Abstract
The spindle pole body (SPB) in Saccharomyces cerevisiae functions to nucleate and organize spindle microtubules, and it is embedded in the nuclear envelope throughout the yeast life cycle. However, the mechanism of membrane insertion of the SPB has not been elucidated. Ndc1p is an integral membrane protein that localizes to SPBs, and it is required for insertion of the SPB into the nuclear envelope during SPB duplication. To better understand the function of Ndc1p, we performed a dosage suppressor screen using the ndc1-39 temperature-sensitive allele. We identified an essential SPB component, Nbp1p. NBP1 shows genetic interactions with several SPB genes in addition to NDC1, and two-hybrid analysis revealed that Nbp1p binds to Ndc1p. Furthermore, Nbp1p is in the Mps2p-Bbp1p complex in the SPB. Immunoelectron microscopy confirmed that Nbp1p localizes to the SPB, suggesting a function at this location. Consistent with this hypothesis, nbp1-td (a degron allele) cells fail in SPB duplication upon depletion of Nbp1p. Importantly, these cells exhibit a “dead” SPB phenotype, similar to cells mutant in MPS2, NDC1, or BBP1. These results demonstrate that Nbp1p is a SPB component that acts in SPB duplication at the point of SPB insertion into the nuclear envelope.
INTRODUCTION
The spindle pole body (SPB) of the budding yeast Saccharomyces cerevisiae is the microtubule-organizing center that duplicates once per cell cycle to serve as the spindle poles during mitosis. The duplication of the SPB (and the centrosome, its mammalian counterpart) is an essential event among many, leading to successful chromosome segregation during each cell cycle. Misregulation of centrosome duplication leads to monopolar or multipolar spindles and aneuploidy, which are frequently observed in cancer cells (Fisk et al., 2002). Even though the SPB and the centrosome are morphologically distinct, they share a number of homologous structural components and regulators critical for function (for reviews, see Jaspersen and Winey, 2004).
The SPB is embedded in the nuclear envelope throughout the cell cycle (reviewed in Adams and Kilmartin, 2000; Jaspersen and Winey, 2004). As viewed by electron microscopy, the core SPB appears as three disklike layers: an outer plaque and an inner plaque on the cytoplasmic and nuclear side of the nuclear envelope, respectively, and a central plaque that spans the nuclear membrane (O'Toole et al., 1999). SPB duplication involves several distinct steps (Adams and Kilmartin, 1999; Jaspersen and Winey, 2004). It begins in G1 with the deposition of amorphous satellite materials containing several SPB components at the distal cytoplasmic tip of the half-bridge, a modified nuclear membrane structure associated with one side of the SPB. As SPB duplication continues, the satellite grows in size to form the duplication plaque that is composed of outer and central plaque components (Adams and Kilmartin, 1999). Insertion of the duplication plaque into the nuclear envelope and assembly of inner plaque components complete SPB duplication. The two SPBs are connected by a full-bridge, which is severed in half as the SPBs separate into each pole of the dividing cell.
The SPB components and their relative position within the organelle were identified using various genetic, biochemical, cytological, and proteomic approaches (for review see Jaspersen and Winey, 2004; Muller et al., 2005). Interestingly, none of the components of the core SPB are integral membrane proteins, raising the question of how the SPB is inserted and tethered in the nuclear envelope. A likely possibility is that one or more of the four integral membrane proteins known to localize to the SPB (Ndc1p, Mps2p, Mps3p, and Kar1p) are involved. Of these four proteins, Ndc1p and Mps2p have been implicated in SPB insertion after the formation of the duplication plaque based on analysis of mutant phenotypes (Winey et al., 1991, 1993). Both proteins localize to the SPB periphery and the central plaque region (Chial et al., 1998; Munoz-Centeno et al., 1999), making them good candidates to form a pore in the nuclear envelope into which the SPB is inserted. Mps2p has previously been shown to bind to Bbp1p, a soluble protein that also binds to the central plaque component Spc29p and to the half-bridge protein Kar1p (Schramm et al., 2000). Thus, Bbp1p is thought to be a protein that connects the core SPB to the nuclear envelope (Schramm et al., 2000). However, it is not known with which protein(s) Ndc1p associates at the SPB to facilitate SPB insertion or how Ndc1p and Mps2p interact. Cells carrying mutations in NDC1 (ndc1-1 and ndc1-39), MPS2 (mps2-1), or BBP1 (bbp1-1) result in duplication plaque that fails to insert into the nuclear envelope (Winey et al., 1991; Winey et al., 1993; Schramm et al., 2000; Lau et al., 2004), but it is not clear if these genes cooperate in one pathway leading to SPB insertion or if they define two partially redundant pathways.
In this study, we searched for NDC1 interacting partner(s) using the ndc1-39 allele in a dosage suppressor screen and identified NBP1 (Nap1-binding protein 1; Shimizu et al., 2000). NBP1 also shows genetic interactions with several other SPB components including MPS2, and we showed that Nbp1p binds to Ndc1p and Mps2p in a two-hybrid assay. Nbp1p is also found to be a component of the Mps2p-Bbp1p complex. By immunoelectron microscopy, Nbp1p localizes to the SPB periphery and to the central plaque of the SPB, where Ndc1p and Mps2p also reside. Consistent with a possible function at the SPB, we found that a “degron” allele of NBP1, nbp1-td, causes defects in SPB duplication. Together, our data support a model that Nbp1p is a SPB component that serves as a molecular linker between the core SPB and the nuclear envelope membrane proteins Ndc1p and Mps2p.
MATERIALS AND METHODS
Yeast Strains and Media
Yeast manipulations were performed using standard techniques (Guthrie and Fink, 2002). Yeast cells were grown in YPD media (1% yeast extract, 2% bactopeptone, and 2% glucose), or synthetic media with 2% glucose and supplemented with the appropriate amino acids. Plates containing 5-fluoroorotic acid (5-FOA) were made as previously described (Boeke et al., 1987).
Polymerase chain reaction (PCR)-based gene deletion (Longtine et al., 1998) was used to create strain 3445. PCR-based gene tagging (Longtine et al., 1998) was used to create strains 3326, 3322, and 3310. Others genes were epitope tagged at the endogenous loci using PCR-based methods (Knop et al., 1999). nbp1-td cells were constructed and grown as described (Labib et al., 2000; Kanemaki et al., 2003). Briefly, cells were synchronized in G1-phase of the cell cycle with 10 μg/ml α-factor in YPA medium (1% yeast extract, 2% peptone, and 0.01% adenine) containing 3% raffinose (YPA-Raf) and 0.1 mM CuSO4 at 23°C. To induce degradation of Nbp1p-td, expression of UBR1 was induced by transferring cells to YP medium with 3% raffinose and 2% galactose (YPA-Raf/Gal) for 45 min at 23°C in the presence of 10 μg/ml α-factor. Cells were then incubated for 50 min at 37°C before removing α-factor by washing cells with YPA-Raf/Gal at 37°C (t = 0). Cells were continuously incubated in YPA-Raf/Gal at 37°C.
Plasmids and Nucleic Acid Techniques
DNA was manipulated using standard techniques as described (Ausubel et al., 1998). Plasmid DNA was prepared using the QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA). PCR products were purified using the QIAEXII Gel Extraction Kit (Qiagen). Plasmids and PCR products were transformed into yeast strains using EZ Transformation Kit (Zymo Research, Orange, CA).
Two-micron URA3 plasmid containing +NBP1/+YLR458W was made by PCR-amplifying YLR458W and the NBP1 regions with primers NBP1-5′KpnI (5′-CGG GGT ACC GGT CAT TTC ACT TAA TGG-3′; KpnI site is underlined) and NBP1-3′XhoI (5′-CCG CTC GAG CGG GCG TCC ATA GTA AAG ATG-3′; XhoI site is underlined). Then, the PCR product was cut with SnaBI-EaeI, and ligated into the XhoI-NotI sites of pRS202 with the use of the XhoI-EcoRV DNA oligonucleotide adaptors (EZ Clone Systems, New Orleans, LA). The CEN-URA3 plasmid containing +NBP1/+YLR458W was created by cutting the PCR product mentioned above with XhoI-SnaBI and ligated into the SalI-SmaI sites of pALR10 (a gift from Alain Camasses). The CEN-TRP1 plasmid containing +NBP1/+YLR458W was generated by cutting the PCR product mentioned above with SnaBI-EaeI and was cloned into the SmaI-NotI sites of pRS314.
Two-micron URA3 and CEN-TRP1 plasmids containing +NBP1/-YLR458W, -NBP1/+YLR458W, and -NBP1/-YLR458W were made by site-directed mutagenesis (Kunkel et al., 1987, and according to the pGEM Single Strand Systems Manual [Promega, Madison, WI]; also see Lau et al., 2004). Single-stranded CEN-TRP1 plasmid (pRS314) containing +NBP1/+YLR458W was isolated from repair-incompetent bacteria CJ236 that has been infected with helper phage R408 (Promega). Then, primers A (GAT CTC TTC TTC AAA ATA CGC GTC ATA TCG CCA CAA ACA TTT TG; MluI site is underlined) and B (CAT CTC TGA TTG TCT GAA AGA TCT TTG TTA ACC TTC; BglII site is underlined) were added for in vitro second-strand synthesis to generate plasmid +NBP1/-YLR458W. Likewise, primers C (CGG ATT TTA GTA TAC TAC CAA TGT CTA TTA TTG; Bst1107I/BstZ17I site is underlined) and D (GTA TTC CAA AAA AAA GCT TTC ACA GTC CCT G; HindIII site is underlined) were used to generate plasmid -NBP1/+YLR458W, and primers A, C, and D were used to generate plasmid -NBP1/-YLR458W. The three plasmids were amplified, and the mutations generated were verified by the presence of the restriction site indicated and also by DNA sequencing. The XhoI-SacI fragments from these plasmids were cloned into the XhoI-SacI sites of pRS202 for further analysis.
