Abstract
Bacillus subtilis exhibits a complex adaptive response to low levels of peroxides. We used global transcriptional profiling to monitor the magnitude and kinetics of changes in the mRNA population after exposure to either hydrogen peroxide (H2O2) or tert-butyl peroxide (t-buOOH). The peroxide stimulons could be largely accounted for by three regulons controlled by the PerR, σB, and OhrR transcription factors. Three members of the PerR regulon (katA, mrgA, and zosA) were strongly induced by H2O2 and weakly induced by t-buOOH. The remaining members of the PerR regulon were only modestly up-regulated by peroxide treatment. Overall, the magnitude of peroxide induction of PerR regulon genes corresponded well with the extent of derepression in a perR mutant strain. The σB regulon was activated by 58 μM H2O2 but not by 8 μM H2O2 and was strongly activated by either t-buOOH or, in a control experiment, tert-butyl alcohol. Apart from the σB regulon there was a single gene, ohrA, that was strongly and rapidly induced by t-buOOH exposure. This gene, controlled by the peroxide-sensing repressor OhrR, was not induced by any of the other conditions tested.
Treatment of bacteria with low levels of oxidants typically results in a classic adaptive response: the treated cells have a greatly enhanced ability to survive subsequent challenge with an otherwise lethal dose of the same oxidant or a related oxidant. This adaptive response is coordinated by the action of transcription factors that sense oxidative stress and regulate the expression of appropriate defensive and repair functions (34).
In Escherichia coli resistance to oxidative stress is largely coordinated by two systems. OxyR activates genes in response to either peroxide stress or changes in the thiol-disulfide status, while the SoxRS system controls genes in response to reactive free radicals, such as superoxide anion and nitric oxide (29, 35). Proteins induced by oxidative stress were initially catalogued by using one- and two-dimensional polyacrylamide gel electrophoresis (PAGE) (12, 15, 26). More recently, the OxyR and SoxRS regulons have been defined by using computational approaches to identify regulator binding sites together with transcriptional profiling (28, 39, 40).
Bacillus subtilis also displays an adaptive response to low levels of oxidants. Enzymatic assays have revealed that the major catalase present in growing cells (KatA) is strongly induced by treatment with peroxides (3, 25). With one-dimensional sodium dodecyl sulfate-PAGE several other proteins were also found to be strongly induced by 50 μM H2O2 (13). The most prominent members of the peroxide stimulon include the two subunits of alkyl hydroperoxide reductase (AhpC and AhpF), KatA, and the DNA-binding protein MrgA (9, 20). Similar conclusions were reached in studies of the protein profile of cells treated with H2O2, as visualized by two-dimensional PAGE (2).
Insight into the mechanisms controlling the inducible peroxide stress response in B. subtilis originally emerged from analysis of mrgA, which encodes a member of the Dps family of protective DNA-binding proteins (7, 9). Expression of mrgA is metalloregulated: repression is elicited by growth with either supplemental manganese or iron (8). This repression is mediated by PerR, a member of the ferric uptake repressor (Fur) family of metal-dependent DNA-binding proteins (5). The PerR regulon is now known to include mrgA, katA, ahpCF, a heme biosynthesis operon (hemAXCDBL), a zinc uptake system (zosA), fur, and perR itself (23). Studies with lacZ reporter fusions have demonstrated that expression of these genes is repressed by manganese and that some, but not all, of the genes are also repressed by iron (18). Some of these genes (mrgA, katA, and zosA) are strongly induced by H2O2, while others (hemA operon, ahpCF, and perR) show less induction. The fur gene is not induced by H2O2; thus, not all members of the PerR regulon are members of the peroxide stimulon (18).
To gain a global perspective on the transcriptional responses to peroxide stress, we monitored gene expression in stressed cells using DNA microarrays. Since many stress responses involve transient changes in gene expression, we isolated RNA at several times following imposition of stress and compared the resulting transcriptional profiles with the profiles of unstressed control cells. In addition, in this paper we describe a direct comparison between wild-type and perR mutant cells. A comparison of the responses to H2O2, tert-butyl peroxide (t-buOOH), and tert-butyl alcohol (t-buOH) and previously reported studies of the heat shock stimulon (22) revealed three distinct classes of induced genes: the PerR, σB, and OhrR regulons.
MATERIALS AND METHODS
Strains and growth conditions.
B. subtilis CU1065 (W168 attSPβ trpC2) was inoculated into 25 ml of prewarmed (37°C) Bacto Mueller-Hinton broth (MHB) (Difco, Detroit, Mich.) from a fresh (overnight) Luria-Bertani agar plate. The flask was shaken at 250 rpm on a rotary platform at 37°C until the optical density at 600 nm (OD600) was 0.5 to 1.0. This seeder flask was used to inoculate a second flask of prewarmed MHB (50 ml in a 500-ml flask) to an OD600 of 0.2. This flask was grown until the OD600 was 1.0, at which time aliquots were removed and used as the time-zero samples. For the H2O2 stress experiments, 100 μl of appropriately diluted H2O2 was added immediately following removal of a time-zero aliquot to give a concentration of either 8 or 58 μM. Water (100 μl) was added to a mock treatment control flask. For the organic peroxide stress experiments, t-buOOH or t-buOH was added to a concentration of 100 μM (total amount added, 100 μl). Each flask was incubated on the shaker, and samples used for RNA isolation were removed at the times indicated below. The initial sampling of a culture was performed rapidly so that cells were frozen in liquid nitrogen within 40 s of removal from the culture.
