Abstract
Neisseria meningitidis is the cause of septicemia and meningococcal meningitis. During the course of infection, N. meningitidis encounters multiple environments within its host, which makes rapid adaptation to environmental changes a crucial factor for neisserial pathogenicity. Employing oligonucleotide-based DNA microarrays, we analyzed the transcriptome of N. meningitidis during two key steps of meningococcal infection, i.e., the interaction with epithelial cells (HeLa cells) and endothelial cells (human brain microvascular endothelial cells). Seventy-two genes were differentially regulated after contact with epithelial cells, and 48 genes were differentially regulated after contact with endothelial cells, including a considerable proportion of well-known virulence genes. While a considerable number of genes were in concordance between bacteria adherent to both cell types, we identified several open reading frames that were differentially regulated in only one system. The data obtained with this novel approach may provide insight into the pathogenicity mechanisms of N. meningitidis and could demonstrate the importance of gene regulation on the transcriptional level during different stages of meningococcal infection.
The gram-negative bacterial pathogen Neisseria meningitidis is a major cause of morbidity and mortality worldwide, with an estimated 500,000 to 1 million cases of septicemia and meningitis reported every year. Invasive meningococcal infections represent a major childhood disease with a mortality of 10% and high morbidity in survivors (3). Vaccines specific for N. meningitidis serogroups A, C, Y, and W135 have been developed, but there is no vaccine for serogroup B, which is responsible for most meningococcal disease in the United States and Europe (23). A thorough understanding of the neisserial virulence mechanisms is a prerequisite for the rational design of novel approaches for the treatment and prevention of disease.
Meningococcal infection starts with colonization of the nasopharyngeal and tonsillar mucosa. Following adherence, meningococci initiate endocytosis, cross the epithelial barrier, and gain access to the bloodstream (29). Meningococci are characterized by a marked tropism towards the central nervous system. As a result of the ability to cross the blood-brain barrier, patients develop purulent meningitis (2). During the different stages of infection, meningococci have to adapt to rapidly changing environments and to escape innate and acquired immune responses (19, 21, 36). N. meningitidis virulence mechanisms employ a variety of strategies for host adaptation. Excessive exchange and mutation of their DNA, including phase variation due to slipped-strand mispairing (9, 14, 16, 27), ensures the existence of a highly diverse bacterial population, which is an important condition for the preadaptation to specific microenvironmental conditions (1). Alternatively, meningococci can directly respond to environmental changes on the transcriptional level. Recently, a transcriptional regulator responsible for the cross talk between meningococci and target cells during infection has been described (5).
Sequencing of the genomes of a serogroup A strain and a serogroup B strain of N. meningitidis (22, 32) provided us with a tremendously broad range of information, but a full understanding of the interaction between meningococci and their human host requires the knowledge of meningococcal gene expression during infection. DNA microarrays offer an ideal tool for the simultaneous transcriptional analysis of all genes present in a bacterial genome (reviewed in references 4 and 25). However, the complex interplay of bacteria and host during infection has been investigated only for the host cell transcriptional response, e.g., for human monocytes infected with different Salmonella strains (6) and meningothelial cells after infection with serogroup B meningococci (38). In the present work we employed oligonucleotide-based DNA microarrays as a technology platform to analyze transcriptional changes in N. meningitidis serogroup B during infection of the two key target cells of meningococcal infection: human epithelial and endothelial cells. Isolation of RNA from cell-adherent serogroup B meningococci and subsequent hybridization to whole-genome DNA microarrays enabled us to characterize the meningococcal transcriptome during the infection of these two cell types. Therefore, this study provides the first analysis of bacterial gene regulation during infection on a global scale.
MATERIALS AND METHODS
Bacterial strains.
The meningococcal strain used in this study is the piliated N. meningitidis serogroup B strain MC58 (belonging to the ET-32 complex) which was isolated in 1983 from a clinical case in the United Kingdom (33). The unencapsulated MC58 siaD mutant strain has been previously described (19). Briefly, the siaD gene encoding the polysialyltransferase as a key enzyme of the capsular polysaccharide biosynthesis machinery was inactivated by the insertion of a chloramphenicol resistance cassette. The resulting capsule-deficient mutant was chosen for the experiments in this study to ensure that sufficient RNA could be isolated; infections with wild-type bacteria resulted in insufficient RNA yields due to the weak adherence of encapsulated bacteria to epithelial or endothelial cells (15, 34). Therefore, the utilization of encapsulated meningococci in the present study was not feasible in our hands.
Cell culture.
The epithelial cell line used in this study, HEp-2, was recently identified as being identical to HeLa cells (as analyzed by the American Type Culture Collection (ATCC) (ATCC catalogue; ATCC, Manassas, Va.). However, for reasons of clarity, the HEp-2 cell nomenclature is used throughout this report. The HEp-2 cells were originally obtained from A. E. Moore. HEp-2 cells were propagated in RPMI 1640 medium plus 10% fetal calf serum (FCS) (both from Gibco Life Technologies, Karlsruhe, Germany) and 2 mM l-glutamine (Biochrom, Berlin, Germany). Human brain microvascular endothelial cells (HBMEC) were described previously (30). HBMEC were cultured in RPMI 1640, supplemented with 10% FCS, 10% Nu Serum IV (Becton Dickinson, Bedford, Mass.), 1% vitamins, 1 mM sodium pyruvate, 2 mM l-glutamine, heparin (5 U/ml) (all reagents were from Biochrom), and 30 μg of endothelial cell growth supplement (Cell Systems Clonetics, St. Katharinen, Germany)/ml in T75 flasks (Noras, Würzburg, Germany) coated with 0.2% gelatin to mediate cell adherence. Both cell lines were grown in a humidified atmosphere at 37°C and 5% CO2.
Bacterial adhesion and invasion assay and isolation of cell-adherent meningococci.
Cells were propagated in T75 flasks to confluent monolayers (1 × 107 cells/flask). All cell infection experiments were done with the unencapsulated strain MC58 siaD since the host cell adherence and infectivity of the encapsulated wild-type strain is insufficient for isolation of amounts of RNA suitable for DNA microarray hybridization. Although a genetically engineered mutant was used in this study, the interaction of capsule-negative bacteria with epithelial cells principally correlates to the physiological conditions on the nasopharyngeal mucosa among meningococcal carriers and individuals with developing meningococcal disease since the bacteria are nonencapsulated during the intimate phase of host cell attachment (16, 33-35). The cells were infected at a multiplicity of infection of 10 with MC58 siaD in RPMI 1640 medium alone (HEp-2) or RPMI 1640 medium with 10% heat-inactivated FCS (HBMEC) at 37°C and 5% CO2 for 6 h before the numbers of nonadherent, total cell-associated, and intracellular bacteria were determined as described previously (19). For assessment of bacterial numbers that could be dissociated from the cell surface, cells were washed three times with phosphate-buffered saline (PBS) followed by treatment with trypsin-EDTA (Gibco Life Technologies) and gentle shaking for 10 min. Mammalian cells were removed by two centrifugation steps at 1,000 × g for 10 min at 4°C, leaving only the bacteria in the supernatant. Bacterial numbers dissociated from the host cells were assessed by plating serial dilutions of the supernatant on GC agar.