Fluorescence Microscopy
Cells containing NBP1-GFP; SPC42-CFP and NBP1-GFP; NDC1-CFP were resuspended in PBSA without fixation. GFP and CFP signals were detected by their autofluorescence, and DNA was stained with 4′,6-diamidono-2-phenylindole (DAPI; Sigma Chemical Co., St. Louis, MO). The cells were visualized using a Zeiss Axioplan 2 deconvolution fluorescence microscope (Thornwood, NY) equipped with a Cooke SensiCam CCD camera (Romulus, MI). The images were processed using SlideBook software (v. 3.0.11, Intelligent Imaging Innovations, Denver, CO).
CFP-, eqFP-, GFP-, and YFP-labeled cells were analyzed by fluorescence microscopy after fixation of cells with 4% paraformaldehyde (150 mM phosphate buffer, pH 6.5) for 10 min at 20°C. For a series of z-focal planes images were collected on a Zeiss Axiophot microscope using a Coolsnap HQ camera (Photometrics, Tucson, AZ) and Metamorph software (Universal Imaging, West Chester, PA). Images in different z-planes were projected and processed in Adobe Photoshop (San Jose, CA).
Electron Microscopy
Nbp1p-GFP cells were high-pressure frozen, freeze-substituted, sectioned, and stained for electron microscopy or for immunoelectron microscopy as previously described (Giddings et al., 2001; Lau et al., 2004). Serial thin sections were viewed on a Philips CM10 electron microscope (Mahwah, NJ), and images were captured with Gatan digital camera and viewed with Digital Micrograph software (Gatan, Pleasanton, CA). Some cells were glutaraldehyde-fixed and embedded into Spurr resin and then prepared for serial section electron microscopy (Byers and Goetsch, 1975).
Two-Hybrid Assay
GAL4 DNA-binding domain and GAL4 DNA-activation domain constructs containing NBP1, NDC1, and ndc1-39 were created by amplified as NcoI-XhoI fragments and inserted into the NcoI-SalI sites of pOBD2 or pOAD1 (Uetz et al., 2000). The remaining two-hybrid plasmids were gifts of Stan Fields, and their construction is described in Uetz et al. (2000). Plasmids were transformed into PJ69-4a for pOAD1 constructs, or PJ69-4α for pOBD2 constructs (James et al., 1996). Two-hybrid interactions were tested as described (Uetz et al., 2000).
Immunoprecipitation
Cells (80 OD600) were lysed with glass beads in L-buffer (50 mM Tris-HCl, pH 7.6, 10 mM EDTA, 1 mM EGTA, 100 mM NaCl, 5% glycerol) in the presence of complete protease inhibition cocktail (Roche Diagnostics, Branchburg, NJ). The lysates were then incubated with 1% Triton X-100 for 30 min at 4°C. The cell extracts were clarified by two consecutive centrifugations (20,000 × g, 5 min). The supernatants were incubated with IgG-Sepharose for 2 h at 4°C with rotation. Beads were washed three times with L-buffer, 1% Triton X-100 and once with TEV cleavage buffer (50 mM HEPES-KOH, pH 7.9, 150 mM KOAc, 0.1% NP40, 0.5 mM EDTA, 1 mM DTT). Proteins were eluate using TEV protease. Samples were analyzed by immunoblotting with the indicated antibodies.
RESULTS
MPS2 and NBP1 Are Dosage Suppressors of ndc1-39
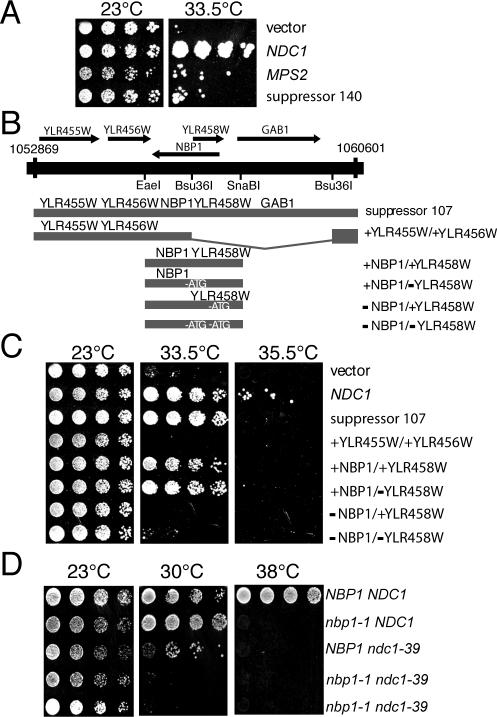
A dosage suppressor screen was performed to identify genes that interact with NDC1. Approximately 18,000 clones from a 2-μm yeast genomic library (Connelly and Hieter, 1996) were screened for their ability to rescue the growth defects of ndc1-39 temperature-sensitive (ts) cells (Lau et al., 2004). Two clones (suppressors 140 and 107) that exhibited the strongest suppression phenotype were isolated and re-transformed into the ndc1-39 strain for further characterization. Suppressor 140 contained four complete open reading frames (ORFs) including MPS2, which encodes an integral membrane protein required for SPB duplication like Ndc1p (Munoz-Centeno et al., 1999; Schramm et al., 2000). Both, the suppressor 140 and MPS2, partially suppress the ts phenotype of ndc1-39 mutants at the restrictive temperature of 33.5°C, although not as well as a plasmid containing wild-type NDC1 (Figure 1A). The other ORFs on suppressor 140 besides MPS2 do not suppress ndc1-39 (unpublished observation). This is the first evidence of a genetic interaction between NDC1 and MPS2.
Figure 1.
MPS2 and NBP1 are dosage suppressors of ndc1-39. (A) Two-micron plasmids containing vector, NDC1 (Chial et al., 1999), MPS2 (McBratney and Winey, 2002), and suppressor 140 (contains DNA from chromosome IV, coordinates 366254-373702, which includes MPS2) were transformed into ndc1-39 cells (3446, Table 1). Cells were grown in YPD media to saturation at 23°C, then diluted to a density of 3 OD600 U/ml, and spotted onto YPD plates in fivefold serial dilutions. Plates were incubated for 4 d at 23 or 33.5°C. (B) A single clone containing DNA from coordinates 1052869-1060601 on chromosome XII was able to partially rescue the temperature sensitivity of ndc1-39 cells. This clone, suppressor 107, contained five ORFs. A clone containing only YLR455W and YLR456W (+YLR455W/+YLR456W) was created by cutting suppressor 107 with Bsu36I and religating the plasmid. A clone containing NBP1 and YLR458W (+NBP1/+YLR458W) was obtained by ligating a PCR-generated EaeI-SnaBI fragment into pRS202 (see Materials and Methods). The start codon of YLR458W was mutated so that only NBP1 is expressed in the +NBP1/-YLR458W plasmid (see Materials and Methods). The start codon of NBP1 was mutated so that only YLR458W is expressed in the -NBP1/+YLR458W plasmid. Both start codons of NBP1 and YLR458W were mutated to create the -NBP1/-YLR458W plasmid. (C) Two-micron URA3-based plasmids containing the indicated ORFs were transformed into ndc1-39 cells (3446, Table 1). Cells were grown and spotted onto YPD plates as in A. Plates were incubated for 4 d at 23, 33.5, or 35.5°C. (D) nbp1-1 cells are synthetically sick with ndc1-39 cells. Heterozygous diploids were generated by crossing nbp1-1 strain (YYS113-71, Table 1) to ndc1-39 strain (2711, Table 1) and then sporulated. Twenty tetrads were dissected and analyzed by spotting fivefold serial dilutions of cells at ∼5 OD600 U/ml on YPD plates. Plates were incubated at 23, 30, and 38°C for 2 d. The nbp1-1 ndc1-39 double mutant (3606, Table 1) was viable at 23°C but was unable to grow at 30°C, a temperature permissive for growth for the single mutant parents. Two isolates of nbp1-1 ndc1-39 double mutant are shown.