Growth of the perR mutant strain.
Cultures were inoculated as described above by using aliquots from an MHB plate containing either CU1065 or an isogenic perR strain (5) that had been grown overnight. The perR inoculum contained only small colonies, which was indicative of a lack of pseudorevertants (6). Each flask was shaken at 250 rpm on a rotary platform at 37°C. Samples used for RNA isolation were collected at an OD600 of 1.0. Prior observation had shown that the cultures exhibited exponential growth at this cell density (data not shown). CU1065 reached an OD600 of 1.0 in ∼3 h, while a similar inoculum of the isogenic perR strain took ∼7.5 h to reach a similar cell density. After the samples used for RNA isolation were removed, the cultures were diluted and plated to obtain single colonies on MHB agar. Following overnight growth the perR strain plate was examined for large pseudorevertant colonies, and individual colonies were tested for catalase activity to confirm that the perR strain culture contained undetectable levels of catalase-negative pseudorevertants (6). RNA was isolated as described above.
Microarray analysis, cDNA labeling, slide hybridization, data collection, and normalization.
The procedures used for microarray analysis, cDNA labeling, slide hybridization, data collection, and normalization have been described previously (22). Time-zero cDNA samples were labeled with Cy3, and samples collected at subsequent times were labeled with Cy5. Slides were scanned and analyzed with a confocal laser scanner and a software package (Axon GenePix 4000A and GenePix Pro 3.0; Axon Instruments, Inc., Foster City, Calif.). Normalization was computed by using the algorithm incorporated into the Stanford Microarray Database (33), and all data are archived in this database. In general, fold induction was determined by comparison of the peroxide-treated samples and the time-zero control samples. However, where indicated below, the experimental data were corrected for time-dependent changes in the samples by using data for the parallel untreated samples. For example, the log-transformed gene induction ratios after mock treatment were subtracted from the log-transformed induction ratios after H2O2 treatment. To identify and display groups of coregulated genes for this set of treatment regimens, we used hierarchical clustering as previously described (14). For the 8 and 58 μM H2O2 regimens, genes were considered to be differentially regulated if after 3, 10, or 20 min (i) the absolute value of the mean of the replicate log2 treatment ratios minus the mean of the log2 mock treatment ratios exceeded 1.585 (corresponding to a threefold difference) and (ii) significance analysis of microarrays (SAM) of all available treatment replicates versus mock replicates in a two-class, unpaired data design indicated statistical significance at the threefold level when the delta parameter was adjusted to give a median number of false significants typically less than 5% of the total number of selected genes but allowing for at least one false significant in a set regardless of its size (36). A similar analysis was carried out for the t-buOOH-versus-t-buOH experiments, with t-buOH being considered the mock treatment in the two-class design. In the perR mutant studies, reciprocal cDNA labeling was conducted with either CU1065 Cy3 versus perR Cy5 or CU1065 Cy5 versus perR Cy3. A total of six slides (three for each labeling regimen) were analyzed. In this case, two-class, unpaired data SAM was performed by using normalized intensity data rather than ratio data, and the delta parameter was adjusted to give a median number of false significants < 2. Supplemental tables and complete data sets are available at http://www.micro.cornell.edu/faculty.JHelmann.html.
RESULTS
Overview of experimental design.
We monitored the global transcriptional profile of B. subtilis under peroxide stress conditions using DNA microarray-based measurements. To monitor changes in gene expression over time, we compared the mRNA profiles obtained 3, 10, 20, and 40 min after stress was imposed with the profile of a time-zero (prestress) control sample. Changes in gene expression were monitored for three different peroxide stress conditions: low (8 μM) and intermediate (58 μM) levels of H2O2 and 100 μM t-buOOH. Note that 58 μM H2O2 is designated an intermediate level of stress to distinguish our results from those of experiments performed by other groups with much higher levels of H2O2 (39). As controls we also monitored gene expression in parallel cultures treated with the corresponding alcohols (H2O and t-buOH). In addition, we compared the transcriptional profile of a strain with a disruption in perR with the profile of the corresponding wild-type control. The majority of comparisons were performed on at least three independent slides, and while the SAM was based on all available replicates, the geometric means of the experimental ratios were, for reasons of convenience, used to generate the figures.
H2O2 stress stimulon.
To define the major groups of genes most affected by peroxide stress, we performed a cluster analysis using the software developed by Eisen et al. (14). For this analysis we clustered the data by using data sets obtained for induction by H2O2 at concentrations of 8 and 58 μM (corrected for changes in gene expression that occurred as a function of time in the parallel untreated cultures), by t-buOOH, and by t-buOH and for the changes noted in the perR mutant compared with data for the wild type. In addition, we included the previously described results obtained for the heat shock stimulon (22) to aid in clustering, although these data are not incorporated in Fig. 1.
