al. (1996) |
| DKY194 (dia2) | As Y80 but dia2Δ::kanMX | Koepp et al. (2001) |
| Y552 (skp1-11) | As Y80 but skp1-11 | Bai et al. (1996) |
| YMT741 (cdc53-1) | MATa cdc53-1 can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 | M. Tyers |
| DKY408 (DIA2-18×myc) | As Y80 but DIA2::DIA2-18×myc-HIS3 | This study |
| W2297-4A (RAD52-YFP) | MATa trp1-1 bar1::LEU2 leu2-3,112 can1-100 ura3-1 his3-11,15 RAD52-YFP | Lisby et al. (2001) |
| W2381-10B (mec1 sml1 RAD52-YFP) | As W2297-4A but mec1Δ::TRP1 sml1Δ::HIS3 | Lisby et al. (2001) |
| DKY443 (dia2 RAD52-YFP) | MATa RAD52-YFP dia2Δ::kanMX ura3-1 his3-11,15 trp1-1 ade2-1 leu2-3,112 | This study |
| DKY399 (clb5 clb6) | As Y80 but clb5Δ::HIS3 clb6Δ::LEU2 | This study |
| DKY444 (clb5 clb6 RAD52-YFP) | MATa RAD52-YFP clb5Δ::HIS3 clb6Δ::LEU2 ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 | This study |
| DKY403 (dia2 clb5 clb6) | As Y80 but dia2Δ::kanMX clb5Δ::HIS3 clb6Δ::LEU2 | This study |
| DKY445 (dia2 clb5 clb6 RAD52-YFP) | MATa RAD52-YFP dia2Δ::kanMX clb5Δ::HIS3 clb6Δ::LEU2 ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 | This study |
| DKY405 (dia2 mrc1) | As Y80 but dia2Δ::kanMX mrc1Δ::HIS3 | This study |
| DKY404 (dia2 chk1) | As Y80 but dia2Δ::kanMX chk1Δ::HIS3 | This study |
| DKY448 (dia2 rad9) | As Y80 but dia2Δ::kanMX rad9Δ::HIS3 | This study |
| DKY449 (dia2 rad17) | As Y80 but dia2Δ::kanMX rad17Δ::kanMX | This study |
| DKY450 (dia2 rad24) | As Y80 but dia2Δ::kanMX rad24Δ::kanMX | This study |
| JBY257 (cdc13-1) | As Y80 but cdc13-1 | Sanchez et al. (1999) |
| DKY396 (dia2 cdc13-1) | As Y80 but dia2Δ::kanMX cdc13-1 | This study |
Strain Construction
CLB5 and CLB6 deletion strains were constructed by standard PCR replacement method using the primers DK133, DK134, DK142, DK143. Double and triple mutant strains were generated using standard S. cerevisiae genetics (Rose et al., 1990). To generate the 18×myc-tagged Dia2 allele, a fragment of the DIA2 gene was amplified using the primers DK176 and DK359, cloned in frame with 18 copies of the myc epitope using XhoI and NcoI sites in plasmid pJBN130. After linearization with EcoRV, the DNA was transformed into Y80 cells and transformants were selected on media lacking histidine. Primer sequences are listed in Supplementary Table 1.
Plasmid Construction
The DIA2 gene was amplified in fragments using primers DK97, DK98, DK99, and DK100. The fragments were cloned into the BamHI and XhoI sites of pRS316 to generate pDMK465. The ΔF-box mutant was generated using a two-step PCR method with the primers DK97, DK9100, DK148, and DK149 and cloned into BamHI and XhoI sites of pRS406 to generate pDMK289. An EcoRI fragment containing the F-box deletion was swapped with the EcoRI fragment of pDMK465 to generate pDMK736. The DIA2 open reading frame was amplified using the primers DK95 and DK96 and cloned in frame with the lox site of pUNI-10 using NdeI and BamHI restriction sites to generate pDMK107. This construct was then recombined with the host vector p1210 to generate an HA3-tagged baculovirus expression construct (pDMK109) using recombinant Cre (New England Biolabs, Beverly, MA) as described by Liu et al., 1998. Primer sequences are listed in Supplementary Table 1.
Baculovirus Production and Protein Expression
Baculovirus was produced in Sf9 cells using Baculogold Baculovirus DNA (BD PharMingen, San Diego, CA) and Lipofectin reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Sf9 cells were cultured in Insect-Xpress (Cambrex, distributed by Fisher Scientific, Pittsburgh, PA) with 10% heat-inactivated fetal calf serum at 27°C. Baculovirus protein expression experiments were performed in HI-Five cells (Invitrogen). Cells were cultured in Excell 405 media (JRH Biosciences, Lenexa, KS) without serum at 27°C. Forty hours after infection, insect cells were collected and lysed in NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% Igepal, 10 mM NaF, 25 mM β-glycerophosphate plus Complete Protease inhibitor cocktail; Roche Applied Science, Indianapolis, IN). Proteins were immunoprecipitated with anti-Flag antibodies (Sigma, St. Louis, MO) or anti-Myc (9E10) antibodies (Covance Research Products, Berkeley, CA) and ProtA/G plus agarose (Santa Cruz Biotechnology, Santa Cruz, CA). Anti-HA (HA.11) antibodies (Covance Research Products) were used for immunoblotting. HRP-conjugated secondary antibodies were from Jackson ImmunoResearch, (West Grove, PA).