Isolation of RNA from host-cell-adherent bacteria.
HEp-2 and HBMEC cells (1 × 107 cells/T75 flask) were infected at a multiplicity of infection of 10 with MC58 siaD in RPMI 1640 medium alone (HEp-2) or RPMI 1640 medium with 10% heat-inactivated FCS (HBMEC) for 6 h before isolation of cell-adherent bacteria. Cells were washed three times with PBS to remove the nonadherent bacteria. The adherent bacteria were subsequently dissociated from host cells by treatment with trypsin-EDTA and harvested by centrifugation. RNA was isolated from the neisserial pellet as described previously (13). Control bacteria were treated identically to bacteria from infections, i.e., all incubations, and wash and centrifugation steps were performed, except that no host cells were present during incubation in RPMI medium with FCS (HBMEC controls) or without FCS (HEp-2 controls). The quality of the RNAs was assessed by formaldehyde-agarose gel electrophoresis. Potential traces of DNA were removed by treatment with RNase-free DNase I (Roche Diagnostics GmbH, Mannheim, Germany). The absence of neisserial DNA was confirmed by PCR with primers specific for the open reading frames (ORFs) NMB0829 (hsdM) and NMB1972 (groEL). The absence of nucleic acids from human cells was confirmed by reverse transcriptase (RT)-PCR with primers specific for the human ORFs XM_004814 (ACTB) and XM_009352 (GAPDS).
Construction of oligonucleotide-based whole-genome DNA microarrays.
For each of the 2,158 ORFs present in the published genome of N. meningitidis serogroup B strain MC58 (32), oligonucleotide-based microarrays were manufactured as previously described (13). Three oligonucleotides (40-mers) per gene comprising gene-specific internal fragments (covering 5′, central, and 3′ parts) were designed. All oligonucleotides (manufactured by MWG Biotech AG, Ebersberg, Germany) carried a C6 amino linker modification at the 5′ end for covalent attachment to the slide surface. The oligonucleotides were spotted by using the Affymetrix 417 Arrayer (MWG Biotech AG) on super aldehyde slides (TeleChem International, Sunnyvale, Calif.), and the slides were processed according to the manufacturer's instructions.
Preparation of labeled cDNA, microarray hybridization, and data analysis.
Equal amounts of the RNAs to be compared were labeled differentially with Cy3-dCTP and Cy5-dCTP (Amersham Pharmacia, Freiburg, Germany) during a first-strand reverse transcription reaction with Superscript II RNase H− RT (Life Technologies) and a balanced mixture (20 pmol each) of C-terminal primers specific for all genes present on the microarrays. The two differentially labeled cDNA samples were combined, brought to 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% (wt/vol) sodium dodecyl sulfate, and hybridized to the array at 50°C for 16 h. Arrays were washed and scanned by using the Affymetrix 418 scanner (MWG-Biotech AG). The average signal intensity and local background measurements were obtained for each spot with ImaGene, version 4.0, software (Biodiscovery Inc., Los Angeles, Calif.). The two channels were normalized with respect to the mean values of all N. meningitidis DNA spots, and the Cy3/Cy5 fluorescence ratios were calculated from the normalized values. The transcript ratios of all ORFs were calculated from the average signal ratios of all oligonucleotides per gene. Biological experiments and subsequent hybridizations were repeated five times, and data of the independent experiments were combined. ORFs, minimally exhibiting a 1.6-fold deregulation on average and being deregulated more than 1.6-fold in at least three experiments, were defined as differentially transcribed; this cutoff is based on previous results which showed a 1.6-fold difference to be detected at a level of confidence of above 99.7% (13).
Real-time RT-PCR.
Before RT reactions were performed, the absence of DNA from RNA samples was verified by PCR amplification of the genes to be assayed with 1 μg of RNA as a template. For quantitative RT-PCR cDNA synthesis, 2 μg of total RNA (experiment and control) and 6 ng of random hexamer primers (Life Technologies) were heat denatured for 5 min at 70°C and chilled on ice before adding the remaining components for the RT reaction: 0.5 mM (each) dATP, dCTP, dGTP, and dTTP, 1 mM dithiothreitol, 200 U of Superscript II RNase H− RT (Life Technologies), and reaction buffer to a final concentration of 1×. Reactions proceeded for 10 min at 25°C and 2 h at 42°C. PCR was done in a final volume of 20 μl in glass capillaries with the LightCycler (Roche Diagnostics). PCR was performed according to the manufacturer's protocol with the FASTStart DNA master kit SYBR green I (Roche Diagnostics). A 4 mM MgCl2 concentration proved to be suitable for all reactions. Cycling conditions were as follows: initial denaturation at 95°C for 8 min and 40 cycles of 95°C for 0 s, 64°C for 5 s, and 72°C for 15 s followed by a melting cycle from 65 to 95°C.
For quantification of RT-PCRs, amplicons corresponding to the genes of interest (mdaB, siaA, ctrC, secY, NMB0200, NMB0985, and NMB2052) were cloned into a PCR II vector (TA cloning kit; Invitrogen, Leiden, The Netherlands). Serial dilutions of the resulting plasmids served as an external standard (corresponding to 107 to 102 copies). RT-PCRs were quantified for experiments and controls with the appropriate standard curve. Normalization for all results was performed with a second quantification in the same run for a gene (NMB2052) with no measurable deregulation. Specificity for all amplicons was confirmed via melting curves and gel analysis.
Transmembrane domain prediction.
The transmembrane regions of the proteins were predicted by use of the PSORT software (Kenta Nakai, Tokyo, Japan).
RESULTS
Isolation of N. meningitidis adherent to human epithelial cells (HEp-2) and HBMEC.