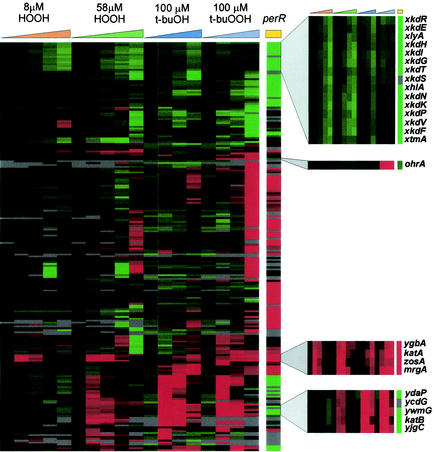
FIG. 1.
Hierarchical cluster analysis (Eisen plot) of selected genes affected by peroxide stress. The hybridization ratios are displayed colorimetrically; genes induced by the experimental treatment are indicated by shades of red, and genes with reduced expression are indicated by shades of green. Genes were selected by being significantly altered in expression under at least one experimental condition (at least threefold induction or repression and SAM positive; the 40-min data were excluded in this analysis), and the data were filtered to remove genes that did not produce a measurable induction value (above the background value) in at least 10 of the 19 columns. Four clusters of genes discussed in the text are highlighted with zoom boxes; from top to bottom these are the xkd cluster of PBSX genes, the single gene strongly and selectively induced by t-buOOH stress (ohrA), the PerR regulon cluster (including ygbA) (see text), and a subset of the genes of the σB general stress regulon.
The cluster analysis clearly grouped a subset of the PerR regulon genes as the genes that were strongly induced by both low and intermediate levels of H2O2 and were derepressed in the perR mutant (Fig. 1). Only four genes (ygbA, katA, zosA, and mrgA) were strongly and rapidly up-regulated by the low level of H2O2 (Fig. 1, third zoom box). Three of these genes are known members of the PerR regulon, whereas peroxide induction of ygbA has not been reported previously. The ygbA gene (also called ssuA) is the second gene in an operon involved in sulfonate transport that is transcribed divergently from the strongly induced katA gene (37). Since other genes in this operon are not peroxide inducible, the apparent induction noted here could have been an artifact (e.g., there could have been cross-hybridization or contamination of the ygbA PCR product with longer products extending into katA). A second, much larger cluster of genes was induced by the intermediate level of H2O2 but not by the low level of H2O2 and was further distinguished by strong and rapid induction in response to t-buOOH and t-buOH. This cluster corresponds to the large σB general stress response regulon (21, 27, 30, 31), and only a subset (ydaP though yjgC) is shown here (Fig. 1, fourth zoom box). Among the genes most dramatically down-regulated following imposition of peroxide stress are the genes comprising the PBSX phage-like element (xkdR through xtmA) (Fig. 1, first zoom box). Expression of these genes is also greatly reduced in a perR mutant (see below).
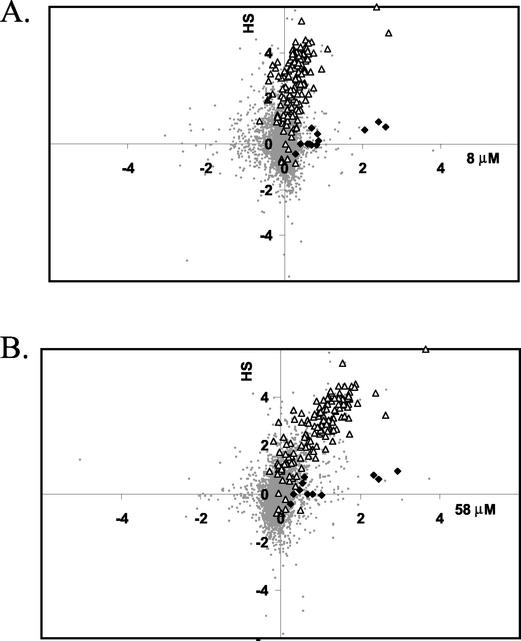
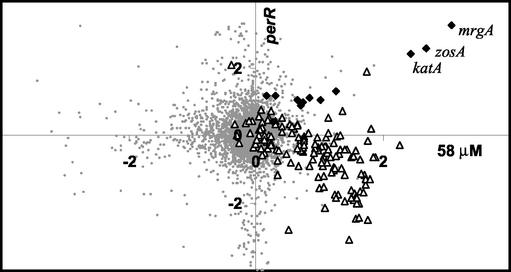
As an alternative method to visualize clusters of coregulated genes, we used a graphical approach to compare changes in gene expression under two different conditions. Since the general stress response is also strongly induced by heat shock, as we documented previously (22), we plotted the log2 values of the ratios of induction by H2O2 (relative to the untreated control at the same time) versus the log2 values of the ratios of induction by heat stress. Since the kinetic analyses revealed that both responses were strongly and selectively induced at the earliest time point (3 min), we focused on these data for comparison. The resulting graphs revealed two clusters of genes induced by both treatments (Fig. 2, upper right quadrants). One set of genes was strongly induced by either 8 or 58 μM H2O2 but only weakly induced by heat stress. These genes include mrgA, katA, and zosA and define a subset of the PerR regulon (23). A second large set of genes, representing the bulk of the heat shock stimulon, was induced by 58 μM H2O2 (Fig. 2B) but was only weakly affected by 8 μM H2O2 (Fig. 2A). This group includes many known and putative members of the large general stress regulon controlled by σB (21, 30).