The chromosomal DNA in suppressor 107 contained five complete ORFs (Figure 1B), and it rescued the growth defect of the ndc1-39 mutant cells at 33.5°C as well as wild-type NDC1, but did not support growth at 35.5°C (Figure 1C). Subclones of the suppressor 107 DNA revealed that the ORFs YLR455W and YLR456W did not suppress the ndc1-39 ts phenotype at 33.5°C, whereas a region containing NBP1 and YLR458W suppressed the ndc1-39 ts phenotype (Figure 1C). NBP1 and YLR458W are two overlapping ORFs transcribed in opposite directions. To definitively show whether NBP1 or YLR458W is the ndc1-39 suppressor, site-directed mutagenesis was used to inactivate NBP1 or YLR458W by mutating the start codon of each gene without affecting the coding sequence or expression of the other gene. A third construct was made where both start codons of NBP1 and YLR458W were mutated. In addition, a stop codon (with a silent mutation on the opposite strand) was also created just downstream of the start codon mutation in each of these clones to prevent read-through of the mutated gene(s). A plasmid containing wild-type NBP1, either in the presence or absence of wild-type YLR458W, suppressed the ndc1-39 mutants as well as suppressor 107 (Figure 1C), suggesting that NBP1 is responsible for the suppression observed with suppressor 107. Consistent with this result, a clone lacking wild-type NBP1, either in the presence or absence of wild-type YLR458W, no longer suppressed the ndc1-39 mutants (Figure 1C). Therefore, we conclude that NBP1 is a dosage suppressor of the ndc1-39 ts mutants.
Suppression of the growth defect of ndc1-39 cells by NBP1 suggests a functional interaction of both genes. To strengthen this conclusion, we examined nbp1-1 cells, which carry a R180W missense mutation in NBP1 (Shimizu et al., 2000) for genetic interactions with mutations in NDC1. Cells containing nbp1-1 exhibit severe growth defects compared with wild-type cells at 38°C (Figure 1D; Shimizu et al., 2000). We reasoned that if NBP1 functions with NDC1, then mutants containing mutations in two of these genes may have synergistic growth defects. Indeed, cells carrying both nbp1-1 and ndc1-39 were unable to grow at 30°C (Figure 1D), a temperature that was permissive for either of the individual mutations. Thus, nbp1-1 cells are synthetically sick in the presence of ndc1-39, further suggesting that NBP1 and NDC1 function together in the cell.
NBP1 Also Suppresses the Growth Defect of Several Mutants Defective in SPB Duplication
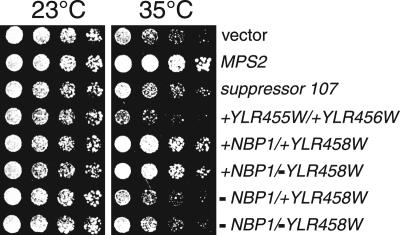
The fact that both MPS2 and NBP1 suppress the growth defect of the ndc1-39 mutants led us to speculate that NBP1 might also interact with MPS2. Therefore, we tested NBP1 suppression on mps2-1 ts mutants using the constructs described above. Two-micron plasmids specifically containing wild-type NBP1 suppressed the mps2-1 ts phenotype as well as wild-type MPS2 at the semirestrictive temperature of 35°C (Figure 2). Intrigued by the ability of increased levels of NBP1 to suppress the ndc1-39 or the mps2-1 mutations, we asked whether NBP1 acts as a dosage suppressor of other known SPB duplication mutations. We found that NBP1 partially suppressed the growth defect of a subset of these mutants, namely spc42-11, cdc31-2, mps1-8, and mps3-1 (but not bbp1-1, cmd1-1, cmn67Δ, kar1Δ17, mps1-1, mps1-737, ndc1-1, nud1-44, spc29-2, spc29-3, spc29-20, spc42-10, spc98-2, spc110-220, tub4-1; unpublished observation). The results suggest that NBP1 is involved in some aspect of SPB function and/or duplication.
Figure 2.
NBP1 also suppresses the growth defect of mps2-1. Two-micron plasmids containing the indicated ORFs (from Figure 1, A and C) were transformed into mps2-1 strain (1210, Table 1). Cells were grown and spotted onto YPD plates as in Figure 1A. Plates were incubated for 4 d at 23 or 35°C. NBP1 was able to rescue the growth defect of mps2-1 at 35°C, the semirestrictive temperature for mps2-1, but not at 37°C.
NBP1 Is an Essential Gene
Nbp1p was originally isolated as a two-hybrid binding partner with Nap1p (Nucleosome Assembly Protein 1; Shimizu et al., 2000). NBP1 encodes a 319-amino acid protein that contains a coiled-coil domain and several potential phosphorylation sites for Cdc28p cyclin-dependent kinase and cAMP-dependent kinase (Shimizu et al., 2000), but lacks significant homology to any characterized protein. Although the authors suggest Nbp1p is required for G2/M transition, the molecular function of Nbp1p remains undefined.
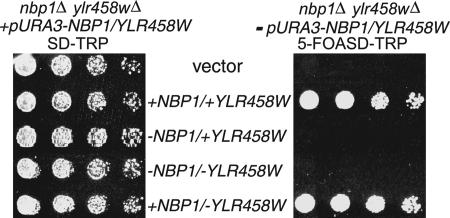
Shimizu et al. (2000) also showed that disruption of the NBP1 region is lethal to yeast cells. However, eliminating the NBP1 ORF also deletes the overlapping ORF YLR458W (Figure 1B). To determine which of the two genes is essential for growth, we tested the ability of plasmids containing NBP1 and/or YLR458W start codon mutations to rescue growth in an nbp1Δ/ylr458wΔ strain. As expected, the nbp1Δ/ylr458wΔ strain with vector alone is not viable, whereas plasmid containing both NBP1 and YLR458W was able to rescue growth (Figure 3). A plasmid lacking NBP1 either in the presence or absence of YLR458W was unable to provide the essential function for growth (Figure 3). Only when the cells were provided with wild-type NBP1 was the lethality of the nbp1Δ/ylr458wΔ strain rescued (Figure 3). These results clearly show that NBP1 is an essential gene in yeast, similar to nearly all genes encoding SPB components and regulators.
Figure 3.
NBP1 is an essential gene. An nbp1Δ/ylr458wΔ strain (3445, Table 1) carrying a genomic deletion of NBP1 and YLR458W originally contained NBP1 and YLR458W on a CEN-URA3 plasmid. It was then transformed with the indicated ORFs (from Figure 1B) on CEN-TRP1 plasmids. The ability of the each transformed construct to rescue the lethality of the nbp1Δ/ylr458wΔ strain was tested by plating fivefold serial dilutions of cells on SD-TRP plates supplemented with 5-fluoroorotic acid (5-FOA; right panel) to select for the TRP1 plasmid and select against the URA3 plasmid. As a growth control, cells were spotted onto SD-TRP plates (left panel). Plates were incubated at 23°C for 4 d.
Nbp1p Localizes to the Periphery and the Central Plaque of SPBs
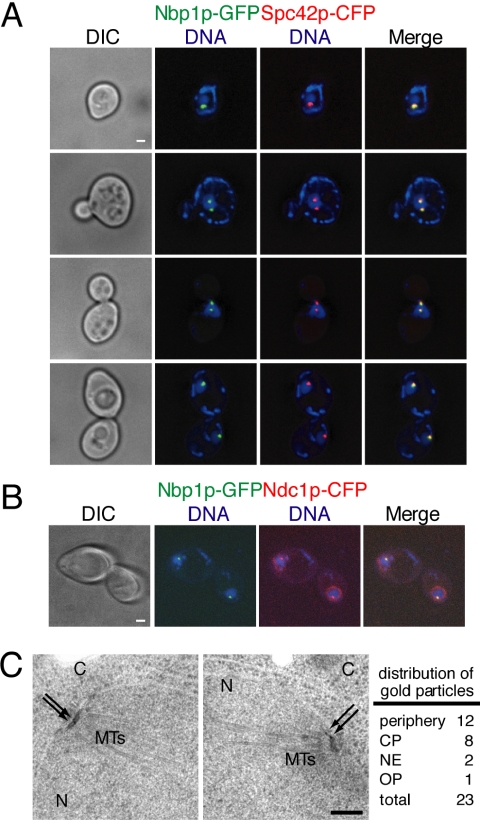
The genetic interactions between NBP1 and the alleles of several genes encoding SPB components suggested that Nbp1p itself may be an SPB component. Previous studies have shown that Nbp1p is present as one or two spots at the nuclear periphery (Shimizu et al., 2000) and colocalizes with the SPB component Spc29p (Sundin et al., 2004). This localization is consistent with an association of Nbp1p either with SPBs or with kinetochores, which in yeast are clustered close to SPBs. Moreover, Ndc1p localizes to both SPBs and nuclear pore complexes (NPCs, Chial et al., 1998). Therefore, we also examined the possibility of Nbp1p localizing to the NPCs using deconvolution fluorescence microscopy. The endogenous copy of NBP1 was fused to green fluorescent protein (GFP), and the strain also contained an allele of the core SPB component Spc42p fused to cyan fluorescent protein (CFP). We consistently found that the number of Nbp1p-GFP spots observed was identical to the number of SPBs expected at the given cell cycle stage; in that one spot in unbudded G1 cells, one or two spots in small budded S-phase cells, and two spots in large budded mitotic cells (Figure 4A). In all of these cells, Nbp1p-GFP colocalized with Spc42p-CFP. This was also the case in metaphase cells when kinetochore proteins show a slightly distinct distribution from SPB components (e.g., Janke et al., 2002). Thus, Nbp1p is associated with the SPB.
Figure 4.