HEp-2 cells were infected at a multiplicity of infection of 10 with the N. meningitidis strain MC58 siaD. At 6 h postinfection (p.i.), meningococci in the supernatant were removed by washing with PBS. At this time point, about 30% of the meningococci present in the flask were cell associated. Of the cell-associated bacteria, only 1% were found to be located intracellularly in gentamicin killing assays, the remaining 99% were localized on the cell surface. The cell-surface-adherent bacteria were dissociated from the HEp-2 cells by treatment with trypsin-EDTA and recovered by centrifugation. By application of this protocol, more than 95% of the bacteria could be removed from the cell surface and RNA could be extracted from over 90% of the bacteria in total. Similar results were obtained during the infection of HBMEC with strain MC58 siaD. The high recovery achieved with this method allowed us to analyze the transcriptome of a representative population of surface-associated bacteria.
Analysis of the transcriptome of meningococci associated with the surface of HEp-2 cells.
Oligonucleotide DNA microarrays were employed which previously exhibited high sensitivity, specificity, and reproducibility, allowing a 2-fold deregulation to be detected at a level of confidence of 99.9% and a 1.6-fold difference at a level of confidence of above 99.7% (13). HEp-2 cells were infected with N. meningitidis strain MC58 siaD, and at 6 h p.i., the cell-adherent bacteria were recovered. Immediately thereafter, the meningococcal RNA was isolated. As a control, MC58 siaD bacteria were incubated in cell culture medium for 6 h. The RNAs of cell-adherent and control meningococci were labeled differentially and hybridized to whole-genome DNA microarrays. Five independent RNA isolations and hybridizations were performed, and ORFs differentially regulated above 1.6-fold on average and in three or more experiments were defined as deregulated.
This experimental approach revealed the differential regulation of 72 genes upon adherence of meningococci to HEp-2 cells, 67 of these were upregulated and 5 were downregulated (Table 1). The degree of differential regulation ranged from 3.7-fold upregulated to 1.8-fold downregulated. The number of differentially regulated genes accounts for 3.3% of the meningococcal genome.
TABLE 1.
N. meningitidis serogroup B ORFs differentially regulated in HEp-2-adherent MC58 siaD meningococci
| Functional category | Serogroup B ORF no.a | Differential regulation (fold) | SD (fold)c | Differential regulation in HBMEC (fold) | Encoded protein | Virulence gened | Serogroup A ORF no.b | Phase variable |
|---|---|---|---|---|---|---|---|---|
| Capsule | NMB0069 | 1.8 | 0.2 | None | Polysialic acid capsule biosynthesis protein SiaB | + | Absent | − |
| NMB0070 | 2.2 | 0.3 | None | Polysialic acid capsule biosynthesis protein SiaA | + | Absent | − | |
| NMB0073 | 2.2 | 0.2 | None | Capsule polysaccharide export inner membrane protein CtrC | + | NMA0196 | − | |
| NMB0074 | 2.4 | 0.2 | None | Capsule polysaccharide export ATP-binding protein CtrD | + | NMA0195 | − | |
| Protein fate | NMB0700 | 2.0 | 0.2 | None | IgA-specific serine endopeptidase | + | NMA0905 | − |
| Membrane | NMB0364/NMB0584/NMB1412/NMB1414 | 2.4 | 0.4 | None | FrpC operon protein | + | NMA2124 | − |
| NMB0442/NMB0926/NMB1465/NMB1636 | 1.9 | 0.2 | None | Opacity proteins, authentic frameshifts, OpaA-D | + | NMA1676/NMA1890/NMA2043 | + | |
| NMB0886 | 1.7 | 0.2 | None | Fimbrial protein FimT | − | NMA1106 | − | |
| NMB0889 | 2.1 | 0.1 | None | Putative membrane protein | − | NMA1109 | − | |
| NMB1415/NMB0585 | 2.3 | 0.3 | None | Iron-regulated protein FrpC | + | NMA1626/NMA0788 | − | |
| NMB1429 | 1.6 | 0.2 | None | Outer membrane protein PorA | + | NMA1642 | + | |
| NMB1651 | 2.6 | 0.4 | None | Alanine racemase Alr | − | NMA1906 | − | |
| NMB1730 | 1.8 | 0.7 | 1.7 | TonB | + | NMA1985 | − | |
| NMB1809 | 1.7 | 0.1 | None | PilN | + | NMA0653 | − | |
| NMB1810 | 2.1 | 0.5 | None | PilO | + | NMA0652 | − | |
| Transport | NMB0177 | −1.6 | 0.1 | None | Putative amino acid transporter | − | NMA0091 | − |
| NMB0633 | 2.0 | 0.6 | None | Iron(III) ABC transporter, permease protein FbpB | + | NMA0843 | − | |
| NMB0787 | 2.1 | 0.4 | None | Amino acid ABC transporter, periplasmic amino acid binding protein | − | NMA0997 | − | |
| NMB0788 | 1.8 | 0.3 | None | Amino acid ABC transporter, permease protein | − | NMA0999 | − | |
| NMB0880 | 1.9 | 0.2 | None | Sulfate ABC transporter, permease protein CysW | − | NMA1098 | − | |
| NMB0881 | 1.6 | 0.1 | None | Sulfate ABC transporter, permease protein CysT | − | NMA1100 | − | |
| NMB1017 | 2.1 | 0.2 | None | Sulfate ABC transporter, periplasmic sulfate-binding protein | − | NMA1243 | − | |
| NMB1728 | 1.8 | 0.4 | None | Biopolymer transporter protein ExbD | + | NMA1983 | − | |
| NMB1888 | 2.3 | 0.3 | None | Protein-export membrane protein SecG | − | NMA0569 | − | |
| NMB1998 | −1.8 | 0.3 | None | Outer membrane protein FetA | − | NMA0453 | + | |
| Transcription | NMB0126 | 2.1 | 0.1 | None | Transcription antitermination protein NusG | − | NMA0147 | − |
| NMB0132 | 1.8 | 0.2 | None | DNA-directed RNA polymerase, beta subunit RpoB | + | NMA0142 | − | |
| NMB0617 | 1.6 | 0.3 | None | Rho transcription termination factor | − | NMA0825 | − | |
| Metabolism | NMB0678 | 2.1 | 0.7 | None | Putative tryptophan synthase alpha chain TrpA | − | NMA0879 | − |
| NMB0994 | 2.0 | 0.3 | None | Acyl-coenzyme A dehydrogenase family protein | − | NMA1202 | − | |
| NMB1377 | 2.1 | 0.2 | None | L-Lactate dehydrogenase LldA | − | NMA1592 | − | |