FIG. 2.
Graphical comparison of the H2O2 and heat shock stimulons. The log2 of the induction ratio at 3 min is compared for a 48°C heat shock (HS) (y axis) and exposure to either 8 μM H2O2 (A) or 58 μM H2O2 (B) (x axis). Members of the PerR (solid diamonds) and σB (open triangles) regulons are highlighted and map, in general, to the upper right quadrant (corresponding to genes that have induction ratios of >1 for both heat shock and H2O2 treatment). All other gene signals are represented by small gray circles. Note that this and other two-dimensional graphical displays were generated without filtration of the data to remove low-quality and nonreproducible signals, so some of the background signals (small gray circles) that appear to represent highly regulated genes are not significant.
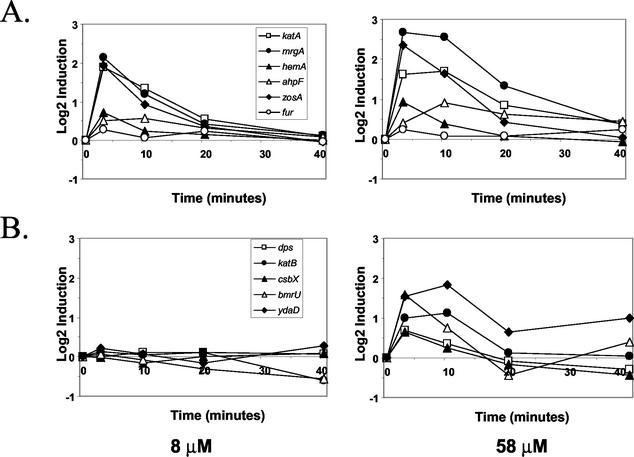
Analysis of the kinetics of the transcriptional responses to H2O2 revealed that both the PerR and σB components of the H2O2 stimulon were induced transiently (Fig. 3). Most σB-dependent general stress genes were maximally induced at 3 min following exposure to 58 μM H2O2, and the mRNA levels declined by 10 min following exposure. This was consistent with the transient induction of the σB regulon in response to heat shock and likely reflected the rapid induction of negative regulatory factors, including RsbX (38). The PerR-controlled stress response was also transient, with the maximal mRNA levels occurring at either 3 or 10 min following exposure.
FIG. 3.
Comparison of the time courses of expression for selected members of the PerR (A) and σB (B) regulons. The log2 fold induction (relative to the untreated control culture at the same time) is plotted for cells exposed to either a low (8 μM) or an intermediate (58 μM) level of H2O2.
PerR regulon.
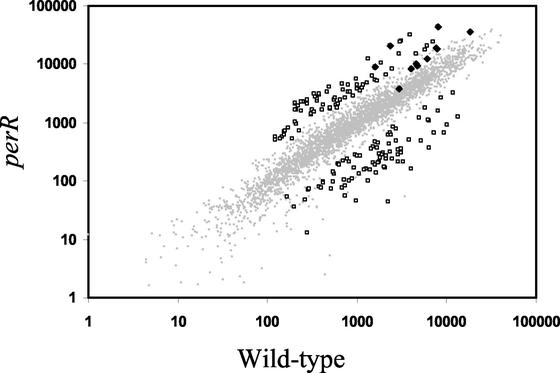
The role of PerR in the regulation of catalase (katA), mrgA, ahpCF, zosA, and the heme biosynthesis operon has been reported previously (4, 9, 19). To identify additional members of the PerR regulon, we compared the transcriptional profiles of perR mutant and wild-type cells grown to the late logarithmic phase (OD600, ∼1.0) (Fig. 4). Our slides contained DNA probes for 90% (3,703 of ∼4,100) of B. subtilis genes (22), and hybridization signals significantly above the background level were detected for 60 to 70% of these genes. In the comparisons of the perR mutant and the wild type, we calculated expression levels for ∼2,400 genes, and 8% of these levels were significantly altered in the mutant strain (Fig. 4). Expression of ∼75 genes (including known PerR regulon members katA, mrgA, and zosA) was significantly elevated in the perR mutant (Table 1), while expression of another ∼120 genes was decreased.
FIG. 4.
Gene expression in the perR mutant compared to gene expression in the isogenic wild-type strain. Relative hybridization intensities (mean normalized hybridization quanta) are plotted for genes as measured for the wild-type (x axis) and perR mutant (y axis) strains. The genes that differ significantly (based on a threefold cutoff and SAM [see Materials and Methods]) in the two strains are indicated by open squares. The members of the PerR regulon are indicated by solid diamonds. Of these, only katA, mrgA, and zosA are derepressed more than threefold. Table 1 provides a summary of the genes that are significantly up-regulated in the perR mutant.
TABLE 1.