Immunoprecipitation of Yeast Proteins
Cells were harvested at OD600 = 1.0 and treated with 1% formaldehyde for 15 min at room temperature to cross-link proteins. Lysates were prepared as for chromatin immunoprecipitation (Ricke and Bielinsky, 2004). Proteins were immunoprecipitated with anti-Skp1 antibodies (Skowyra et al., 1997) and ProtA/G plus agarose (Santa Cruz Biotechnology). Samples were boiled for 30 min before SDS-PAGE to reverse cross-links. Anti-myc (9E10) antibodies (Covance Research Products) were used for immunoblotting. HRP-conjugated secondary antibodies were from Jackson ImmunoResearch.
Flow Cytometry
Cells were collected and fixed in 70% ethanol for at least 30 min and then resuspended in phosphate-buffered saline (PBS; 140 mM NaCl, 3 mM KCl, 5 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) + 0.02% sodium azide. To prepare for flow cytometry, fixed cells were treated with FACS buffer (200 mM Tris-HCl, 20 mM EDTA + 0.1% RNAse A) overnight at 37°C and then stained in 50 μg/ml propidium iodide in PBS. Before analysis, cells were diluted 10-fold in PBS and sonicated for 5 s at 15% efficiency using a Sonic Dismembrator (Fisher Scientific, Pittsburgh, PA). Data analysis was performed with the FlowJo v6.3.3 software and graphs generated using Deltagraph 4.0.
Chromatin Immunoprecipitation
All assays followed the procedure of Ricke and Bielinsky (2004). ARS305 and ARS603 primers are described (Aparicio et al., 1999). ATP11 primers are listed in Supplementary Table 1. Anti-Myc (9E10) antibodies were from Covance Research Products and anti-Skp1 antibodies were a gift from J. Wade Harper (Skowyra et al., 1997).
Fluorescence Microscopy
Cells were sonicated briefly as described for flow cytometry before observation. YFP signals were observed using a Zeiss Axioscop 2 microscope equipped with a Zeiss Axiocam R2 digital camera and 100× objective with DIC filter. Images were captured using Zeiss Axiovision software release 3.1 (Carl Zeiss, Thornwood, NY).
RESULTS
Dia2 Assembles with SCF Components and Is Required for Normal Growth
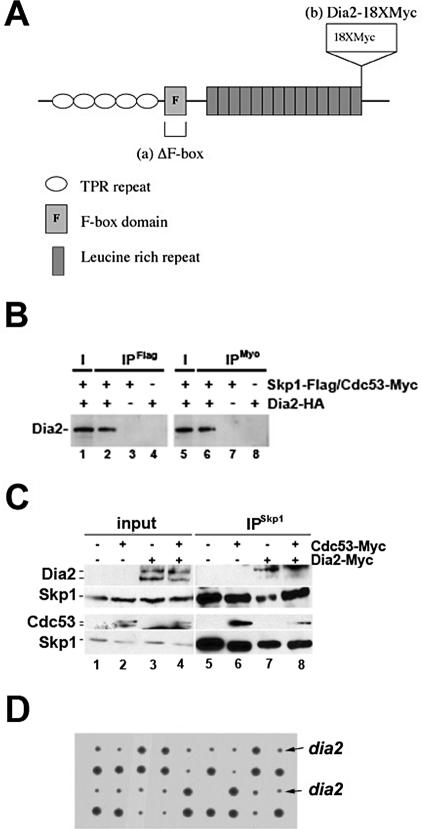
Dia2 is an F-box protein with TPR repeats in the N-terminus and leucine-rich repeats in the C-terminus (Figure 1A). It has previously been shown that recombinant Dia2 can assemble with SCF components (Kus et al., 2004) and that overexpressed Dia2 assembles with Skp1 and Cdc53 in yeast cells (Ho et al., 2002). We also observed Dia2 assembly with SCF components using an HA3-tagged Dia2 baculovirus for expression in insect cells. As shown in Figure 1B, HA3-Dia2 coimmunoprecipitates with both Flag-tagged Skp1 and Myc-tagged Cdc53 when coexpressed in insect cells. We examined whether Dia2 associates with SCF components under endogenous expression levels in yeast. To facilitate this experiment, we generated a strain that expressed a Dia2 fusion protein tagged at the C-terminus with 18 copies of the myc (9E10) epitope. The Dia2 18×myc-tagged strain complements a deletion strain (Supplementary Figure 1), indicating that the tagged protein is functional. We observe two major forms of Dia2; the larger form migrates at the predicted size for full-length Dia2. The larger form of Dia2 preferentially coprecipitates with Skp1 (Figure 1C). We also observe Cdc53 coprecipitating with Skp1 when a myc-tagged version of Cdc53 is expressed in the Dia2 18×myc-tagged strain. Consistent with what has been previously observed, we find that Cdc53 exists as two forms in yeast cells (Mathias et al., 1996; Willems et al., 1996); presumably the larger form that we observe in Figure 1C is modified by Rub1 as it preferentially interacts with Skp1 (Lammer et al., 1998). We conclude that Dia2 is a bona fide F-box protein and assembles into an SCF complex in yeast cells.
Figure 1.
Dia2 is an F-box protein required for normal growth. (A) Diagram of Dia2 domain structure. Structure adapted from (Patton et al., 1998; Katayama et al., 2002). Modified forms described in the text are shown in a and b. (B) Recombinant Dia2 assembles with SCF components. Insect cells were coinfected with HA3-tagged Dia2 baculovirus and Flag-tagged Skp1 or Myc-tagged Cdc53 baculoviruses. After 40 h, cells were harvested and lysates were subjected to immunoprecipitation with anti-Flag or anti-myc antibodies. Immunoblots were probed with anti-HA antibodies. I, input; IP, immunoprecipitate. (C) Dia2 assembles with SCF components in yeast cells. Skp1 was immunoprecipitated using anti-Skp1 antibodies (Skowyra et al., 1997) from the indicated strains. Immunoblots were probed with anti-myc and anti-Skp1 antibodies. Because of significant differences in signal intensity between Dia2 and Cdc53, two separate blots are shown. (D) The dia2 deletion exhibits a slow growth phenotype at room temperature. Colonies from tetrad dissection of DIA2/dia2 diploid induced to undergo meiosis are shown. Arrows point to representative small dia2 colonies, whereas large colonies are DIA2.