| NMB1527 | 2.1 | 0.9 | 1.6 | ADP-heptose:lipopolysaccharide heptosyltransferase II RfaF | + | NMA1727 | − | |
| NMB1541 | 1.9 | 0.1 | None | Lactoferrin-binding protein LbpB | + | NMA1740 | − | |
| NMB1574 | 2.2 | 0.3 | 1.7 | Ketol acid reductoisomerase IIvC | + | NMA1763 | − | |
| NMB1576 | 2.3 | 0.9 | 1.7 | Acetolactate synthase isozyme III small subunit IIvH | + | NMA1765 | − | |
| NMB1577 | 2.7 | 0.7 | 1.6 | Acetolactate synthase isozyme III large subunit IIvI | + | NMA1766 | − | |
| NMB1581 | 2.1 | 0.4 | None | Histidinol dehydrogenase HisD | − | NMA1770 | − | |
| NMB1845 | 2.7 | 0.6 | 2.0 | Putative periplasmic thioredoxin | − | NMA0612 | − | |
| NMB1857/NMB0977 | 2.2 | 0.2 | 2.4 | Modulator of drug activity B, putative MdaB | − | NMA0600/NMA1174 | − | |
| NMB1861 | 1.9 | 0.3 | None | Putative acetyl-coenzyme A carboxylase biotin carboxylase component AccC | − | NMA0596 | − | |
| NMB1887 | 2.1 | 0.5 | None | Putative triosephosphate isomerase TpiA | − | NMA0570 | − | |
| NMB1940 | 2.2 | 0.3 | None | ATP synthase A chain AtpB | − | NMA0513 | − | |
| NMB2154 | 1.8 | 0.2 | None | Electron transfer flavoprotein alpha-subunit EtfA | − | NMA0241 | − | |
| NMB2155 | 1.9 | 0.4 | None | Electron transfer flavoprotein beta-subunit EtfB | − | NMA0242 | − | |
| Translation | NMB0124 | 2.2 | 0.2 | None | Elongation factor TU TufA1 | + | NMA0134 | − |
| NMB0145 | 1.8 | 0.4 | None | 50S ribosomal protein L2 RplB | − | NMA0126 | − | |
| NMB0159 | 1.7 | 0.3 | 1.6 | 30S ribosomal protein S5 RpsE | − | NMA0112 | − | |
| NMB0167 | 1.7 | 0.3 | 2.0 | 30S ribosomal protein S4 RpsD | − | NMA0104 | − | |
| NMB0591 | 1.7 | 0.2 | None | 16S rRNA processing protein RimM | − | NMA0794 | ||
| DNA | NMB0210 | 2.9 | 0.5 | None | Pseudogene (modification methylase) | − | NMA0059 | − |
| Phage related | NMB0101 | 1.6 | 0.3 | None | Putative transposase for IS1016 | − | NMA0175 | − |
| NMB0240 | 1.9 | 0.2 | None | Putative IS1016 transposase | − | NMA0022 | −/PICK> | |
| Hypothetical | NMB0065 | 1.8 | 0.2 | None | Hypothetical protein | − | Absent | + |
| NMB0129 | 1.8 | 0.3 | None | Hypothetical protein | − | Intergenic between NMA0144 and NMA0145 | − | |
| NMB0200 | 2.1 | 0.3 | 2.1 | Hypothetical protein | − | Absent | − | |
| NMB0239 | 2.1 | 0.4 | None | Hypothetical protein | − | NMA0021 | − | |
| NMB0328 | 1.7 | 0.2 | None | Hypothetical protein | − | Absent | − | |
| NMB0429 | −1.8 | 0.4 | None | Hypothetical protein | − | NMA2055 | − | |
| NMB0451 | 2.3 | 0.4 | None | Hypothetical protein Tou2 | − | NMA2035 | − | |
| NMB0475 | 1.6 | 0.2 | None | Hypothetical protein | − | NMA2010 | − | |
| NMB0476 | 2.0 | 0.3 | None | Hypothetical protein | − | NMA2009 | − | |
| NMB0517 | 2.1 | 0.3 | None | Hypothetical protein | − | Absent | − | |
| NMB0744 | 3.7 | 0.9 | 1.6 | Hypothetical protein | − | NMA0957 | − | |
| NMB0995 | 2.1 | 0.2 | None | Hypothetical protein Mip | + | NMA1203 | − | |
| NMB1016 | 1.8 | 0.3 | None | Hypothetical protein | − | NMA1237 | − | |
| NMB1452 | 2.6 | 1.4 | None | Conserved hypothetical protein | − | NMA1666 | − | |
| NMB1575 | 2.1 | 0.5 | 1.6 | Conserved hypothetical protein | − | NMA1764 | − | |
| NMB1578 | 1.7 | 0.2 | None | Conserved hypothetical protein | − | NMA1767 | − | |
| NMB1632 | −1.6 | 0.1 | None | Hypothetical protein | − | NMA0780 | − | |
| NMB1889 | 1.6 | 0.2 | 1.6 | Hypothetical protein | − | Absent | − | |
| NMB2000 | −1.6 | 0.3 | None | Hypothetical HsIO | − | NMA0441 | − | |
| NMB2140 | 2.1 | 0.3 | None | Conserved hypothetical protein | − | NMA0226 | − |
ORF numbers from the published N. meningitidis serogroup B genome sequence (32) are given. ORF numbers from the published N. meningitidis serogroup A genome sequence (22) are given. Standard deviations of five independent experiments are given. +, yes; −, no.
ORF numbers from the published N. meningitidis serogroup A genome sequence (22) are given.
Standard deviations of five independent experiments are given.
+, yes; −, no.
The meningococcal genes differentially regulated during HEp-2-cell interaction can be clustered in several functional groups: genes encoding membrane proteins and transporters, gene expression regulators, and genes involved in protein biosynthesis and general metabolism. Furthermore, we found a wide range of hypothetical ORFs and ORFs with unknown function to be differentially regulated. A high proportion (fifteen) of the ORFs upregulated after interaction with HEp-2 cells represent previously identified virulence genes: genes encoding capsule synthesis and export functions (siaB, siaA, ctrC, and ctrD), immunoglobulin A (IgA) protease (iga), lipooligosaccharide biosynthesis enzymes (rfaF), several iron uptake systems (tonB, exbD, lbpB, fbpB), type IV pilus assembly proteins (pilN and pilO), invasins (opa andporA), and potential toxins (frpC). Interestingly, all differentially regulated virulence-associated genes were found to be induced. A total of 20 genes encoding membrane proteins and transporters were differentially regulated. Of these, 18 were upregulated and 2 were downregulated. Twenty-four differentially regulated genes were involved in transcription, translation, and metabolism. All of these were induced upon meningococcal adherence to HEp-2 cells. Twenty differentially transcribed ORFs had no known function. Of these, 17 were upregulated and 3 were downregulated. Eleven of the hypothetical ORFs had a size of >200 amino acids, and 5 ORFs encoded proteins for which transmembrane regions were predicted by the PSORT software (Kenta Nakai).