Genes derepressed in the perR mutant strain
| Gene(s) | Foldinductiona | Regulator(s) |
|---|---|---|
| comER | 9.90 | ComK |
| yybF | 9.81 | |
| mrgA | 9.64 | PerR |
| ywfM | 8.94 | |
| comEA | 8.27 | ComK |
| comG(ABCDEFG) | 7.93 ± 1.74 | ComK |
| melA | 7.31 | |
| cwlJ | 7.64 | σE |
| gbsAB | 7.62 ± .52 | Osmotic stress |
| msmRE amyDC | 7.44 ± 1.67 | |
| dppABCDE | 7.04 ± .81 | CodY |
| zosA | 5.97 | PerR |
| appDF | 5.53 ± .34 | Hpr |
| katA | 5.34 | PerR |
| ilvBC leuBC | 5.01 ± .71 | |
| ureABC | 4.96 ± 1.4 | CodY, GlnR, TnrA |
| yckDE | 4.96 ± .10 | |
| yybl | 4.92 | |
| appBC | 4.89 ± .90 | |
| yxbC | 4.60 | |
| yxbBA | 4.50 ± .10 | |
| yufN | 4.44 | |
| yvaWY | 4.36 ± .20 | |
| yhdG | 4.27 | |
| sipT | 4.25 | |
| ygbA | 4.18 | |
| ppsABCDE | 3.93 ± .52 | |
| yuiA | 3.90 | |
| ykvKLM | 3.83 ± .60 | |
| yheK | 3.76 | |
| yisS | 3.67 | |
| ykfBCD | 3.67 ± .34 | |
| ywcE | 3.50 | |
| yoeB | 3.29 | |
| yhdC | 3.27 | |
| bioAFDB | 3.26 ± .11 | |
| rapH | 3.21 | |
| yybG | 3.14 | |
| opuBB | 3.14 | Osmotic stress |
| ytpQ | 3.10 | |
| gltB | 3.07 | |
| ywfH | 3.07 | |
| yqzE | 2.88 |
Induction values for genes in the same operon were used to determine the mean and standard deviation for the operon. All individual gene values are averages of induction ratios computed from six separate slides (see Materials and Methods).
To identify additional members of the PerR regulon, we compared the effects of 58 μM H2O2 and the perR mutation in a two-dimensional plot (Fig. 5). Known members of the PerR regulon were induced by H2O2 and derepressed by the perR mutation (Fig. 5, upper right quadrant). Remarkably, there was an excellent correlation between induction by 58 μM H2O2 (at 3 min) and the extent of derepression in the perR mutant (correlation coefficient [R2], 0.9). This suggests that PerR repression was completely relieved under these conditions, and the difference in the magnitude of induction may have resulted primarily from differences in the extent to which PerR affected gene expression. Several other genes (including ygbA, yerL, opuBC, and ureB) also mapped to this region of the graph, but these genes were not associated with Per boxes and the effects may have been indirect. Note that the majority of the genes up-regulated in the perR mutant were not peroxide inducible (Fig. 5). Furthermore, the expression of many genes that were induced by 58 μM H2O2 (including many members of the σB regulon, as noted above) was decreased in the perR mutant, and these genes clustered in the lower right quadrant in Fig. 5.
FIG. 5.
Graphical comparison of the H2O2 stimulon and the effects of the perR mutation. The log2 of the induction ratio at 3 min after exposure to 58 μM H2O2 (x axis) is compared with the log2 of the ratio of induction in the perR mutant to induction in the wild type (y axis). Members of the PerR and σB regulons are indicated by solid diamonds and open triangles, respectively. The three most strongly regulated members of the PerR regulon are labeled. Note that PerR-regulated genes are in the upper right quadrant (these genes derepressed in the perR mutant and peroxide inducible in the wild type).
Many of the observed changes in the perR mutant are likely to be indirect effects (Table 1). For example, the perR mutant may be stressed by the production of high levels of catalase, which could deplete heme pools or affect global iron homeostasis. In addition, the lower growth rate of the mutant affects the expression of many metabolic genes. Many of the 75 genes up-regulated in the perR mutant, including genes associated with competence, nutrient uptake, and the CodY regulon, are associated with the transition phase. The perR mutant strain also had significantly decreased expression of many genes, including the σB regulon genes (Fig. 5), several genes of the Fur regulon (ykuNO, yxeB, ywbL, and ywbN) (1), and the PBSX defective prophage (24). While the origins of the latter effect are unclear, PBSX is known to be induced by 100 μM H2O2 (12). Our results suggest that PerR or a PerR-regulated gene may act as a regulator of gene expression for PBSX.
t-buOOH stimulon.
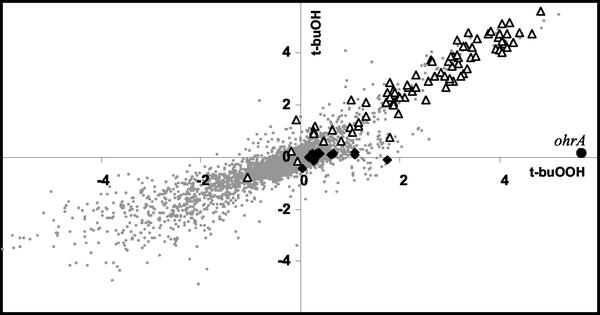
We next investigated the transcriptional response elicited by exposure of cells to t-buOOH. Within 3 min of exposure to t-buOOH, more than 100 genes were induced at least threefold compared to the expression in the time-zero sample. At subsequent times many more changes in gene expression became apparent (Fig. 1), but these appeared to be secondary effects and were not analyzed in detail. As a control, we also determined the time course of transcriptional changes elicited by exposure to the corresponding alcohol, t-buOH. A two-dimensional comparison of the responses revealed a remarkable pattern: the vast majority of the genes induced by t-buOOH were induced equally by t-buOH (Fig. 6). Not surprisingly, most of these genes belong to the σB regulon, which is strongly induced by ethanol (31).