Nbp1p localizes to the periphery and the central plaque of SPBs. (A and B) Cells containing Nbp1p-GFP and Spc42p-CFP fusion proteins (3326, Table 1) or Nbp1p-GFP and Ndc1p-CFP (3322, Table 1) were grown in YPD media at 23°C, and processed for fluorescence microscopy the following day. DIC images are shown on the left column. Nbp1p-GFP (green) and Spc42p-CFP or Ndc1p-CFP (red) signals were detected by autofluorescence, and DNA was stained with DAPI (blue). Yellow color corresponds to colocalization signals. Examples of cells in various stages of the cell cycle are shown in A. Note that in B, Ndc1p-CFP has additional perinuclear signal corresponding to its known localization at the NPCs. Each fluorescent image shown was projected from 10 consecutive images taken at 0.1-μm intervals along the z-axis that have been deconvolved. Bar, 1 μm. (C) Localization of Nbp1p-GFP was examined by immunoelectron microscopy using anti-GFP antibodies and colloidal gold-conjugated secondary antibodies. Nbp1p-GFP cells (3310, Table 1) were grown to log-phase in YPD at 23°C and processed for immunoelectron microscopy the following day. The SPB is embedded in the nuclear envelope, which separates the nucleus (N) and cytoplasm (C). Microtubules (MTs) can be seen on the nuclear side of the SPB. Arrows indicate the positions of the gold particles that recognize Nbp1p-GFP. The two SPBs shown are from the same cell. A total of 11 SPBs were analyzed, and the distribution of gold particles to the central plaque region (CP), the outer plaque region (OP), the periphery of the SPB, and the nuclear envelope (NE) is indicated. Bar, 0.2 μm.
In addition, we fused Ndc1p with CFP in the Nbp1p-GFP strain, and we found that Nbp1p-GFP also colocalized with the Ndc1p-CFP at the SPBs, but it did not colocalize with Ndc1p to the nuclear periphery at NPCs (Figure 4B), suggesting that Nbp1p functions at the SPB and not with Ndc1p at the NPC.
To confirm the localization of Nbp1p to the SPB, a strain containing Nbp1p-GFP was examined by immunoelectron microscopy using anti-GFP primary antibodies and secondary antibodies conjugated to colloidal gold. Detailed examination of the collodial gold particles that decorated 11 SPBs revealed that about half of the Nbp1p-GFP signal was at the periphery of the SPB, one quarter was at the central plaque, and a few were at the nuclear envelope or at the outer plaque (Figure 4C). Consistent with our fluorescence microscopy analysis, Nbp1p-GFP was not observed elsewhere in the cell away from the SPB. These data demonstrate that Nbp1p is a SPB component predominantly found at the SPB periphery and the central plaque similar to the localization of Mps2p and Ndc1p at the SPBs (Chial et al., 1998; Munoz-Centeno et al., 1999).
Nbp1p Binds to Both Ndc1p and Mps2p
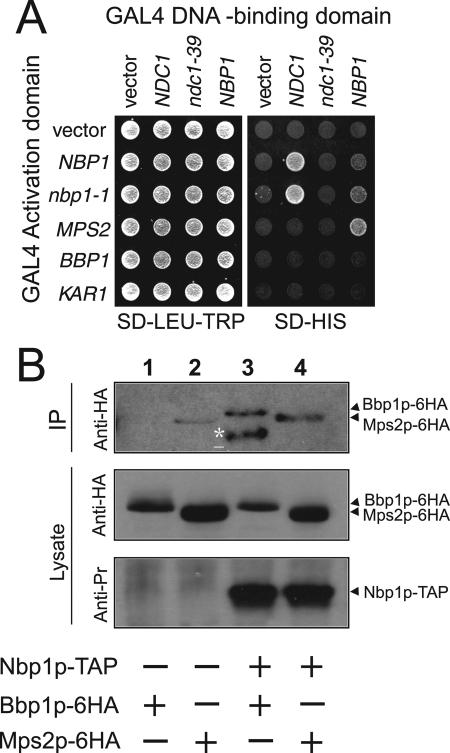
Localization of Nbp1p at the SPB and its genetic interactions with NDC1 and MPS2 prompted us to investigate whether Nbp1p physically associates with Ndc1p and Mps2p. Initially, we conducted two-hybrid assays and found interactions between Nbp1p and Ndc1p, and Nbp1p and Mps2p (Figure 5A). Interestingly, Nbp1p binding was compromised when the mutated ndc1-39 two-hybrid construct was used (Figure 5A). The Nbp1p-Ndc1p interaction is therefore likely to be important for SPB duplication. In addition, Ndc1p did not bind to Mps2p (Figure 5A), suggesting Nbp1p may mediate the interaction between Ndc1p and Mps2p as revealed by genetics. Importantly, Nbp1p showed specificity for a subset of membrane proteins at the SPB, as we did not observe Nbp1p binding to Kar1p (Figure 5A) or to Mps3p (unpublished observation). Finally, Nbp1p failed to interact with Bbp1p (Figure 5A), which is known to bind to Mps2p (Schramm et al., 2000). Thus, the interaction between Nbp1p and Mps2p may be direct and not mediated by Bbp1p. In conclusion, these results suggest that Nbp1p physically interacts with Ndc1p and Mps2p.
Figure 5.
Nbp1p binds to both Ndc1p and the Mps2p-Bbp1p complex. (A) Two-hybrid analysis was done with PJ69-4a cells (Table 1) containing the indicated GAL4 activation domain fusions (rows) were crossed to PJ69-4α cells (Table 1) containing the indicated GAL4 DNA-binding domain fusions (columns), and diploids were selected on SD-LEU-TRP plates at 30°C (left). The ability of fusion proteins to interact was assayed by plating cells, which contain a version of HIS3 driven by the GAL1 promoter, on SD-HIS plates at 30°C (right). 30°C is the semipermissive temperature for strains containing ndc1-39 allele. Similar results were obtained at 23 and 37°C. (B) Coimmunoprecipitation of Bbp1p and Mps2p with Nbp1p. Lysates of BBP1-6HA (lane 1), MPS2-6HA (lane 2), NBP1-TAP BBP1-6HA (lane 3), or NBP1-TAP MPS2-6HA cells (lane 4) were subjected to TAP purification using IgG Sepharose (strains listed in Table 1, YAY167-170). IgG bound complexes were eluted with Tev protease. The eluates were analyzed by immunoblotting with anti-HA (12CA5) and anti-PrA antibodies. Lane 3 contains a degradation product of Bbp1p-6HA (asterisks). Mps2p-6HA (lane 2) bound weakly to IgG beads. However, the Mps2p-6HA signal in lane 4 was repeatedly stronger, indicating coimmunoprecipitation of Mps2p-6HA and Nbp1p-TAP.
We sought to pursue the interaction of Nbp1p with Mps2p to ask if Nbp1p can bind to the Bbp1p-Mps2p complex (Schramm et al., 2000). Nbp1p-TAP was bound to IgG beads and then eluted by Tev protease as described in Materials and Methods. Using this condition, we could show coimmunoprecipitation between Nbp1p-TAP and Bbp1p-6HA (Figure 5B). Extracts from the control NBP1 MPS2-6HA strain revealed a small amount of Mps2p-6HA in the Tev eluate (Figure 5B). However, we repeatedly recovered 2-3 times more Mps2p-6HA when extracts from NBP1-TAP MPS2-6HA cells were used (Figure 5B), consistent with the two-hybrid result. Therefore, we conclude that Nbp1p interacts with the Bbp1p-Mps2p complex.
Cells Lacking Nbp1p Fail in SPB Duplication and Exhibit a Mitotic Cell Cycle Arrest
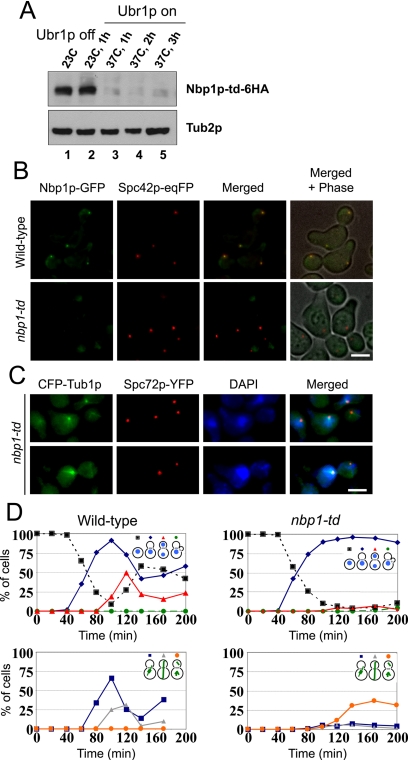
NBP1's genetic interactions and the finding that Nbp1p is in complex with Mps2p and Bbp1p suggests that Nbp1p is required for SPB duplication, which would be revealed by the phenotypes of conditional alleles of the gene. However, several attempts to obtain a temperature-sensitive nbp1(ts) allele failed. We therefore constructed temperature-sensitive NBP1-degron (nbp1-td) cells. In these cells, NBP1 was fused to the degron element, which targets proteins for degradation at 37°C (Dohmen et al., 1994). To ensure efficient degradation, UBR1, encoding a ubiquitin-protein ligase, was expressed from the inducible GAL1 promoter (Kanemaki et al., 2003). In the presence of Ubr1p almost all of the Nbp1p-td protein became degraded when cells were shifted to 37°C (Figure 6A). Moreover, analysis of nbp1-td-GFP cells by fluorescence microscopy revealed that upon shifting cells to 37°C, the Nbp1p-td-GFP signal was no longer detected and therefore did not colocalize with the Spc42-eqFP-marked SPBs (Figure 6B). This data suggests that most of the cellular Nbp1p-td is targeted for degradation at 37°C.