Analysis of the transcriptome of HBMEC cell-surface-associated meningococci.
HBMEC were infected with N. meningitidis strain MC58 siaD, and RNA was isolated from cell-adherent bacteria 6 h p.i. Again, five independent RNA isolations and hybridizations were performed, and the data of the independent experiments were combined. ORFs were defined as deregulated if they exhibited a differential transcript level of above 1.6-fold on average and in at least three experiments. Comparison of RNA isolated from HBMEC-associated bacteria with RNA derived from control meningococci revealed significant transcriptional changes for 48 ORFs (Table 2), equivalent to 2.2% of all ORFs present in strain MC58. Forty-one of these were induced, and seven were repressed. The degree of differential regulation ranged from 4.5-fold upregulated to 1.9-fold downregulated (Table 2).
TABLE 2.
N. meningitidis serogroup B ORFs differentially regulated in HBMEC-adherent MC58 siaD meningococci
| Functional category | Serogroup B ORF no.a | Differential regulation (fold) | SD (fold)c | Differential regulation in HEp-2 (fold) | Encoded protein | Virulence gened | Serogroup A ORF no.b | Phase variable |
|---|---|---|---|---|---|---|---|---|
| Membrane | NMB1468 | 2.4 | 0.4 | None | Putative membrane protein | − | NMA1680 | − |
| NMB1646 | 1.7 | 0.2 | None | Putative hemolysin | + | NMA1900 | − | |
| NMB1730 | 1.7 | 0.2 | 1.8 | TonB | + | NMA1985 | − | |
| Transport | NMB0162 | 1.9 | 0.2 | None | Preprotein translocase SecY subunit | − | NMA0109 | − |
| Transcription | NMB1843 | 1.6 | 0.2 | None | Transcriptional regulator MarR family | − | NMA0613 | − |
| Metabolism | NMB0546 | −1.8 | 0.2 | None | Alcohol dehydrogenase, propanol-preferring AdhA | − | NMA0725 | − |
| NMB0950 | −1.8 | 0.3 | None | Putative succinate dehydrogenase flavoprotein subunit | − | NMA1145 | − | |
| NMB0951 | −1.6 | 0.3 | None | Putative succinate dehydrogenase iron-sulphur protein | − | NMA1146 | − | |
| NMB1527 | 1.6 | 0.2 | 2.1 | ADP-heptose:lipopolysaccharide heptosyltransferase II RfaF | + | NMA1727 | − | |
| NMB1574 | 1.7 | 0.1 | 2.2 | Ketol acid reductoisomerase IIvC | + | NMA1763 | − | |
| NMB1576 | 1.7 | 0.1 | 2.3 | Acetolactate synthase isozyme III small subunit IIvH | + | NMA1765 | − | |
| NMB1577 | 1.6 | 0.2 | 2.7 | Acetolactate synthase isozyme III large subunit IIvI | + | NMA1766 | − | |
| NMB1710 | 1.7 | 0.2 | None | Glutamate dehydrogenase, NADP-specific GdhA | + | NMA1964 | − | |
| NMB1845 | 2.0 | 0.4 | 2.7 | Putative periplasmic thioredoxin | − | NMA0612 | − | |
| NMB1857/NMB0977 | 2.4 | 0.2 | 2.2 | Modulator of drug activity B, putative MdaB | − | NMA0600/NMA1174 | − | |
| Translation | NMB0143 | 1.6 | 0.1 | None | 50S ribosomal protein L4 RplD | − | NMA0128 | − |
| NMB0144 | 1.7 | 0.2 | None | 50S ribosomal protein L23 RplW | − | NMA0127 | − | |
| NMB0148 | 1.6 | 0.3 | None | 30S ribosomal protein S3 RpsC | − | NMA0123 | − | |
| NMB0149 | 2.3 | 0.9 | None | 50S ribosomal protein L16 RplP | − | NMA0122 | − | |
| NMB0150 | 1.7 | 0.5 | None | 50S ribosomal protein L29 RpmC | − | NMA0121 | − | |
| NMB0151 | 1.7 | 0.1 | None | 30S ribosomal protein S17 RpsQ | − | NMA0120 | − | |
| NMB0159 | 1.6 | 0.2 | 1.7 | 30s ribosomal protein S5 RpsE | − | NMA0112 | − | |
| NMB0160 | 1.6 | 0.3 | None | 50S ribosomal protein L30 RpmD | − | NMA0111 | − | |
| NMB0161 | 1.6 | 0.3 | None | 50S ribosomal protein L15 RplO | − | NMA0110 | − | |
| NMB0163 | 2.0 | 0.2 | None | Translation initiation factor IF-1 InfA | − | NMA0108 | − | |
| NMB0164 | 1.7 | 0.1 | None | 50S ribosomal protein L36 RpmJ | − | NMA0107 | − | |
| NMB0167 | 2.0 | 0.1 | 1.7 | 30S ribosomal protein S4 RpsD | − | NMA0104 | − | |
| NMB0169 | 1.6 | 0.1 | None | 50S ribosomal protein L17 RplQ | − | NMA0102 | − | |
| NMB0941 | 2.4 | 0.2 | None | Putative 50S ribosomal protein L36 RpmJ2 | − | NMA1137 | − | |
| NMB0942 | 4.5 | 0.2 | None | 50S ribosomal protein L31, putative RpmE2 | − | NMA1138 | − | |
| DNA | NMB0232 | 2.6 | 1.9 | None | DNA helicase II UvrD | − | NMA0027 | − |
| Phage related | NMB0987 | 1.8 | 0.9 | None | Putative N-acetylmuramoyl-l-alanine amidase | − | NMA1303/NMA1188 | − |
| NMB0961 | 2.1 | 0.7 | None | FunZ | − | Absent | + | |
| Hypothetical | NMB0091 | −1.7 | 0.3 | None | Hypothetical protein | − | Absent | − |
| NMB0092 | −1.8 | 0.2 | None | Hypothetical protein | − | Absent | − | |
| NMB0093 | −1.8 | 0.3 | None | Hypothetical protein | − | Absent | − | |
| NMB0200 | 2.1 | 0.4 | 2.1 | Hypothetical protein | − | Absent | − | |
| NMB0267 | 1.7 | 0.3 | None | Putative periplasmic protein | − | NMA2219 | − | |
| NMB0555 | −1.9 | 0.4 | None | Hypothetical protein NMA0737 | − | NMA0737 | − | |
| NMB0744 | 1.6 | 0.2 | 3.7 | Hypothetical protein | − | NMA0957 | − | |
| NMB0985 | 2.1 | 0.6 | None | E16-related protein | − | NMA1301/NMA1186 | − | |
| NMB0986 | 1.8 | 0.9 | None | Hypothetical protein | − | NMA1187/NMA1302 | − | |
| NMB1009 | 1.7 | 0.2 | None | Conserved hypothetical protein | − | NMA1197 | − | |
| NMB1010 | 1.6 | 0.2 | None | Hypothetical protein | − | Absent | − | |
| NMB1330 | 1.7 | 0.3 | None | Hypothetical protein | − | Absent | − | |
| NMB1575 | 1.6 | 0.1 | 2.1 | Conserved hypothetical protein | − | NMA1764 | − | |
| NMB1844 | 1.6 | 0.2 | None | Hypothetical protein | − | Absent | − | |
| NMB1889 | 1.6 | 0.2 | 1.6 | Hypothetical protein | − | Absent | − |
Most of the differentially regulated genes were involved in virulence, transport, protein biosynthesis, and metabolism or comprise a range of hypothetical ORFs. The virulence-associated genes were responsible for lipooligosaccharide biosynthesis (rfaF), iron uptake (tonB), a putative hemolysin (NMB1646), and an NADP-specific glutamate dehydrogenase (NMB1710), and they encoded essential metabolic functions (ilvC, ilvH, and ilvI). Interestingly, all virulence genes were again induced upon host cell interaction of the meningococci. Likewise, all four membrane proteins and transporters are upregulated. Of the 26 deregulated genes involved in metabolism, transcription, and translation, 23 were induced and 3 were repressed. The differentially regulated genes encompassed 15 hypothetical ORFs, of which 12 were upregulated and 3 were downregulated. Seven of them had a size of >100 amino acids, and one gene encoded a putative transmembrane protein (predicted by the PSORT software).