FIG. 6.
Graphical comparison of the t-buOOH and t-buOH stimulons. The log2 of the induction ratio at 3 min after exposure to 100 μM t-buOOH (x axis) is compared with the log2 of the induction ratio after exposure to 100 μM t-buOH (y axis). Members of the PerR and σB regulons are indicated by solid diamonds and open triangles, respectively. The ohrA gene (indicated by a solid circle) is strongly and specifically induced by t-buOOH. Note that the σB regulon defines a line with a slope near 1, which is indicative of equivalent induction by t-buOOH and t-buOH.
There are also several genes that were induced by t-buOOH but not by t-buOH, and these genes are therefore candidates for an organic peroxide stimulon. Most notably, the ohrA gene was strongly and specifically induced by t-buOOH (Fig. 1, second zoom box, and Fig. 6). Expression of ohrA is regulated by the peroxide-sensing transcription factor, OhrR (16, 17). Other genes that were selectively induced by t-buOOH (although no gene was induced as dramatically as ohrA) include the PerR regulon member mrgA, the thioredoxin B gene (trxB), and at least two genes of the ArsR-regulated ars operon (32).
Three peroxide-induced regulons.
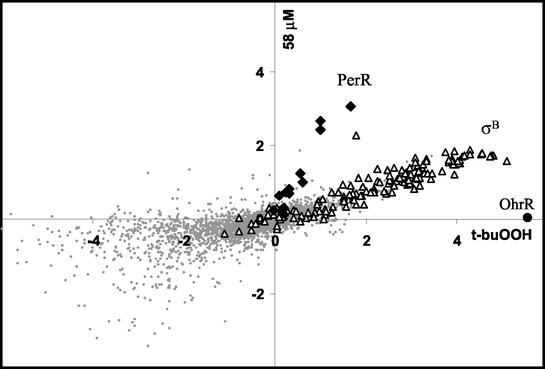
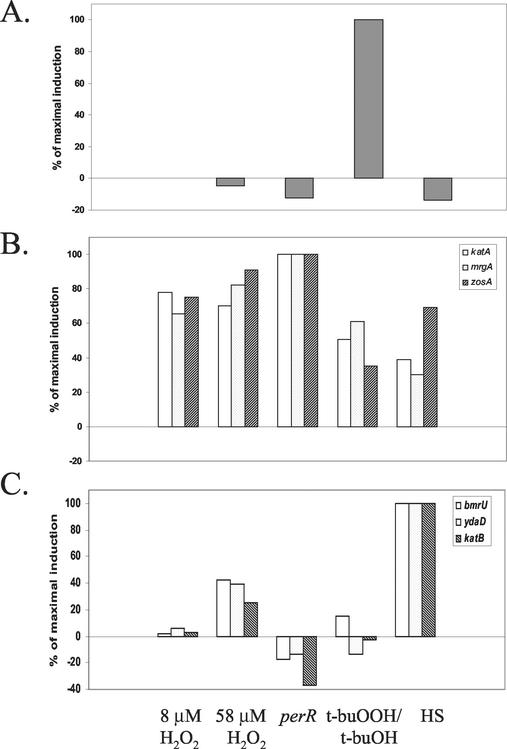
Overall, our analyses led us to conclude that the peroxide stimulon has three major components: the PerR, σB, and OhrR regulons. These three regulons can be clearly visualized by comparing the transcriptional responses (after 3 min) to 58 μM H2O2 with the responses elicited by 100 μM t-buOOH (Fig. 7). Three distinct sets of genes clearly emerged, as follows: the OhrR-regulated gene, ohrA, was strongly induced by t-buOOH but not by H2O2; the σB regulon was induced by t-buOOH more than it was induced by H2O2; and the PerR-regulated genes were induced by H2O2 more than they were induced by t-buOOH. We summarized the selectivity of these transcriptional responses by comparing the efficacies of various inducers for representative members of each regulon (Fig. 8).
FIG. 7.
Graphical comparison of the t-buOOH and H2O2 stimulons. The log2 of the induction ratio at 3 min after exposure to 100 μM t-buOOH (x axis) is compared with the log2 of the induction ratio after exposure to 58 μM H2O2 (y axis). Genes of the PerR and σB regulons are indicated by solid diamonds and open triangles, respectively, and ohrA is indicated by a solid circle. The regulators responsible for controlling the clusters of genes (PerR, σB, and OhrR) are indicated.
FIG. 8.