Figure 6.
nbp1-td cells are defective in mitotic spindle formation and chromosome segregation. (A) nbp1-td-6HA cells (YAY144, Table 1) were first grown at 23°C without expression of UBR1 (lane 1). Induction of UBR1 at 23°C did not affect Nbp1p-td-6HA protein levels as determined by immunoblotting with anti-HA (12CA5) antibodies (lane 2). However, upon shifting the cells to 37°C Nbp1p-td-6HA was rapidly degraded after 1, 2, or 3 h in the presence of UBR1 (lanes 3-5, respectively). An immunoblot using anti-Tub2p antibodies is shown as a protein loading control. (B) Nbp1p-td-GFP is not associated with SPBs. NBP1-GFP SPC42-eqFP (YAY157, Table 1) and nbp1-td-GFP SPC42-eqFP (YAY158, Table 1) cells were synchronized at 23°C using α-factor. Cells were then shifted to 37°C with UBR1 induction. We confirmed that Nbp1p-td-GFP was degraded upon shifting cells to 37°C by immunoblotting with anti-GFP antibodies. Consistently, no Nbp1p-td-GFP fluorescence signal was observed at SPBs. Bar, 5 μm. (C) nbp1-td cells fail to assemble a mitotic spindle. Wild-type and nbp1-td cells with CFP-TUB1 SPC72-YFP (SHM1097 and SHM1098, Table 1) were synchronized with α-factor in G1-phase of the cell cycle. α-factor was removed by washing the cells (t = 0). Samples were taken every 20 min. Fixed wild-type and nbp1-td cells were stained with DAPI and analyzed 140 min after release by fluorescence microscopy. (D) Quantification of C. More than 100 cells were counted per time point. nbp1-td cells arrested with a large bud with only one DAPI-staining region located in the mother cell body close to the bud neck (top panel, blue line). CFP-tubulin fluorescence revealed that >95% of the large budded nbp1-td cells failed to form a proper mitotic spindle. About 40% of nbp1-td cells had a SPB signal in the bud (bottom panel, orange line). This SPB organized 1-2 cytoplasmic but no nuclear microtubules. In these panels, percentages of total cells (budded or not) are reported.
To examine the effects of Nbp1p depletion, nbp1-td cells were first synchronized in G1-phase by the addition of α-factor. Cells were then washed to remove α-factor and shifted to 37°C in the presence of UBR1 induction to trigger Nbp1p-td degradation. Analysis of the DNA content by flow cytometry (unpublished observations) and of the budding index indicated that nbp1-td cells replicated the DNA as wild-type cells but then were arrested in the first cell cycle with a 2C DNA content and a large bud (Figure 6D). We went onto to ask whether nbp1-td cells have a defect in spindle formation. Microtubules were visualized by expression of CFP-α-tubulin (CFP-TUB1). In addition, SPC72-YFP was expressed in order to mark the outer plaque of the SPB, which is assembled after the duplication plaque is inserted into the nuclear envelope. Most nbp1-td CFP-TUB1 SPC72-YFP cells (>95%) failed to assemble a bipolar spindle (Figure 6, C and D). About 60% of these cells arrested with a single Spc72p-YFP signal, indicating a defect in SPB duplication or separation. The remainder ∼40% of cells showed two Spc72p-YFP signals of equal intensity. In these cells, the SPB in the mother cell body carried a thick bundle of nuclear microtubules, whereas the SPB in the bud was only associated with 1-2 microtubules. All microtubules of the bud SPB were directed toward the cell cortex and probably represent cytoplasmic microtubules. The daughter SPB is therefore devoid of most nuclear microtubules. Consistently, the nuclear DNA was still within the mother cell. Thus, Nbp1p has an essential function during the SPB duplication process.
Nbp1p Is Required to Insert the Duplication Plaque into the Nuclear Envelope
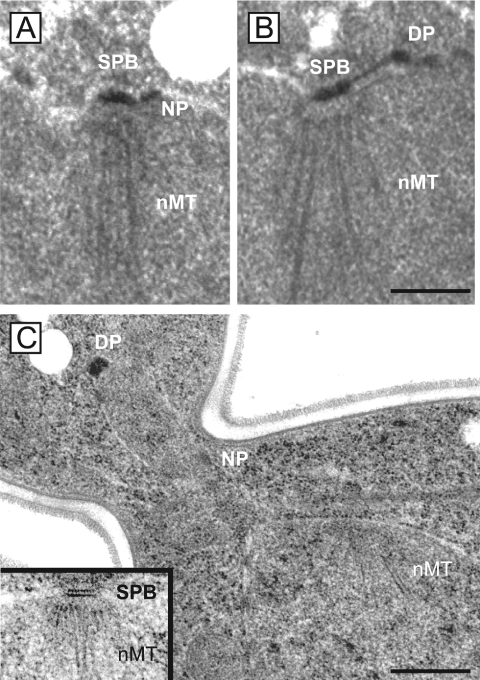
Electron microscopy was performed in order to better understand the SPB defect of nbp1-td cells. SPB substructures, such as the bridge, satellite, and the half-bridge can be visualized this way (Byers and Goetsch, 1975). Synchronized wild-type and nbp1-td cells were prepared for thin-section electron microscopy after 1 h at 37°C. All analyzed wild-type cells had fully duplicated the SPB and formed a short metaphaselike bipolar spindle. In contrast, nbp1-td cells showed the mother SPB still associated via the bridge structure with the extended duplication plaque (Figure 7B). The duplication plaque was not embedded into the nuclear envelope, and it did not contain nuclear microtubules, which were only associated with the mother SPB. Strikingly, in some of the nbp1-td cells, the duplication plaque was found on the cytoplasmic side of the nuclear envelope in the daughter cell away from the mother SPB (Figure 7C). The observed “dead” pole phenotype is reminiscent of conditional lethal ndc1, bbp1, and mps2 cells. These gene products have an essential function in insertion of the duplication plaque into the nuclear envelope (Winey et al., 1991, 1993; Munoz-Centeno et al., 1999; Schramm et al., 2000). Thus, it appears that Nbp1p is also required for insertion of the duplication plaque into the nuclear envelope, most likely as a component of the Mps2p-Bbp1p complex and in cooperation with Ndc1p.
Figure 7.
nbp1-td cells fail to insert the new SPB into the nuclear envelope. Synchronized wild-type and nbp1-td cells (GPY658 and SHM1043, Table 1) were prepared for thin-section electron microscopy 1 h after shifting cells to 37°C. Serial sections of 20 cells were analyzed which spanned the entire nucleus. (A) All wild-type cells had a duplicated and separated SPB (only one of the two SPB is shown). (B) In contrast, 19/20 nbp1-td cells showed the mother SPB (SPB) associated through the bridge structure with the duplication plaque (DP). Nuclear microtubules (nMT) were only associated with the mother SPB but not the duplication plaque. One nbp1-td cell assembled a bipolar spindle. In this cell, nbp1p-td was probably not completely degraded. (C) A nbp1-td cell with the “dead pole” in the bud on an extension of the nuclear envelope. The nuclear microtubules mark the position of the mother SPB, which was found in a different section and shown in the inset at the same magnification as the cell. The cells in A and B were prepared by glutaraldehyde fixation, and the cell in C was prepared by high-pressure freezing and freeze substitution (see Materials and Methods). DP, duplication plaque (or “dead pole” in C); nMT, nuclear microtubules; NP, nuclear pore. Bars, 0.3 μm.
The Defective SPB in nbp1-td Cells Is Devoid of Bbp1p, Mps2p, and Spc110p
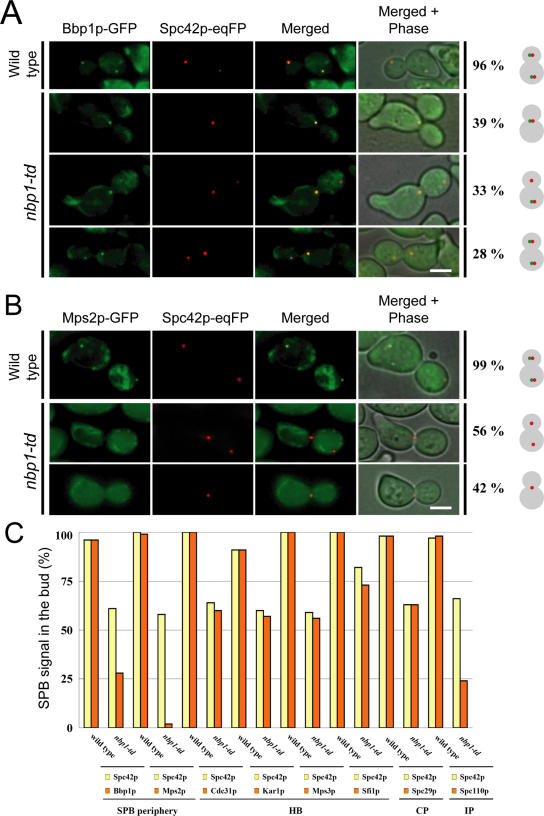
SPB duplication is a hierarchical process that involves a number of SPB-associated proteins such as Cdc31p, Kar1p, Sfi1p, Bbp1p, and Mps2p (Jaspersen and Winey, 2004). We reasoned that proteins interacting with or functioning downstream of Nbp1p may not be associated with the defective SPB in nbp1-td cells. We first analyzed synchronized nbp1-td SPC42-eqFP cells either expressing BBP1-GFP or MPS2-GFP. At 135 min after the release from the α-factor block, 96% of large-budded wild-type cells were in anaphase and showed colocalization of Spc42p-eqFP and Bbp1p-GFP (Figure 8A). In contrast, 61% of nbp1-td cells were large-budded and contained two Spc42-eqFP signals. Of these cells, about half of them did not have Bbp1p-GFP associated with the defective new SPB located in the bud (Figure 8A). However, Bbp1p-GFP was still associated with the mother SPB. This suggests that in a high proportion of nbp1-td cells, Bbp1p fails to associate with the newly formed SPB, whereas it remains associated with the mother SPB. The localization dependency was even more pronounced in the case of Mps2p. In nbp1-td cells, Mps2p-GFP was neither associated with the mother nor the daughter SPB (Figure 8B). Furthermore, Mps2-GFP is degraded upon inactivation of Nbp1-td (unpublished observation). This result is consistent with the notion that Mps2p binds to SPBs through Nbp1p and that Mps2p is not stable when not at the SPB.