Global analysis of genes differentially regulated during HEp-2 and HBMEC interaction.
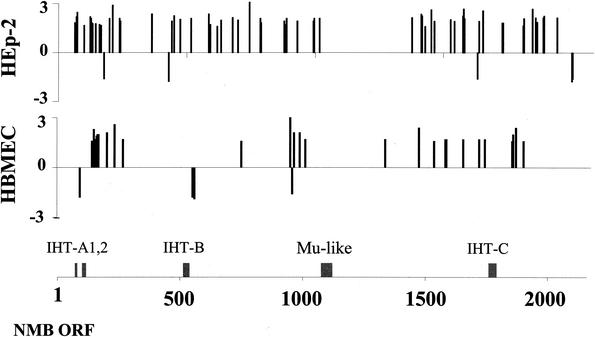
We analyzed the genomic localization of the genes regulated differentially during interaction with HEp-2 and HBMEC cells. These genes exhibit a mainly random distribution throughout the meningococcal genome (Fig. 1). Thirteen ORFs are differentially regulated in both HEp-2- and HBMEC-adherent meningococci (Table 3). The majority of the differentially regulated genes are also present in the genome of N. meningitidis serogroup A strain Z2491. However, 14 differentially transcribed ORFs are absent from this strain (Tables 1 and 2).
FIG. 1.
Genomic distribution of differentially regulated genes throughout the N. meningitidis strain MC58 genome. Differential regulation of genes of N. meningitidis adherent to HEp-2 cells and HBMEC is shown. The positions of differentially transcribed ORFs within the MC58 genome (32) are shown on the x axis, and the level of deregulation is shown on the y axis. NMB, Neisseria meningitidis serogroup B; IHT, islands of horizontal DNA transfer.
TABLE 3.
N. meningitidis serogroup B ORFs differentially regulated in HEp-2- and HBMEC-adherent MC58 siaD meningococci
| Functional category | Serogroup B ORF no.a | HEp-2 regulation (fold) | HBMEC regulation (fold) | Encoded protein | Virulence genec | Serogroup A ORF no.b |
|---|---|---|---|---|---|---|
| Membrane | NMB1730 | 1.8 | 1.7 | TonB | + | NMA1985 |
| Metabolism | NMB1527 | 2.1 | 1.6 | ADP-heptose:lipopolysaccharide heptosyltransferase II, RfaF | + | NMA1727 |
| NMB1574 | 2.2 | 1.7 | Ketol acid reductoisomerase, IIvC | + | NMA1763 | |
| NMB1576 | 2.3 | 1.7 | Acetolactate synthase isozyme III small subunit, IIvH | + | NMA1765 | |
| NMB1577 | 2.7 | 1.6 | Acetolactate synthase isozyme III large subunit, IIvI | + | NMA1766 | |
| NMB1845 | 2.7 | 2.0 | Putative periplasmic thioredoxin | − | NMA0612 | |
| NMB1857/NMB0977 | 2.2 | 2.4 | Modulator of drug activity B, MdaB | − | NMA0600/NMA1174 | |
| Translation | NMB0159 | 1.7 | 1.6 | 30S ribosomal protein S5, RpsE | − | NMA0112 |
| NMB0167 | 1.7 | 2.0 | 30S ribosomal protein S4, RpsD | − | NMA0104 | |
| Hypothetical | NMB0200 | 2.1 | 2.1 | Hypothetical protein | − | Absent |
| NMB0744 | 3.7 | 1.6 | Hypothetical protein | − | NMA0957 | |
| NMB1575 | 2.1 | 1.6 | Conserved hypothetical protein | − | NMA1764 | |
| NMB1889 | 1.6 | 1.6 | Hypothetical protein | − | Absent |
A total of 5 differentially regulated ORFs are phase variable; of these, opa, porA, fetA, and NMB0065 were induced in HEp-2-associated bacteria and funZ was induced during HBMEC interaction. Interestingly, none of these were regulated differentially in meningococci during interaction with both cell types (Table 3). Phase variation occurs due to variation in the length of simple sequence repeats caused by slipped-strand mispairing. If the repeat is present within an ORF, sequence variation leads to an alteration of the translational reading frame. If the repeat is present within the promoter sequence, it can influence the transcription of the ORF. While for opa, fetA, NMB0065, and funZ, the repeats are localized within the ORFs, porA carries the sequence repeat within the promoter sequence. The ORFs opa, porA, fetA, and NMB0065 are all present in serogroup A strain Z2491, and funZ is absent from this strain.
Analysis of specific changes by RT-PCR.