The dia2 null mutant grows slower at low temperatures. When individual spores from a dissection of a dia2 null heterozygous diploid are grown on rich media at room temperature, a pattern of 2 slow-growing to 2 fast-growing colonies is seen (Figure 1D). The slow-growing colonies are resistant to G418, indicating that they are deletions of DIA2 as the kanMX gene (Wach et al., 1994), which confers G418 resistance, was used to replace the DIA2 gene in the genome (Koepp et al., 2001). The slow growth defect of dia2 mutant is rescued by introducing a CEN plasmid carrying the DIA2 gene (see Figure 3A). Thus, Dia2 is required for normal cell growth in S. cerevisiae.
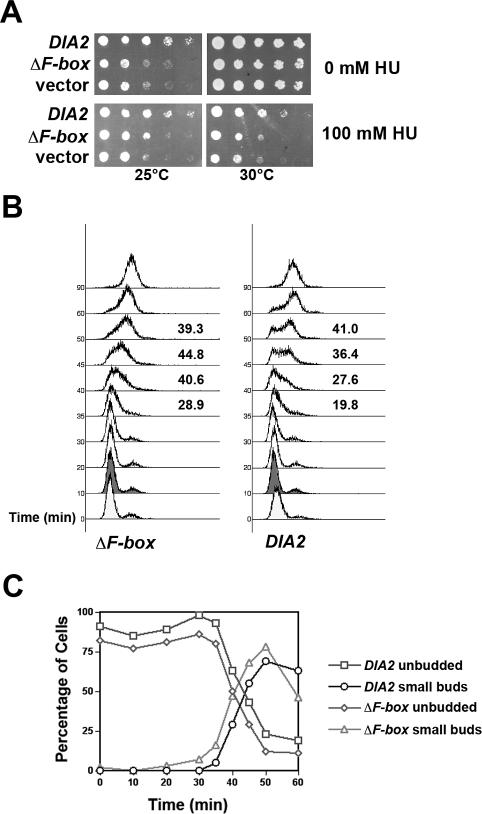
Figure 3.
The dia2 ΔF-box mutant exhibits growth and S-phase defects. (A) The ΔF-box mutant is cold- and HU-sensitive. The dia2 strain was transformed with DIA2, ΔF-box and empty vector DNA. Serial dilutions of each were then spotted on rich media containing the indicated amount of HU and incubated at either 25 or 30°C. (B) The ΔF-box mutant enters S-phase earlier than WT cells. WT and dia2 ΔF-box cells were arrested in G1 with alpha factor and then released from arrest at 25°C. Samples were taken at the indicated time points and used for flow cytometry. The percentage of S-phase cells was calculated for the 35-, 40-, 45-, and 50-min time points using FlowJo software and is shown next to the profile for each strain. (C) The dia2 ΔF-box mutants exhibit early bud emergence. Cells were treated as in B and budding morphology was assayed by light microscopy. At least 100 cells were counted per time point and the experiment was repeated three times. Representative results from a single experiment are shown. The identity of each strain and bud stage is indicated next to the graph.
dia2 Mutants Enter S-Phase Prematurely
Previous work has established a role for Dia2 in G2/M progression as a significant fraction of dia2 cells accumulate as large-budded cells with the nucleus at the bud neck (Sarin et al., 2004). However, we also observed defects that hinted at a role for DIA2 in S-phase. As shown in Figure 2A, dia2 mutants are sensitive to hydroxyurea (HU) as are skp1-11 and cdc53-1 mutants. In addition, overexpression of the S-phase cyclins Clb5 and Clb6 from a galactose-inducible promoter is toxic to dia2 cells but not wild-type (WT) cells (Supplementary Figure 2).
Figure 2.
The dia2 mutant exhibits S-phase defects. (A) SCF mutants are HU-sensitive. Serial dilutions of WT, dia2, skp1-11, and cdc53-1 strains were spotted onto rich media containing indicated amounts of HU and grown at 30°C. (B) The dia2 mutant enters S-phase earlier than WT. WT and dia2 cells were arrested in G1 with alpha factor and then released from arrest at 25°C. Samples were taken at the indicated time points and used for flow cytometry. The percentage of S-phase cells was calculated for the 30-, 40-, and 50-min time points using FlowJo software and is shown next to the profile for each strain. (C) The dia2 mutants exhibit early bud emergence. Cells were treated as in B and budding morphology was assayed by light microscopy. At least 100 cells were counted per time point and the experiment was repeated three times. Representative results from a single experiment are shown. The identity of each strain and bud stage is indicated next to the graph.
To test whether DIA2 might have role in S-phase, we synchronized dia2 and WT cells in G1 with alpha factor, released them from arrest at 25°C, and prepared samples at 10-min intervals for flow cytometry. We observed that the dia2 mutant cells entered S-phase 10 min earlier than WT cells under these conditions (Figure 2B, compare the 40- and 50-min time points). We calculated the percentage of cells in S-phase for both strains at the 30-, 40-, and 50-min time points; these numbers are shown next to the profiles in Figure 2B. Similar premature S-phase entry was observed in dia2 cells when we counted bud emergence as a measure for S-phase entry (Figure 2C).