Figure 8.
Bbp1p and Mps2p are not associated with the new SPB of nbp1-td cells. (A and B) Wild-type and nbp1-td cells with SPC42-eqFP and GFP-tagged SPB components (Table 1) were synchronized with α-factor and analyzed by fluorescence microscopy 135 min after release. Images of large-budded wild-type and nbp1-td cells expressing either BBP1-GFP (A; YAY075 and YAY074, Table 1) or MPS2-GFP (B; YAY093 and YAY101, Table 1) are shown. (A) Most large budded NBP1 (96%) and nbp1-td cells (61%) showed one SPB signal in the mother cell and one in the bud (n > 100). The remainder of large-budded nbp1-td cells showed only one Spc42p-eqFP SPB signal in the mother cell. Only about half of the large-budded nbp1-td cells that had two SPB signals contained Bbp1p in the new SPB in the bud. (B) In all nbp1-td cells, Mps2p failed to bind to the motherand daughter SPBs. Bar, 5 μm. (C) Wild-type and nbp1-td cells were synchronized as in A and B. Large-budded cells (n > 100) were scored for two SPB signals. On average, ∼60% of large budded nbp1-td cells showed Spc42p-eqFP signal in the mother cell and in the bud. Of these cells, for example, only 28% of cells carried a Bbp1p-GFP signal in the bud. An even stronger reduction of the SPB signal in the bud was observed for Spc110p and Mps2p, whereas Cdc31p, Kar1p, Mps3p, Sfi1p, and Spc29p behaved similar to Spc42p. HB, half-bridge; CP, central plaque; IP, inner plaque.
Analysis of additional SPB components involved in the duplication process showed that SPB binding of Cdc31p, Kar1p, Mps3p, Sfi1p, and Spc29p were not affected by the lack of Nbp1p (Figure 8C). In contrast, binding of the inner plaque protein Spc110p to the newly formed SPB was strongly reduced in nbp1-td cells (Figure 8C). This result is in agreement with Nbp1p functioning in duplication plaque insertion because the nuclear inner plaque with Spc110p only assembles to the newly formed SPB after the insertion of the central plaque into the nuclear envelope (Adams and Kilmartin, 1999; Elliott et al., 1999).
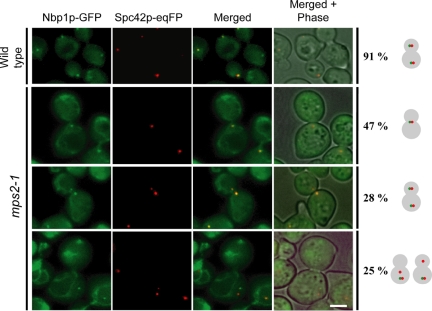
Nbp1p Binds with Reduced Efficiency to the New SPB of mps2-1 Cells
Binding of Mps2p to the SPB was most dramatically affected in nbp1-td cells (Figure 8), raising the possibility that Mps2p binds to SPBs via Nbp1p. Inactivation of Mps2p may therefore also affect SPB association of Nbp1p. To test this possibility, we examined the localization of Nbp1p-GFP in mps2-1, mps2-2, or bbp1-1 cells, which are mutated in the genes encoding components of the Mps2p-Bbp1p complex. Analysis of synchronized wild-type, mps2-1, mps2-2, and bbp1-1 cells showed that SPB localization of Nbp1p-GFP was only affected in mps2-1 cells (Figure 9). Of the 53% of large-budded mps2-1 cells with two Spc42-eqFP signals, about half of them carried only one Nbp1p-GFP signal. In most of these cells, the defective new SPB in the bud was devoid of an Nbp1p-GFP signal. Thus, SPB localization of Nbp1p is in part dependent on Mps2p, reinforcing the importance of the interaction of Nbp1p and Mps2p in SPB duplication.
Figure 9.
Localization of Nbp1p with SPBs is dependent on Mps2p. MPS2 (wild-type) and mps2-1 cells with NBP1-GFP SPC42-eqFP (YAY133 and YAY120, Table 1) were synchronized with α-factor at 23°C. After washing (t = 0), cells were shifted to 37°C and analyzed after 135 min by fluorescence microscopy for localization of Nbp1p-GFP at SPBs (marked by Spc42p-eqFP). Bar, 5 μm.
DISCUSSION
NBP1 Encodes a Newly Identified SPB Component
We identified NBP1 as a dosage suppressor of the conditional lethal ndc1-39 allele, and NBP1 also suppresses mutations in a number of genes that act in SPB duplication. This suggests that Nbp1p itself is a SPB component with a function in SPB duplication. Indeed, the localization of Nbp1p was placed at SPBs in a genome-wide screen of GFP-tagged ORFs (Huh et al., 2003), even though Nbp1p was not identified in a previous mass spectrometry study aimed at revealing SPB components (Wigge et al., 1998). However, the mass spectrometry study did not identify several peripheral SPB components including Mps3p, Sfi1p, Kar1p, and Ndc1p, possibly because of the nature of the SPB preparation and/or abundance of these components. Nonetheless, we show here that Nbp1p is a new part of the Mps2p-Bbp1p complex (Schramm et al., 2000) and that Nbp1p is required to insert the nascent SPB into the nuclear envelope, similar to Ndc1p, Mps2p, and Bbp1p (Winey et al., 1991, 1993; Schramm et al., 2000).
Besides SPB duplication defects, ndc1-39 mutants also fail in NPC assembly (Lau et al., 2004). It is likely that extra copies of NBP1 suppress only the SPB duplication defects in ndc1-39 mutants, because Nbp1p only localizes to the SPBs and not to the NPCs. The assertion that Nbp1p is at the SPB is based on fluorescence microscopy and immunoelectron microscopy with tagged alleles of the gene. However, both of these approaches have detection limits making it impossible to rule out other cellular localizations for Nbp1p. Furthermore, the very useful nbp1-td allele may not reveal other non-SPB-related functions of NBP1 because of the first cell cycle arrest of cells depleted for Nbp1p. The identification of other Nbp1p functions, if any, is likely to require new alleles and/or the identification and analysis of additional binding partners.
NBP1-like Genes
Using a BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/), we identified clear Nbp1p homologues in other budding yeast species (see Supplementary Figure S1) but not in fission yeast or higher eukaryotes, which is also true for Bbp1p. In addition, the protein encoded by S. cerevisiae YPR174C displays significant homology to the C-terminal half (residues 188-304) of Nbp1p (35% identical, 52% similar). Like Nbp1p, the protein encoded by YPR174C also contains a coiled-coil domain and has been localized to the SPB and nuclear periphery by fluorescence microscopy (Huh et al., 2003; Sundin et al., 2004). Both promoters of NBP1 and YPR174C contain a MluI “cell cycle box” and in both cases the mRNA expression peaks during G1 (Spellman et al., 1998; Shimizu et al., 2000), consistent with playing a role during SPB duplication. Furthermore, both Nbp1p and Ypr174p contain cyclin-dependent kinase consensus phosphorylation sites and they have been shown to function as in vitro substrates of the cyclin-dependent kinase Cdc28p (Ubersax et al., 2003). Interestingly, Nbp1p becomes phosphorylated in mid-S-phase (H. Maekawa, unpublished observations), consistent with the finding that Cdc28p-Clb5p may perform this phosphorylation. However, unlike NBP1, YPR174C is not essential and cell cycle analysis of ypr174cΔ cells did not show a SPB duplication defect (H. Maekawa, unpublished observations). Thus, the YPR174C gene product cannot be required for SPB duplication. Although NBP1 is involved in SPB duplication, the cellular function of YPR174C remains unknown.