Selectivity of induction of different genes by various stress conditions. The extent of induction (percentage of the maximal value) is compared for ohrA (A), three members of the PerR regulon (katA, mrgA, and zosA) (B), and three members of the σB regulon (bmrU, ydaD, and katB) (C). The conditions tested included 8 and 58 μM H2O2, the perR mutant strain, t-buOOH (induction by t-buOH was first subtracted), and heat shock (HS).
DISCUSSION
Here we provide a global overview of the major transcriptional changes elicited by various peroxidative stress conditions. Our findings indicate that PerR is the major regulator of genes induced by low levels of H2O2, while OhrR regulates the single gene that is most rapidly and selectively induced by the model organic peroxide t-buOOH. However, the largest single group of genes induced by these treatments is the σB-dependent general stress regulon (30). Indeed, knowledge about the general stress response regulon, as deduced both from previous studies of the heat shock stimulon (22) and from extensive characterization of the σB regulon (21, 30), provided important background for our analysis.
Our work also illustrates the value of monitoring the kinetics of global changes in gene expression following application of a stress. Many of the most dramatic transcriptional effects were maximal within 3 min of application of the stress conditions, and there was an often rapid return to lower expression levels within 10 to 20 min. The transient nature of the σB-dependent general stress response is well known, but transient induction of the PerR and OhrR regulons has not been well documented previously. Thus, studies based on a single time point may miss major parts of the transcriptional response.
Our results emphasize the importance of control experiments for interpreting transcriptional profiles. We found, for example, that the vast majority of the transcriptional changes noted 40 min after H2O2 treatment were also observed in mock (H2O)-treated samples and were due to a growth phase transition rather than any effect of the peroxide treatment. Similarly, the comparison of the t-buOOH and t-buOH stimulons (Fig. 6) highlighted the need to use appropriate controls when the effects of particular classes of compounds are investigated. Indeed, we cannot exclude the possibility that t-buOOH is an inducer of the σB regulon because it is reduced in vivo to t-buOH. However, the rapid kinetics of these transcriptional responses make it seem unlikely that reduction to the alcohol is necessary for induction.
While transcriptional profiling is an extremely powerful tool for monitoring changes in gene expression after a change in culture conditions, the comparison of mutant and wild-type strains presented additional challenges. In this case, the perR mutant and wild-type strains differed significantly in growth rate and physiological state. Despite these differences, the perR mutant clearly revealed derepression of known PerR-repressed target genes. Remarkably, the extent of derepression in the samples corresponded very well with the fold induction 3 min after treatment with H2O2 (Fig. 5). This correlation suggests that the magnitude of peroxide induction of different genes reflects the extent to which PerR represses gene expression. As noted elsewhere, the fur gene is unusual among PerR-repressed genes in that it is not peroxide inducible under these or any other conditions tested (18). While not apparent from the transcriptional profiling data, PerR does mediate an approximately fourfold repression of fur in response to Mn(II) (18).
It is instructive to compare the B. subtilis and E. coli peroxide stress responses. In E. coli, transcriptional profiling was performed with cells treated with much higher levels of H2O2 (1 mM), in part because lower levels led to only transient changes in gene expression (39). Under these conditions 140 genes were induced approximately fourfold in the wild-type strain, and an even greater number of genes were induced in an oxyR mutant strain. The OxyR regulon includes genes with protective and detoxification functions, such as katG (hydroperoxidase I), ahpCF (alkyl hydroperoxide reductase), and dps (DNA-binding protein). In addition, OxyR activates transcription of genes that maintain intracellular thiols in their reduced states, including gorA (gluthathione reductase), grxA (glutaredoxin), and trxA (thioredoxin 2). In aerobically growing E. coli the concentration of intracellular H2O2 is maintained at levels near 20 nM by the potent peroxidase activity of Ahp (10, 11). Catalase peroxidase appears to be most important in detoxifying higher levels of peroxides and has the added advantage of being active even in energy-depleted cells that may lack sufficient reducing capacity to maintain optimal Ahp activity. Ahp itself was originally characterized as the major resistance factor protecting cells against alkyl peroxides. However, Ahp is also active with H2O2, and this may be the physiologically relevant substrate (10).
The PerR regulon differs from the regulon controlled by OxyR in several respects. PerR does not appear to control functions that might be involved in maintaining intracellular thiols in a reduced state. Although B. subtilis encodes several predicted peroxiredoxins, including a thiol-dependent peroxidase (tpx) and the product of ygaF (the gene immediately upstream of perR), only the ahpCF genes are members of the peroxide stimulon characterized in this study. In future work, it will be important to define the roles of catalase, Ahp, peroxiredoxins, and the organic hydroperoxide resistance proteins in B. subtilis. While all of these enzymes may act to detoxify reactive oxygen species in the cell, genetic studies indicate that they are not redundant.
Acknowledgments
This work was supported by grant MCB-9983656 from the National Science Foundation to J.D.H.