To determine if the S-phase defects associated with the dia2 deletion mutant were a result of the absence of an SCFDia2 complex, we generated an in-frame deletion of the F-box domain in Dia2 (ΔF-box) to prevent association with SCF components. This mutant was placed under the control of the DIA2 promoter on a CEN plasmid and transformed into the dia2 deletion strain. As shown in Figure 3A, the ΔF-box mutant exhibits a growth defect and HU sensitivity similar to the dia2 deletion mutant covered by an empty vector. In addition, the dia2 cells covered by the ΔF-box mutant show an early S-phase entry phenotype when cells are synchronized in G1, released at 25°C, and then examined for DNA content and bud emergence (Figure 3, B and C). We calculated the percentage of cells in S-phase for both strains at the 35-, 40-, 45-, and 50-min time points in the samples analyzed by flow cytometry; these numbers are listed next to the profiles in Figure 3B. As shown, the defect in the ΔF-box mutant is not as dramatic as the dia2 deletion strain, but a significant fraction of the cells do reproducibly enter S-phase earlier. A smaller fraction of the dia2 cells covered by the DIA2 plasmid also enter S-phase early. We speculate that the null strain covered by the DIA2 plasmid is not entirely equivalent to WT cells; this may be because Dia2 protein levels are tightly regulated (A. C. Kile and D. M. Koepp, unpublished observations). Nevertheless, the ΔF-box mutant clearly exhibits S-phase defects. Taken together, these data indicate that Dia2 plays a role in S-phase timing and that this role is dependent on its function as a component of an SCF ubiquitin ligase.
Dia2 Binds Replication Origins
A large-scale proteomic screen determined that a number of DNA replication proteins copurified with the Dia2 protein (Ho et al., 2002). Given this observation and the links to S-phase with the dia2 mutant, we wanted to determine if the Dia2 protein might be localized to origins of replication. To test this possibility, we used chromatin immunoprecipitation to assay for Dia2 binding to both an early and a late-firing origin. To facilitate this experiment, we used the Dia2 18×myc-tagged strain.
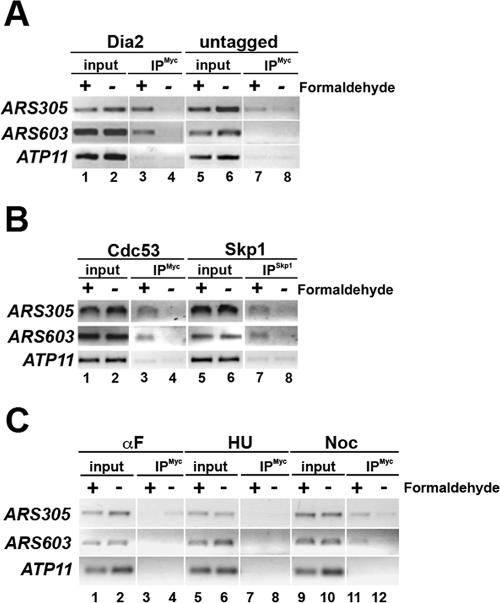
We observed that the Dia2 protein binds both ARS305 (early-firing) and ARS603 (late-firing) sequences (Aparicio et al., 1999) but not to the nonorigin DNA near ATP11, which is 20 kb away from an origin in both directions (Figure 4, lanes 1–4). This binding is dependent on the addition of the cross-linker formaldehyde and is not observed in immunoprecipitations with a strain lacking the epitope-tagged Dia2 fusion (Figure 4A, lanes 5–8). We conclude that Dia2 is an origin-binding protein, which is capable of binding both early and late-firing origins.
Figure 4.
The Dia2 protein binds replication origins. (A) Chromatin immunoprecipitation was performed on the Dia2-18×myc–tagged strain and an untagged strain. After formaldehyde cross-linking, cells were harvested and lysates prepared. Equal amounts of total protein were subjected to immunoprecipitation with anti-myc antibodies. Chromatin bound during the reaction was isolated and cross-links were reversed before PCR analysis. Primers specific to ARS305 (an early-firing origin), ARS603 (a late-firing origin), and ATP11 (a nonorigin control) were used for PCR. (B) Skp1 and Cdc53 bind origins. Chromatin immunoprecipitation was performed as in A using anti-Skp1 and anti-myc antibodies in a WT strain carrying a Cdc53-myc–tagged vector. (C) Dia2 binds replication origins in a cell cycle–dependent manner. Chromatin immunoprecipitation was performed as in A except that cells were arrested in G1-, S-, and M-phase by treatment with alpha factor, HU, and nocodazole, respectively.
To test whether other SCF components localized to origins, we performed chromatin immunoprecipitations using anti-Skp1 antibodies (Skowyra et al., 1997) and a myc-tagged version of Cdc53 (Willems et al., 1996). As shown in Figure 4B, both Skp1 and Cdc53 coprecipitated with ARS305 and ARS603 DNA in a cross-linker dependent manner. These results strongly suggest that an SCFDia2 complex is present at replication origins.