Quantitative RT-PCR with the LightCycler was employed as an independent method to confirm the differential regulation of selected ORFs. These experiments were done with RNAs isolated in three independent infections of HEp-2 and HBMEC cells infected with strain MC58 siaD. In this experiment, we analyzed ORFs that had been identified as differentially regulated in both systems (NMB0200 and mdaB) only during interaction with HEp-2 cells (siaA and ctrC) or HBMEC (secY and NMB0985). The RT-PCR data confirmed the microarray-observed regulation of mdaB, siaA, ctrC, secY, NMB0200, and NMB0985 during the interaction with both host cell types (Table 4). In addition, the stable transcription of NMB2052 during host cell interaction was confirmed and served to normalize the increased transcript levels of induced genes. Hence, all genes detected as induced by microarrays were also found to be upregulated by the RT-PCR analysis. Likewise, the results for the genes identified as nonderegulated (i.e., being below the 1.6-fold cutoff) were almost identical for both technology platforms (Table 4). This close correlation of the RT-PCR data to the microarray-assessed transcription profile of the genes investigated underlines the relevance of the microarray data for both experimental conditions.
TABLE 4.
Assessment of differential regulation of N. meningitidis serogroup B ORFs during interaction with HEp-2 cells and HBMEC by RT-PCR and comparison to array data
| ORF no.a (gene or protein) | Differential regulation (fold) by test with cell type:
|
|||
|---|---|---|---|---|
| HEp-2
|
HBMEC
|
|||
| RT-PCR | Array | RT-PCR | Array | |
| NMB0070 (siaA) | 2.0 | 2.2 | 1.2 | 1.4 |
| NMB0073 (ctrC) | 3.0 | 2.2 | 0.9 | 1.2 |
| NMB0162 (secY) | 1.2 | 1.0 | 2.0 | 1.9 |
| NMB0200 (hypothetical protein) | 1.9 | 2.1 | 1.7 | 2.1 |
| NMB0985 (E16-related protein) | 1.4 | 1.4 | 1.8 | 2.1 |
| NMB1857 (mdaB) | 2.6 | 2.2 | 2.4 | 2.4 |
ORF numbers from the published N. meningitidis serogroup B genome sequence (32) are given.
In our experimental design, a capsule-deficient mutant with an insertionally inactivated polysialyltransferase gene (siaD) was chosen to increase the adhesion of meningococci to epithelial and endothelial cells. Since we observed an increased transcriptional activity in the sia operon, in which siaD is one of four genes (8), we had to exclude effects of the insertional inactivation of siaD on the transcription rate of the entire sia operon. For this purpose, HEp-2 cells were infected with encapsulated MC58 wild-type bacteria under the same experimental conditions described above for the siaD mutant. After 6 h of infection, mRNA was prepared and the siaA cDNA was amplified according to the quantitative RT-PCR LightCycler protocol. For siaA, a 1.6-fold increase in transcription was observed, thus confirming the results from the microarray experiments with the siaD mutant.
DISCUSSION
In the present study, we characterized for the first time the transcriptome of a bacterial pathogen (N. meningitidis) during interaction with human host cells. As a technology platform, oligonucleotide-based DNA microarrays encompassing the entire genome of N. meningitidis serogroup B strain MC58 (2,158 ORFs) were employed. The same arrays had recently been used for the elucidation of the heat shock transcriptome of N. meningitidis (13). In those studies, the oligonucleotide arrays exhibited high sensitivity, specificity, and reproducibility. They allow a 1.6-fold deregulation to be detected at a level of confidence of 99.7%, clearly demonstrating the suitability of the oligonucleotide DNA arrays for transcriptional profiling in N. meningitidis (13). Here, we describe the use of these arrays to analyze the transcriptional profile of meningococci adherent to human epithelial and endothelial cells.
The present study revealed a wide range of ORFs which are transcriptionally deregulated during different stages of meningococcal infection of the human host. Upon adherence to human epithelial cells, 72 genes were found to be differentially regulated. In bacteria adherent to brain endothelial cells, 48 ORFs were deregulated. The differentially transcribed genes could be classified in several categories: genes encoding membrane proteins and transporters, gene expression regulators, and genes involved in protein biosynthesis and general metabolism. Furthermore, we found a wide range of hypothetical ORFs and ORFs with unknown function to be differentially regulated. Several of the differentially regulated genes are known to contribute to the virulence of meningococci, and their corresponding gene products are involved in the interaction of pathogenic neisseriae with epithelial and endothelial cells (7, 16, 18, 24, 28, 33, 35). In addition, NMB1857 exhibits high homology to a gene encoding the drug modulator protein MdaB present in a wide range of pathogenic bacteria, and NMB0995 codes for a putative FK506-inhibitable rotamase which is homologous to mip, encoding the macrophage infectivity potentiator of Legionella and Streptococcus (26). Of the genes differentially regulated during interaction with HEp-2 and HBMEC cells, five (rfaF, ilvI, tonB, exbD, and gdhA) were recently identified as being essential for meningococcal pathogenesis in the infant rat model (31). Interestingly, while transcription of exbD was induced only upon HEp-2 contact and gdhA was upregulated only upon HBMEC adherence, three ORFs (rfaF, ilvI, and tonB) were upregulated upon contact with both HEp-2 and HBMEC cells.
Of particular interest for the pathogenesis of meningococcal disease are the genes involved in the synthesis and export of the polysaccharide capsule (10). The current concept of the role of capsule expression for the invasion of epithelial cells suggests that the capsule inhibits meningococcal uptake by the epithelial cell, a process known to be mediated by the subcapsularly located membrane proteins Opa and Opc (34). Previously, it was shown that capsule expression undergoes phase variation either by a slipped-strand mispairing-based mechanism within the siaD gene (16) or, alternatively, by a reversible insertion of an insertion element (IS1301) in other genes of the capsular polysaccharide biosynthesis pathway (15). In contrast to a recent report of a negative transcriptional control mechanism of capsule expression in serogroup C meningococci controlled by the transcription factor CrgA (5), we could not confirm this by use of transporter gene systems (37). The data of the present study even indicate a slight increase in transcription of the capsule biosynthesis genes after contact with HEp-2 cells and are therefore in accordance with the view that capsule expression is not negatively regulated at the transcriptional level. It should be noted, however, that the meningococcal strain used in this study is nonencapsulated. Wild-type meningococci expressing the polysaccharide capsule exhibit weak adherence to both HEp-2 cells and HBMEC, which in turn does not allow isolation of sufficient amounts of RNA for microarray hybridization.