Nbp1p Functions in SPB Duplication
As mentioned above, we used the nbp1-td allele to show that cells depleted of Nbp1p fail in SPB duplication and come to a mitotic arrest as is common for SPB mutants (reviewed in Jaspersen and Winey, 2004). Importantly, the terminal phenotype observed in these cells indicated that an aberrant, or “dead” SPB had been assembled, and this SPB morphology was confirmed by electron microscopy. Strains mutant in MPS2, NDC1, or BBP1 exhibit a similar defect in SPB duplication (Winey et al., 1991, 1993; Schramm et al., 2000), which has been interpreted as a defect in the insertion of the nascent SPB into the nuclear envelope. Our finding that Nbp1p is in a complex with Mps2p and Bbp1p and interacts with Ndc1p is consistent with the common phenotype of nbp1-td and conditional lethal bbp1, mps2, and ndc1 cells.
The failure to insert the nascent SPB results in the formation of a “dead” SPB that resides on the cytoplasmic face of the nuclear envelope. It is not thought to be a normal intermediate in SPB duplication, which can be assembled to the point of forming cytoplasmic microtubules (Winey et al., 1991, 1993; Schramm et al., 2000). To have this function, the outer plaque must be assembled, and the failure to form microtubules in the nucleus arises from the fact that “dead” SPB does not have access to the nucleus and lacks an inner plaque as revealed by electron microscopy (Winey et al., 1991, 1993; Schramm et al., 2000). The “dead” SPB formed in nbp1-td cells migrates into the new cell like the “dead” pole in other mutants that form this structure (Jaspersen and Winey, 2004), whereas the new SPB generally stays in the mother cell in wild-type cells (Pereira et al., 2001). This could arise from unusual mobility of the “dead” SPB because it not restrained by nuclear microtubules. Furthermore, the inner plaque component, Spc110p is absent from “dead” SPBs in bbp1-1 mutants (Schramm et al., 2000). We extend that analysis here to show that the “dead” SPBs in nbp1-td cells contain the central plaque components Spc29p and Spc42p, but lack the inner plaque component Spc110p, as expected. The “dead” SPBs have reduced levels of Nbp1p's binding partners, Mps2p and Bbp1p. Interestingly, the bridge components Cdc31p, Kar1p, Mps3p, and Sfi1p are all present at the “dead” SPB. The reduced levels of the Mps2p-Bbp1p complex may be sufficient to retain the half-bridge, and/or Ndc1p may mediate the interaction. Also, the fact that half-bridge is associated with the “dead” SPB at all may be a remnant of the attachment of the duplication plaque to the half-bridge such that the half-bridge is broken as the cytoplasmic microtubules pull on the “dead” SPB. Alternatively, the bridge may go through its normal separation process that requires mitotic CDK activity in the absence of SPB insertion. It would be intriguing if normal bridge severing occurred, because such a finding would indicate that some steps in the SPB cycle do not depend on completion of earlier steps. Furthermore, it would be interesting to analyze the composition of the “dead” SPBs in the different mutants that form these aberrant structures.
Although the precise mechanism of SPB insertion into the nuclear envelope is unknown, Adams and Kilmartin (1999) suggested fusion of the inner and outer nuclear membranes at the SPB-distal end of the bridge. We propose that during SPB duplication, Nbp1p interacts with the Mps2p-Bbp1p complex and likely with Ndc1p to promote fusion of the inner and outer nuclear membranes for the insertion of the duplication plaque into the nuclear envelope.
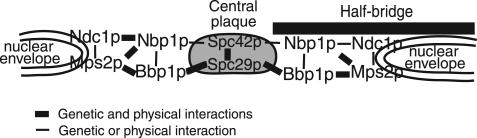
Beyond functioning in SPB insertion, Ndc1p, Mps2p, Bbp1p, and Nbp1p all remain at the SPB after assembly. It has been proposed that the Mps2p-Bbp1p complex serves as a tether to hold the SPB in place, where Mps2p is the membrane-bound component of the complex and Bbp1p is the bridge to the SPB via interaction with Spc29p (Schramm et al., 2000). It is possible that Nbp1p has a similar bridging function. Beyond being part of the Mps2p-Bbp1p complex, NBP1 displayed a genetic interaction with SPC42, and both genetic and two-hybrid interactions with NDC1. These interactions suggest that Nbp1p may bridge between the membrane proteins, Mps2p and Ndc1p, and/or bridge from the membrane proteins to Spc42p in the central plaque of the SPB (Figure 10). It is tempting to speculate that a complex including Nbp1p and Bbp1p form the “hooklike” structure between the central plaque and the nuclear envelope observed by high-voltage electron tomography (O'Toole et al., 1999; Schramm et al., 2000).
Figure 10.
Model of Nbp1p interactions at the SPB. The relative positions of Nbp1p, Ndc1p, Mps2p, and Bbp1p between the nuclear envelope and the SPB's central plaque. The cytoplasm and the nucleoplasm are oriented on the top and the bottom of the nuclear envelope, respectively. For simplicity, only the central plaque components Spc42p and Spc29p and the half-bridge of the SPB are shown. Ndc1p and Mps2p are integral membrane proteins at the nuclear periphery of the SPB where they interact with Nbp1p. Nbp1p also binds to Mps2p-Bbp1p complex.
Interestingly, we found that depletion of Nbp1p lead to the loss of Mps2p from already inserted SPBs, suggesting that Nbp1p is important in maintaining Mps2p at the SPB and that Bbp1p cannot retain Mps2p by itself. Moreover, because the already inserted SPBs of nbp1-td cells appear intact when examined by electron microscopy, it is likely that Mps2p and Nbp1p are only essential for duplication plaque insertion but not for maintaining SPBs in the nuclear envelope.
Finally, NBP1 is somewhat unusual among genes encoding SPB components in that it acts as a dosage suppressor of mutations in a variety of genes encoding SPB components required of SPB duplication. Furthermore, although Nbp1p is clearly part of the Mps2p-Bbp1p complex, the immunoelectron microscopy data suggests that it could be at other SPB substructures. Taken together, these results suggest Nbp1p could have a very central role in SPB duplication, possibly through interactions with multiple SPB components. New alleles of NBP1 and/or the identification and analysis of additional binding partners in the SPB will be required in order to reveal other non-SPB functions and additional SPB functions of Nbp1p.
Supplementary Material
Table 1.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| 1210 | MATamps2-1 his3Δ200 trp1Δ1 ura3-52 leu2,3-112 | McBratney and Winey (2002) |
| 2711 | MATandc1Δ::KanMX-ndc1-39-TRP1 ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 | Lau et al. (2004) |
| 3310 | MATa/α NBP1-GFP-His3MX6/NBP1-GFP-His3MX6 ade2Δ::hisG/ade2Δ::hisG his3Δ200/his3Δ200 leu2Δ0/leu2Δ0 lys2Δ0/lys2Δ0 met15Δ0/met15Δ0 trp1Δ63/trp1Δ63 ura3Δ0/ura3Δ0 | This study |
| 3322 | MATa/α NBP1-GFP-His3MX6/NBP1-GFP-His3MX6 NDC1-CFP-KanMX/NDC1-CFP-KanMX ade2/ade2 his3/his3 leu2/leu2 trp1/trp1 ura3/ura3 | This study |
| 3326 | MATa/α NBP1-GFP-His3MX6/NBP1-GFP-His3MX6 SPC42-CFP-KanMX/SPC42-CFP-KanMX ade2/ade2 his3/his3 leu2/leu2 lys2Δ0/+ trp1Δ63/+ ura3/ura3 | This study |
| 3445 | MATanbp1Δ::His3MX6 ade2Δ::hisG ade3Δ his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 + pALR10(ADE3-URA3)-NBP1 | This study |
| 3446 | MATα ndc1Δ::KanMX-ndc1-39-TRP1 ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 | Lau et al. (2004) |
| 3606 | MATanbp1-1 ndc1Δ::KanMX-ndc1-39-TRP1 ade2 his3 leu2 trp1 ura3 | This study |
| GPY658 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 | This study |
| PJ69-4a | MATatrp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. (1996) |
| PJ69-4α | MATα trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. (1996) |
| SHM1043 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 nbp1-td-hph | This study |
| SHM1097 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 CFP-TUB1-URA3 SPC72-YFP-kanMX4 | This study |
| SHM1098 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 nbp1-td-hph CFP-TUB1-URA3 SPC72-YFP-kanMX4 | This study |
| YAY072 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 nbp1-td-hph SPC42-eqFP611-nat SPC110-GFP-kanMX4 | This study |
| YAY073 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 SPC42-eqFP611-hph SPC110-GFP-kanMX4 | This study |
| YAY074 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 nbp1-td-hph SPC42-eqFP611-nat BBP1-GFP-kanMX4 | This study |
| YAY075 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 SPC42-eqFP611-hph BBP1-GFP-kanMX4 | This study |
| YAY078 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 nbp1-td-hph SPC42-eqFP611-nat SFI1-GFP-kanMX4 | This study |
| YAY079 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 SPC42-eqFP611-hph SFI1-GFP-kanMX4 | This study |
| YAY081 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 nbp1-td-hph SPC42-eqFP611-nat MPS3-GFP-kanMX4 | This study |
| YAY082 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 SPC42-eqFP611-hph MPS3-GFP-kanMX4 | This study |
| YAY091 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 SPC42-eqFP611-nat SPC29-GFP-kanMX4 | This study |
| YAY093 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 SPC42-eqFP611-nat MPS2-GFP-kanMX4 | This study |
| YAY098 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 nbp1-td-hph SPC42-eqFP611-nat SPC29-GFP-kanMX4 | This study |
| YAY101 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 nbp1-td-hph SPC42-eqFP611-nat MPS2-GFP-kanMX4 | This study |
| YAY103 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 nbp1-td-hph SPC42-eqFP611-nat GFP-KAR1-URA3 | This study |