We next examined the association of Dia2 with origins throughout the cell cycle using cells that had been arrested in G1-, S- and M-phases by treatment with alpha factor, HU, and nocodazole, respectively. We observe specific association of Dia2 with both ARS305 and ARS603 only in nocodazole-arrested cells (Figure 4C), indicating that Dia2 likely binds origins after origin firing. In addition, Skp1 also associates with ARS603 in nocodazole-arrested cells (Supplementary Figure 3), consistent with the assembly of an SCF-Dia2 complex at origins at this time during the cell cycle.
dia2 Mutants Accumulate DNA Damage in S and G2/M
To better understand the role of Dia2 during the cell cycle and because Schizosaccharomyces pombe pof3 exhibits synthetic lethality with several checkpoint mutants, we tested for genetic interactions between the dia2 mutant and checkpoint mutants. We observed synthetic interactions between the dia2 deletion mutant and a number of DNA damage and DNA replication checkpoint mutants, including mec1, mrc1, chk1, rad9, rad17, rad24, pds1, and cdc20-1 (Table 2). The synthetic lethal interactions with rad9 and pds1 have been previously identified (Sarin et al., 2004). These interactions suggest that the dia2 mutant accumulates damaged DNA and requires functional checkpoint pathways for viability.
Table 2.
The dia2 mutant interacts genetically with checkpoint mutants
| dia2 Double mutants | Role | Phenotype |
|---|---|---|
| DNA damage | ||
| rad9 | Adaptor | Slower growth at 16 and 25°C |
| rad17 | Clamp loader | Slower growth at 30°C |
| rad24 | Clamp loader | Slower growth at 30°C |
| Kinases | ||
| mec1 sml1 | Transducer kinase | Inviable |
| chk1 | Transducer kinase | Slower growth at 25°C |
| DNA replication | ||
| mrc1 | Adaptor | Slower growth at 25°C, inviable at 37°C |
| Targets | ||
| pds1 | Cohesin | Inviable |
| cdc20-1 | APC activator | Inviable |
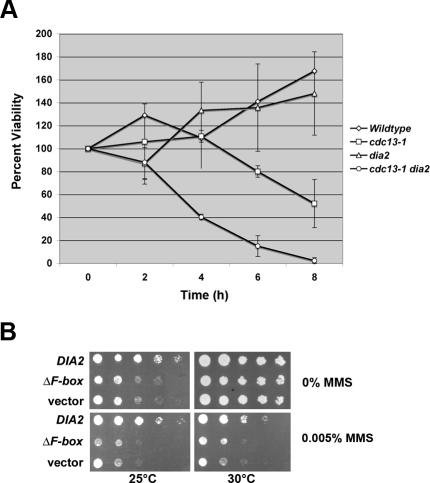
Consistent with this possibility, we found that the dia2 mutant is hypersensitive to both exogenous and endogenous DNA damage. To test the ability of dia2 cells to respond to endogenous DNA damage, we used the temperature-sensitive cdc13-1 strain, which accumulates single-stranded DNA at the nonpermissive temperature. As shown in Figure 5A, the dia2 cdc13-1 double mutant exhibits a faster loss of viability after exposure to 37°C than the cdc13-1 mutant does. We observed no significant loss of viability in either the dia2 or WT strains after incubation at 37°C. To examine the ability of dia2 cells to an exogenous DNA damaging agent, we used MMS (methyl methanesulfonate), a DNA methylating agent. As shown in Figure 5B, both the dia2 null and ΔF-box mutants are sensitive to 0.005% MMS, whereas dia2 cells expressing DIA2 are not.
Figure 5.
dia2 mutants are hypersensitive to DNA damage. (A) The dia2 cdc13-1 strain exhibits greater loss of viability than the cdc13-1 strain. The indicated strains were incubated at 37°C for the indicated times. Cells were then plated on rich media and incubated at room temperature until colonies formed. Colony counts are expressed as a percentage using the 0 time point as 100%. Error bars indicate the SD of at least three experiments. (B) dia2 mutants are sensitive to MMS. Serial dilutions of the indicated strains were spotted onto selective media containing the indicated amounts of MMS and incubated at either 25 or 30°C.
To examine the possibility that dia2 cells accumulate spontaneous DNA damage, we used a Rad52-YFP reporter strain. Rad52 is involved in recombinational repair and has been shown to localize to discrete nuclear foci when cells are treated with DNA-damaging agents (Lisby et al., 2001). We generated a dia2 Rad52-YFP strain and examined the cells using fluorescence microscopy. A deletion mutant of the checkpoint kinase Mec1 (suppressed by deletion of the ribonucleotide reductase inhibitor Sml1) was used as a positive control (Lisby et al., 2001). The dia2 mutant showed an increased number of Rad52-YFP foci over WT. Strikingly, the dia2 mutant showed nearly as many Rad52-YFP foci as the mec1 sml1 mutant. Representative examples are shown in Figure 6A.
Figure 6.
The dia2 mutant accumulates Rad52-YFP foci. (A) WT, dia2, and mec1 sml1 strains containing Rad52-YFP were grown to log phase in rich media and then examined by fluorescence and DIC (differential interference contrast) microscopy. Arrows point to examples of foci. Equal exposure times were used for all strains. Scale bar, 5 μm. (B) The dia2 mutant accumulates Rad52-YFP foci in S- and M-phases. The number of Rad52 foci and bud morphology as a measure of cell cycle stage were counted in cells treated as in A. At least 150 cells were counted per strain. Error bars indicate the SD of at least three experiments. (C) The ΔF-box mutant accumulates Rad52 foci in S- and G2/M-phases. The number of Rad52 foci and bud morphology as a measure of cell cycle stage were counted in cells treated as in A, except that the cells were grown in selective media to maintain the appropriate plasmid. At least 150 cells were counted per strain. Error bars indicate the SD of at least three experiments.