REFERENCES
- 1.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. [DOI] [PubMed] [Google Scholar]
- 2.Bernhardt, J., U. Volker, A. Volker, H. Antelmann, R. Schmid, H. Mach, and M. Hecker. 1997. Specific and general stress proteins in Bacillus subtilis—a two-dimensional protein electrophoresis study. Microbiology 143:999-1017. [DOI] [PubMed] [Google Scholar]
- 3.Bol, D. K., and R. E. Yasbin. 1994. Analysis of the dual regulatory mechanisms controlling expression of the vegetative catalase gene of Bacillus subtilis. J. Bacteriol. 176:6744-6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bsat, N., L. Chen, and J. D. Helmann. 1996. Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J. Bacteriol. 178:6579-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. [DOI] [PubMed] [Google Scholar]
- 6.Casillas-Martinez, L., A. Driks, B. Setlow, and P. Setlow. 2000. Lack of a significant role for the PerR regulator in Bacillus subtilis spore resistance. FEMS Microbiol. Lett. 188:203-208. [DOI] [PubMed] [Google Scholar]
- 7.Chen, L., and J. D. Helmann. 1995. Bacillus subtilis MrgA is a Dps(PexB) homologue: evidence for metalloregulation of an oxidative stress gene. Mol. Microbiol. 18:295-300. [DOI] [PubMed] [Google Scholar]
- 8.Chen, L., L. P. James, and J. D. Helmann. 1993. Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially repressed by metal ions. J. Bacteriol. 175:5428-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, L., L. Keramati, and J. D. Helmann. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. USA 92:8190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa Seaver, L., and J. A. Imlay. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa Seaver, L., and J. A. Imlay. 2001. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J. Bacteriol. 183:7182-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowds, B. C. A. 1994. The oxidative stress response in Bacillus subtilis. FEMS Microbiol. Lett. 124:255-264. [DOI] [PubMed] [Google Scholar]
- 13.Dowds, B. C. A., P. Murphy, D. J. McConnell, and K. M. Devine. 1987. Relationship among oxidative stress, growth cycle, and sporulation in Bacillus subtilis. J. Bacteriol. 169:5771-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farr, S. B., and T. Kogoma. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55:561-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuangthong, M., S. Atichartpongkul, S. Mongkolsuk, and J. D. Helmann. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183:4134-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuangthong, M., and J. D. Helmann. 2002. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl. Acad. Sci. USA 99:6690-6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuangthong, M., A. F. Herbig, N. Bsat, and J. D. Helmann. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184:3276-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaballa, A., and J. D. Helmann. 2002. A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis. Mol. Microbiol. 45:997-1005. [DOI] [PubMed] [Google Scholar]
- 20.Hartford, O. M., and B. C. A. Dowds. 1994. Isolation and characterization of a hydrogen peroxide resistant mutant of Bacillus subtilis. Microbiology 140:297-304. [DOI] [PubMed] [Google Scholar]
- 21.Hecker, M., and U. Volker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 22.Helmann, J. D., M. F. Wu, P. A. Kobel, F. J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbig, A., and J. D. Helmann. 2002. Metal ion uptake and oxidative stress, p. 405-414. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives, 2nd ed. ASM Press, Washington, D.C.
- 24.Krogh, S., M. O'Reilly, N. Nolan, and K. M. Devine. 1996. The phage-like element PBSX and part of the skin element, which are resident at different locations on the Bacillus subtilis chromosome, are highly homologous. Microbiology 142:2031-2040. [DOI] [PubMed] [Google Scholar]
- 25.Loewen, P. C., and J. Switala. 1987. Multiple catalases in Bacillus subtilis. J. Bacteriol. 169:3601-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan, R. W., M. F. Christman, F. S. Jacobson, G. Storz, and B. N. Ames. 1986. Hydrogen peroxide-inducible proteins in Salmonella typhimurium overlap with heat shock and other stress proteins. Proc. Natl. Acad. Sci. USA 83:8059-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pomposiello, P. J., M. H. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pomposiello, P. J., and B. Demple. 2001. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19:109-114. [DOI] [PubMed] [Google Scholar]
- 30.Price, C. W. 2002. General stress response, p. 369-384. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives, 2nd ed. ASM Press, Washington, D.C.
- 31.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 32.Sato, T., and Y. Kobayashi. 1998. The ars operon in the skin element of Bacillus subtilis confers resistance to arsenate and arsenite. J. Bacteriol. 180:1655-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherlock, G., T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J. C. Matese, S. S. Dwight, M. Kaloper, S. Weng, H. Jin, C. A. Ball, M. B. Eisen, P. T. Spellman, P. O. Brown, D. Botstein, and J. M. Cherry. 2001. The Stanford microarray database. Nucleic Acids Res. 29:152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 35.Storz, G., and M. Zheng. 2000. Oxidative stress, p. 47-59. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 36.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Ploeg, J. R., M. Barone, and T. Leisinger. 2001. Expression of the Bacillus subtilis sulphonate-sulphur utilization genes is regulated at the levels of transcription initiation and termination. Mol. Microbiol. 39:1356-1365. [PubMed] [Google Scholar]
- 38.Voelker, U., T. Luo, N. Smirnova, and W. Haldenwang. 1997. Stress activation of Bacillus subtilis sigma B can occur in the absence of the sigma B negative regulator RsbX. J. Bacteriol. 179:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng, M., X. Wang, B. Doan, K. A. Lewis, T. D. Schneider, and G. Storz. 2001. Computation-directed identification of OxyR DNA binding sites in Escherichia coli. J. Bacteriol. 183:4571-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]