| YAY104 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 SPC42-eqFP611-nat GFP-KAR1-URA3 | This study |
| YAY120 | MATaura3-52 leu2-3,112 his3Δ200 trp1Δ63 mps2-1 SPC42-eqFP611-hph NBP1-GFP-KanMX4 | This study |
| YAY133 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 NBP1-GFP-KanMX6 SPC42-eqFP611-His3MX6 | This study |
| YAY134 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 nbp1-td-hph SPC42-eqFP611-nat GFP-CDC31-LEU2 | This study |
| YAY135 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 SPC42-eqFP611-nat GFP-CDC31-LEU2 | This study |
| YAY143 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 NBP1-6HA-KanMX4 | This study |
| YAY144 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 nbp1-td-6HA-hph-KanMX4 | This study |
| YAY157 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 NBP1-GFP-hph SPC42-eqFP611-nat | This study |
| YAY158 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 pGal1-HA-UBR1-HIS3 nbp1-td-GFP-hph-klTRP1 SPC42-eqFP611-nat | This study |
| YAY167 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 bar1Δ::HisG pep4Δ::hphMX4 BBP1-6HA-klTRP1 | This study |
| YAY168 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 bar1Δ::HisG pep4Δ::hphMX4 MPS2-6HA-klTRP1 | This study |
| YAY169 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 bar1Δ::HisG pep4Δ::hphMX4 BBP1-6HA-klTRP1 NBP1-TAP-KanMX4 | This study |
| YAY170 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 bar1Δ::HisG pep4Δ::hphMX4 MPS2-6HA-klTRP1 NBP1-TAP-KanMX4 | This study |
| YYS113-71 | MATα leu2 his3 trp1 ura3 ade2 nbp1-1 | Shimizu et al. (2000) |
Acknowledgments
We thank Tony Hazburn, Stan Fields, and the Yeast Resource Center, an NCRR facility (P. I. Trisha Davis, P41 RR11823), for two-hybrid materials; Akihiko Kikuchi for providing the nbp1-1 strain, Lorraine Pillus for discussions and reagents, and Michele Jones and Sandi Jacobson for critical reading of this manuscript. We thank S. Murray for the help with the electron microscopy analysis of nbp1-td strains. This work was supported by National Institutes of Health (NIH) training grant GM-07135 (C.K.L.), a Leukemia and Lymphoma Society Fellowship (S.L.J.), a program grant of CR UK (E.S.), and by the NIH, having been initiated under GM-59992 and completed under GM-51312 (both to M.W.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-07-0668) on January 25, 2006.
Abbreviations used: CFP, cyan fluorescent protein; DAPI, 4′,6-diamidono-2-phenylindole; GFP, green fluorescent protein; NPC, nuclear pore complex; ORF, open reading frame; SPB, spindle pole body; ts, temperature-sensitive.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Adams, I. R., and Kilmartin, J. V. (1999). Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 145, 809-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, I. R., and Kilmartin, J. V. (2000). Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol. 10, 329-335. [DOI] [PubMed] [Google Scholar]
- Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (1998). Current Protocols in Molecular Biology, New York: John Wiley & Sons.
- Boeke, J. D., Trueheart, J., Natsoulis, G., and Fink, G. R. (1987). 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154, 164-175. [DOI] [PubMed] [Google Scholar]
- Byers, B., and Goetsch, L. (1975). Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124, 511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial, H. J., Giddings, T. H., Jr., Siewert, E. A., Hoyt, M. A., and Winey, M. (1999). Altered dosage of the Saccharomyces cerevisiae spindle pole body duplication gene, NDC1, leads to aneuploidy and polyploidy. Proc. Natl. Acad. Sci. USA 96, 10200-10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial, H. J., Rout, M. P., Giddings, T. H., and Winey, M. (1998). Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol. 143, 1789-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly, C., and Hieter, P. (1996). Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86, 275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen, R. J., Wu, P., and Varshavsky, A. (1994). Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 263, 1273-1276. [DOI] [PubMed] [Google Scholar]
- Elliott, S., Knop, M., Schlenstedt, G., and Schiebel, E. (1999). Spc29p is a component of the Spc110p-subcomplex and is essential for spindle pole body duplication. Proc. Natl. Acad. Sci. USA 96, 6205-6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk, H. A., Mattison, C. P., and Winey, M. (2002). Centrosomes and tumour suppressors. Curr. Opin. Cell Biol. 14, 700-705. [DOI] [PubMed] [Google Scholar]
- Giddings, T. H., Jr., O'Toole, E. T., Morphew, M., Mastronarde, D. N., McIntosh, J. R., and Winey, M. (2001). Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol. 67, 27-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G. R. (2002). Guide to yeast genetics and molecular and cell biology. Methods Enzymol. 350, 1-623. [PubMed] [Google Scholar]
- Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686-691. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., Oritz, J., Tanaka, T. U., Lechner, J., and Schiebel, E. (2002). Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21, 181-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen, S. L., and Winey, M. (2004). The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 20, 1-28. [DOI] [PubMed] [Google Scholar]
- Kanemaki, M., Sanchez-Diaz, A., Gambus, A., and Labib, K. (2003). Functional proteomics by induced proteolysis in vivo identifies novel DNA-replication proteins. Nature 423, 720-724. [DOI] [PubMed] [Google Scholar]
- Knop, M., Siegers, K., Pereira, G., Zachariae, W., Winsor, B., Nasmyth, K., and Schiebel, E. (1999). Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963-972. [DOI] [PubMed] [Google Scholar]
- Kunkel, T. A., Roberts, J. D., and Zakour, R. A. (1987). Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154, 367-382. [DOI] [PubMed] [Google Scholar]
- Labib, K., Tercero, J. A., and Diffley, J.F.X. (2000). Uninterrupted MCM2-7 function required for DNA replication fork progression. Science 288, 1643-1647. [DOI] [PubMed] [Google Scholar]
- Lau, C. K., Giddings, T. H., Jr., and Winey, M. (2004). A novel allele of Saccharomyces cerevisiae NDC1 reveals a potential role for the spindle pole body component Ndc1p in nuclear pore assembly. Eukaryot. Cell 3, 447-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- McBratney, S., and Winey, M. (2002). Mutant membrane protein of the budding yeast spindle pole body is targeted to the endoplasmic reticulum degradation pathway. Genetics 162, 567-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, E. G., Snydsman, B. E., Novik, I., Hailey, D. W., Gestaut, D. R., Niemann, C. A., O'Toole, E., T., Giddings, T. H., Jr., Sundin, B. A., and Davis, T. N. (2005). The organization of the core proteins of the yeast spindle pole body. Mol. Biol. Cell 16, 3341-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Centeno, M. C., McBratney, S., Monterrosa, A., Byers, B., Mann, C., and Winey, M. (1999). Saccharomyces cerevisiae MPS2 encodes a membrane protein localized at the spindle pole body and the nuclear envelope. Mol. Biol. Cell 10, 2393-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, E. T., Winey, M., and McIntosh, J. R. (1999). High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 10, 2017-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., Tanaka, T. U., Nasmyth, K., and Schiebel, E. (2001). Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 20, 6359-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm, C., Elliott, S., Shevchenko, A., Shevchenko, A., and Schiebel, E. (2000). The Bbp1p-Mps2p complex connects the SPB to the nuclear envelope and is essential for SPB duplication. EMBO J. 19, 421-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, Y., Akashi, T., Okuda, A., Kikuchi, A., and Fukui, K. (2000). NBP1 (Nap1 binding protein 1), an essential gene for G2/M transition of Saccharomyces cerevisiae, encodes a protein of distinct sub-nuclear localization. Gene 246, 395-404. [DOI] [PubMed] [Google Scholar]
- Spellman, P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., Brown, P. O., Botstein, D., and Futcher, B. (1998). Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9, 3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin, B. A., Chiu, C. H., Riffle, M., Davis, T. N., and Muller, E. G. (2004). Localization of proteins that are coordinately expressed with Cln2 during the cell cycle. Yeast 21, 793-800. [DOI] [PubMed] [Google Scholar]
- Ubersax, J. A., Woodbury, E. L., Quang, P. N., Paraz, M., Blethrow, J. D., Shah, K., Shokat, K. M., and Morgan, D. O. (2003). Targets of the cyclin-dependent kinase Cdk1. Nature 425, 859-864. [DOI] [PubMed] [Google Scholar]
- Uetz, P. et al. (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623-627. [DOI] [PubMed] [Google Scholar]
- Wigge, P. A., Jensen, O. N., Holmes, S., Soues, S., Mann, M., and Kilmartin, J. V. (1998). Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141, 967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., Goetsch, L., Baum, P., and Byers, B. (1991). MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 114, 745-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., Hoyt, M. A., Chan, C., Goetsch, L., Botstein, D., and Byers, B. (1993). NDC1: a nuclear periphery component required for yeast spindle pole body duplication. J. Cell Biol. 122, 743-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.