We correlated the number of cells exhibiting Rad52-YFP foci with bud morphology as a measure of cell cycle stage. Approximately 40% of both small-budded (S-phase) and large-budded (G2/M phase) dia2 cells exhibit Rad52-YFP foci, whereas only a small number are seen in unbudded (G1-phase) cells (Figure 6B). The percentage of dia2 cells showing Rad52-YFP foci was similar to the percentage of mec1 sml1 cells exhibiting foci. As expected, a small fraction of WT cells exhibit Rad52-YFP foci in S-phase, but almost none in G1 and G2/M phases (Lisby et al., 2001).
To confirm that the formation of Rad52-YFP foci resulted from a defect in an SCF-dependent role of Dia2, we repeated these experiments with the dia2 Rad52-YFP strain covered by an empty vector, a plasmid expressing the ΔF-box mutant or WT DIA2 (Figure 6C). Both the empty vector and ΔF-box samples showed a high percentage of S and G2/M cells with Rad52-YFP foci, whereas the strain covered with the DIA2 plasmid showed nearly WT levels of foci formation. From these data, we conclude that the dia2 mutant accumulates DNA damage in S and G2/M phases of the cell cycle. Furthermore, this damage is a result of the absence of SCFDia2 activity.
Delaying S-Phase Entry Alleviates S-Phase DNA Damage in dia2 Cells
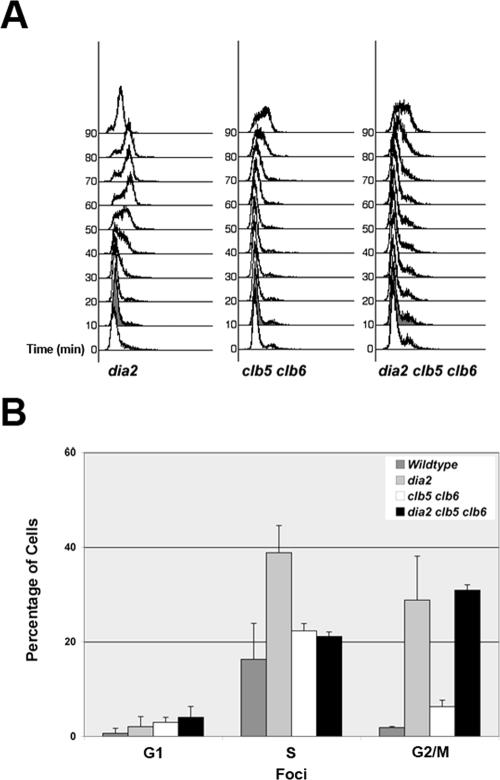
One possible explanation for the accumulation of DNA damage in dia2 cells is that it is a result of the premature entry into S-phase. Similar precocious S-phase defects linked to DNA damage and chromosomal instability have been observed in sic1 mutants as well as in cells overexpressing members of the Swi/Snf family of transcription factors (Lengronne and Schwob, 2002; Sidorova and Breeden, 2002). To address this possibility, we generated a dia2 clb5 clb6 triple deletion mutant. Deletion of CLB5 and CLB6 has been shown to prolong G1-phase (Schwob and Nasmyth, 1993), and we reasoned that the premature S-phase entry in dia2 cells might be dependent on the presence of S-phase cyclins. As predicted, the early S-phase entry from an alpha factor–induced G1 arrest in dia2 cells is abolished in the dia2 clb5 clb6 mutant (Figure 7A).
Figure 7.
Deletion of CLB5 and CLB6 alleviates the premature S-phase in dia2 cells and leads to decreased S-phase damage. (A) The early S-phase entry in dia2 is dependent on Clb5 and Clb6. dia2, clb5 clb6, and dia2 clb5 clb6 cells were arrested in G1 with alpha factor and then released from arrest at 25°C. Samples were taken at the indicated time points and used for flow cytometry. (B) The S-phase damage in dia2 cells is dependent on Clb5 and Clb6. The number of Rad52 foci and bud morphology as a measure of cell cycle stage were counted in cells treated as in Figure 6. At least 150 cells were counted per strain. Error bars indicate the SD of at least three experiments.
We then examined the formation of Rad52-YFP foci in the dia2 clb5 clb6 background. As a control, the clb5 clb6 Rad52-YFP strain showed a slightly elevated percentage of S-phase cells with foci compared with WT (Figure 7B). Strikingly, we found that deletion of CLB5 and CLB6 in the dia2 strain decreased the percentage of S-phase cells showing Rad52-YFP foci to nearly the same levels as the clb5 clb6 strain. However, the percentage of G2/M dia2 clb5 clb6 cells exhibiting foci was not significantly different from dia2 cells alone. These results suggest that prolonging G1-phase in dia2 cells can prevent most of the S-phase–specific DNA damage that occurs in these cells.
DISCUSSION
A Role for Dia2 in Regulating DNA Replication
We propose that Dia2 is an origin-binding protein that plays a role in regulating DNA replication. This is supported by the premature S-phase entry in dia2 cells as well as the rescue of S-phase DNA damage in dia2 cells by deletion of CLB5 and CLB6. The role of Dia2 in the regulation of DNA replication is most likely the result of its function as part of an SCF complex. Dia2 assembles with Skp1 and Cdc53 in yeast cells and the Dia2 ΔF-box mutant shows phenotypes similar to complete deletion of the DIA2 gene. Furthermore, Dia2, Skp1, and Cdc53 all associate with replication origins as measured by chromatin immunoprecipitation. A simple model then would be that an SCFDia2 complex targets an unknown factor that promotes DNA replication. The ubiquitination of this substrate prevents premature S-phase entry and likely leads to the substrate's degradation via the proteasome.
Because Dia2 assembles at origins of replication, one intriguing possibility is that Dia2 regulates DNA replication by facilitating the ubiquitination of another origin-binding protein. In this way, Dia2 might influence the make-up and/or activity of replication components found at origins. Our chromatin immunoprecipitation results suggest that Dia2 associates with replication origins after origin firing. Consistent with this, we observe Dia2 association with origins only after bud emergence in cells that have been released from a G0/G1 arrest (S. Swaminathan and D. M. Koepp, unpublished observations). One possibility is that Dia2 targets an origin-binding protein for ubiquitination after origins fire and in this way, helps to “reset” the origins for use in the next S-phase. In the absence of Dia2, this substrate protein promotes premature onset of S-phase in the next cell cycle. Examining in more detail how Dia2 associates with origins and determining which other proteins are found at origins at the same time will provide additional insight into the functional consequences of SCFDia2 activity.
Precocious S-phase entry has been linked to genomic instability in a number of mutants that affect the cell cycle (Sidorova and Breeden, 2003). Our results are consistent with these observations, in that the premature S-phase entry in dia2 mutants leads to at least some of the DNA damage observed in these cells. Our results predict that the pathway that leads to premature S-phase in dia2 cells is dependent on Clb5 and Clb6 activity, although whether they act upstream or downstream of the SCFDia2 substrate is unclear. A straightforward explanation for the rescue of DNA damage in dia2 cells by the clb5 clb6 mutant might be that Clb5 and Clb6 are themselves the targets of an SCFDia2 complex. However, we have not observed any changes in the protein stability of either Clb5 or Clb6 in dia2 cells (D. M. Koepp and S. J. Elledge, unpublished observations), making this scenario unlikely. Taken together, the results presented here provide an important link between SCF-mediated ubiquitination and S-phase entry that has implications for understanding genomic stability.
Other Roles for SCFDia2
There are almost certainly other pathways in addition to DNA replication in which the SCFDia2 complex acts. For example, the dia2 clb5 clb6 mutant still accumulates DNA damage foci in large-budded cells, indicating that there is a Clb5 and Clb6–independent pathway that is disrupted in dia2 cells, which also causes DNA damage. Like pof3 cells, the dia2 mutant exhibits a chromosome loss phenotype (Katayama et al., 2002; note that Fcl1 is an alternate name for Dia2). However, this phenotype is not rescued in the dia2 clb5 clb6 mutant (D. M. Koepp and S. J. Elledge, unpublished observations), further suggesting that there is also a Clb5 and Clb6–independent role for Dia2.
The dia2 mutant was initially identified because it exhibits elongated cells with a bipolar budding pattern with enhanced invasiveness (Palecek et al., 2000). It has recently been proposed that Tec1, a pheromone-responsive transcription factor involved in the filamentous growth pathway, is a substrate of SCFDia2 (Bao et al., 2004). Although it is formally possible that the role of Dia2 in the filamentation pathway is linked to the DNA replication timing defects in dia2 mutants, we think these pathways are independent as the dia2 clb5 clb6 mutant still exhibits the elongated cell/bipolar budding pattern phenotype. Furthermore, the dia2 tec1 double mutant exhibits the same growth and S-phase defects as the dia2 mutant (A. C. Kile, P. Midthun, and D. M. Koepp, unpublished observations). If Tec1 were the substrate responsible for the premature S-phase entry in dia2 cells, we would expect the dia2 tec1 strain to exhibit normal S-phase timing.
The most likely explanation for the pleiotropic phenotypes in the dia2 deletion is that the SCFDia2 complex controls the ubiquitination of more than one substrate. Many F-box proteins regulate the turnover of multiple substrates. Thus it is possible that one SCFDia2 substrate is involved in DNA replication, whereas another is important for filamentous growth. Future studies identifying SCFDia2 substrates will shed light on the interplay between these pathways. The results presented here are an essential step in providing a biological framework in which the role of any Dia2 substrate may be assessed.
Supplementary Material
Acknowledgments
We are indebted to Stephen J. Elledge for insightful discussions throughout this project. We are grateful to Stephen J. Elledge, Mike Tyers, and Rodney Rothstein for gifts of strains and plasmids. We acknowledge J. Wade Harper for gifts of antibodies and baculovirus and helpful discussions at the early stages of this work. We thank Elizabeth MacDonald and Mai Thao for technical assistance. This work was supported in part by a V Foundation for Cancer Research Scholar Award to D.M.K. V.R.R. was supported by a National Science Foundation-Research Experience for Undergraduates grant (DBI-0139450) in Molecular Biology to the University of Minnesota.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-09-0884) on January 18, 2006.
Abbreviations used: SCF, Skp1-Cdc53-F-box protein; HU, hydroxyurea; WT, wild type; YFP, yellow fluorescent protein.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Aparicio, O. M., Stout, A. M., and Bell, S. P. (1999). Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci USA 96(16), 9130–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J. W., and Elledge, S. J. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86(2), 263–274. [DOI] [PubMed] [Google Scholar]
- Bao, M. Z., Schwartz, M. A., Cantin, G. T., Yates, J. R. 3rd, and Madhani, H. D. (2004). Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in; Yeast. Cell 119(7), 991–1000. [DOI] [PubMed] [Google Scholar]
- Bell, S. P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374. Epub 2001 Nov 9. [DOI] [PubMed] [Google Scholar]
- Carrano, A. C., Eytan, E., Hershko, A., and Pagano, M. (1999). SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1(4), 193–199. [DOI] [PubMed] [Google Scholar]
- Chou, S., Huang, L., and Liu, H. (2004). Fus3-regulated Tec1 degradation through SCF(Cdc4) determines MAPK signaling specificity during mating in yeast. Cell 119(7), 981–990. [DOI] [PubMed] [Google Scholar]
- Ciechanover, A., Elias, S., Heller, H., and Hershko, A. (1982). “Covalent affinity” purification of ubiquitin-activating enzyme. J. Biol. Chem. 257(5), 2537–2542. [PubMed] [Google Scholar]
- Deshaies, R. J. (1999). SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Feldman, R. M., Correll, C. C., Kaplan, K. B., and Deshaies, R. J. (1997). A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91(2), 221–230. [DOI] [PubMed] [Google Scholar]
- Hershko, A., Heller, H., Elias, S., and Ciechanover, A. (1983). Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 258(13), 8206–8214. [PubMed] [Google Scholar]
- Ho, Y. et al. (2002). Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415(6868), 180–183. [DOI] [PubMed] [Google Scholar]
- Katayama, S., Kitamura, K., Lehmann, A., Nikaido, O., and Toda, T. (2002). Fission yeast F-box protein Pof3 is required for genome integrity and telomere function. Mol. Biol. Cell 13(1), 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp, D. M., Harper, J. W., and Elledge, S. J. (1999). How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell 97(4), 431–434. [DOI] [PubMed] [Google Scholar]
- Koepp, D. M., Schaefer, L. K., Ye, X., Keyomarsi, K., Chu, C., Harper, J. W., and Elledge, S. J. (2001). Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294(5540), 173–177. [DOI] [PubMed] [Google Scholar]
- Kus, B. M., Caldon, C. E., Andorn-Broza, R., and Edwards, A. M. (2004). Functional interaction of 13 yeast SCF complexes with a set of yeast E2 enzymes in vitro. Proteins 54(3), 455–467. [DOI] [PubMed] [Google Scholar]
- Lammer, D., Mathias, N., Laplaza, J. M., Jiang, W., Liu, Y., Callis, J., Goebl, M., and Estelle, M. (1998). Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 12(7), 914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne, A., and Schwob, E. (2002). The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol. Cell 9(5), 1067–1078. [DOI] [PubMed] [Google Scholar]
- Lisby, M., Rothstein, R., and Mortensen, U. H. (2001). Rad52 forms DNA repair and recombination centers during S-phase. Proc. Natl Acad Sci USA 98(15), 8276–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Li, M. Z., Leibham, D., Cortez, D., and Elledge, S. J. (1998). The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 8(24), 1300–1309. [DOI] [PubMed] [Google Scholar]
- Mathias, N., Johnson, S. L., Winey, M., Adams, A. E., Goetsch, L., Pringle, J. R., Byers, B., and Goebl, M. G. (1996). Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol. Cell Biol. 16(12), 6634–6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek, S. P., Parikh, A. S., and Kron, S. J. (2000). Genetic analysis reveals that FLO11 upregulation and cell polarization independently regulate invasive growth in Saccharomyces cerevisiae. Genetics 156(3), 1005–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, E. E., Willems, A. R., and Tyers, M. (1998). Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 14(6), 236–243. [DOI] [PubMed] [Google Scholar]
- Pickart, C. M. (1997). Targeting of substrates to the 26S proteasome. FASEB J. 11(13), 1055–1066. [DOI] [PubMed] [Google Scholar]
- Ricke, R. M., and Bielinsky, A. K. (2004). Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol. Cell 16(2), 173–185. [DOI] [PubMed] [Google Scholar]
- Rose, M. D., Winston, F., and Hieter, P. (1990). Methods in Yeast Genetics: A Laboratory Manual, New York: Cold Spring Harbor Laboratory Press.
- Sanchez, Y., Bachant, J., Wang, H., Hu, F., Liu, D., Tetzlaff, M., and Elledge, S. J. (1999). Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286(5442), 1166–1171. [DOI] [PubMed] [Google Scholar]
- Sarin, S., Ross, K. E., Boucher, L., Green, Y., Tyers, M., and Cohen-Fix, O. (2004). Uncovering novel cell cycle players through the inactivation of securin in budding yeast. Genetics 168(3), 1763–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob, E., and Nasmyth, K. (1993). CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 7, 1160–1175. [DOI] [PubMed] [Google Scholar]
- Sidorova, J. M., and Breeden, L. L. (2003). Precocious G1/S transitions and genomic instability: the origin connection. Mutat. Res. 532(1–2), 5–19. [DOI] [PubMed] [Google Scholar]
- Sidorova, J. M., and Breeden, L. L. (2002). Precocious S-phase entry in budding yeast prolongs replicative state and increases dependence upon Rad53 for viability. Genetics 160(1), 123–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra, D., Craig, K. L., Tyers, M., Elledge, S. J., and Harper, J. W. (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91(2), 209–219. [DOI] [PubMed] [Google Scholar]
- Skowyra, D., Koepp, D. M., Kamura, T., Conrad, M. N., Conaway, R. C., Conaway, J. W., Elledge, S. J., and Harper, J. W. (1999). Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284(5414), 662–665. [DOI] [PubMed] [Google Scholar]
- Strohmaier, H., Spruck, C. H., Kaiser, P., Won, K. A., Sangfelt, O., and Reed, S. I. (2001). Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413(6853), 316–322. [DOI] [PubMed] [Google Scholar]
- Tsvetkov, L. M., Yeh, K. H., Lee, S. J., Sun, H., and Zhang, H. (1999). p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 inp27. Curr. Biol. 9(12), 661–664. [DOI] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Pohlmann, R., and Philippsen, P. (1994). New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Willems, A. R., Lanker, S., Patton, E. E., Craig, K. L., Nason, T. F., Mathias, N., Kobayashi, R., Wittenberg, C., and Tyers, M. (1996). Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86(3), 453–463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.