The upregulation of the iga gene is in accordance with the requirements for the IgA protease within the epithelial cell. Here, the IgA protease cleaves Lamp1 (17), and the resulting alterations of the lysosomes are thought to play a significant role in intracellular survival and trafficking (18). Among the genes upregulated upon cell adherence, we also found multiple known iron acquisition systems, suggesting iron limitation in close vicinity to the host cell surface. Of particular interest we found tonB and exbD to be upregulated. TonB is a conserved macromolecule of the inner membrane which, together with ExbB and ExbD, triggers conformational changes in TonB-dependent outer membrane receptors, including iron-uptake receptors. Recently, it was shown that tonB-dependent iron-uptake is essential for intracellular multiplication of meningococci (20). Thus, the significant upregulation of genes encoding the TonB complex and also the elevated expression rates of the igaA gene are in concordance with the requirements of the bacterium for the intracellular environment and therefore form part of the bacterial adaptation process. Interestingly, this adaptation of the meningococcus for the intracellular lifestyle already occurs upon close contact of the bacteria with the cell surface before meningococci enter the intracellular compartments.
At a first glance, the high proportion of the differentially regulated genes involved in metabolism seems to be intriguing, since it is generally assumed that virulence factors are the main determinants of the outcome of bacterial infection and that the human host cell represents a milieu rich in nutrients for bacterial growth. However, proper adaptation of bacterial metabolism to the host environment seems to be essential for in vivo replication (11). In addition, a wide range of genes encoding metabolic functions has recently been shown to be crucial for meningococcal pathogenicity in the infant rat model (31).
A substantial proportion of the ORFs in the meningococcal genome have no known function. A high number of ORFs for which no specific role has been assigned so far are transcribed differentially upon cell contact, suggesting that they may contribute to meningococcal host cell interaction. DNA microarrays may therefore provide a suitable technology platform to unravel the role of ORFs for which no biological function has previously been found.
The endothelial cells employed in this study are HBMEC and should therefore be representative for meningococcal infection of the human blood-brain barrier. The epithelial cell line used, HEp-2, was recently shown to be identical to HeLa cells. Ideally, one would use nasopharyngeal epithelial cells for modeling the first step of the infectious cycle of meningococci, adherence of the nasopharyngeal mucosa. However, the use of HEp-2 cells may nevertheless allow the identification of ORFs which are differentially regulated upon meningococcal infection of epithelial cells in general, thus allowing conclusions for nasopharyngeal epithelial cells to be drawn, at least to some extent. This was also confirmed by a recently published study on meningococcal interaction with epithelial cells (12). In that study, MC58 bacteria were used to infect 16HBE14 cells, a human bronchial epithelial cell line, and the transcriptional profiles were analyzed at 0.5, 1, 2, and 3 h p.i. by cDNA microarrays. Twenty-one ORFs were indeed found to be differentially regulated in both studies: NMB0124, NMB0129, NMB0159, NMB0167, NMB0177, NMB0328, NMB0517, NMB0617, NMB0700, NMB0787, NMB0788, NMB0880, NMB0881, NMB0977, NMB0994, NMB0995, NMB1017, NMB1377, NMB1845, NMB1857, and NMB1940. The genes differentially regulated after the adherence to 16HBE14 cells could also be classified as virulence genes (cell adherence and host-pathogen cross talk), various transporters (for iron, amino acids, and sulfur), genes encoding metabolic functions (like amino acid biosynthesis), and a variety of hypotheticals. These similarities clearly show the relevance of the data obtained by the meningococcal transcriptome analysis after infection of HEp-2 cells. Nevertheless, there is also a number of nonconcordant ORFs found to be differentially regulated in the two studies. However, this may be due to the use of different cell lines, bacterial strains, infection times, RNA isolation methods, microarray types, and stringencies of cutoffs chosen for the definition of differential regulation.
In our study, 13 of the ORFs (27%) differentially regulated upon cell contact show high agreement in HEp-2- and HBMEC-adherent meningococci. Interestingly, most virulence-associated genes differentially regulated in HBMEC-adherent meningococci are also induced during HEp-2 interaction, suggesting that they have a general function for host cell interaction. The nonconcordant differentially regulated genes may in contrast be specifically required for infection of epithelial or endothelial cells. For example, most iron-regulated genes, pilus and capsule synthesis genes, and iga are induced only in HEp-2-adherent bacteria. Potentially, the different transcription patterns of HEp-2- and HBMEC-adherent meningococci may be caused by bacterial interaction with the host cells through different molecules, e.g., while Opa proteins play a major role in the infection of epithelial cells (39), Opc seems to be more important for the infection of endothelial cells (35). Furthermore, the differences may also be caused by binding to different receptors on the surface of the host cells. While most of the differentially regulated genes are also present in the genome of N. meningitidis serogroup A strain Z2491, 14 deregulated ORFs are absent from this strain and may be specific for serogroup B meningococci. Interestingly, most of these comprise hypothetical ORFs. Future experiments should reveal whether these ORFs either contribute to specific virulence mechanisms of serogroup B strains or are just dispensable for meningococcal pathogenicity.
Our data support the model of dynamic interactions between meningococci and their human hosts during infection. They add a further level of meningococcal gene regulation and immune evasion to the already identified mechanisms based on mutation, horizontal transfer of DNA, and phase variation. The differentially expressed genes identified in this study are important candidates for further studies to unravel the pathogenicity mechanisms and the interaction of meningococci with their human hosts and will form the basis for the development of novel strategies to cure or prevent infection by these important pathogens.
Acknowledgments
G.D., S.K., and C.H. contributed equally to this paper.
We thank A. Unkmeir and U. Vogel for helpful discussions and M. Dietrich, I. Gentschev, and I. Metcalfe for critically reading the manuscript. We are very grateful to A. Glück and M. Dümig for expert technical assistance, and we thank R. Moxon for strain MC58 and K. S. Kim for HBMEC. A special thanks goes to 3D.
This work was supported by a grant within Sonderforschungsbereich 479 (Erregervariabilität und Wirtsreaktion bei infektiösen Krankheitsprozessen), project B2, by EU grant QLK2-CT-2001-01436, and by the Bayerische Forschungsstiftung grant “Molecular biological analysis for the development of novel antiinfectives.”
REFERENCES
- 1.Achtman, M. 1994. Clonal spread of serogroup A meningococci. A paradigm for the analysis of microevolution in bacteria. Mol. Microbiol. 11:15-22. [DOI] [PubMed] [Google Scholar]
- 2.Cartwright, K. A., and D. A. Ala'Aldeen. 1997. Neisseria meningitidis: clinical aspects. J. Infect. 34:15-19. [DOI] [PubMed] [Google Scholar]
- 3.Connolly, M., and N. Noah. 1999. Is group C meningococcal disease increasing in Europe? A report of surveillance of meningococcal infection in Europe 1993-1996. Epidemiol. Infect. 122:41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cummings, C. A., and D. A. Relman. 2000. Using DNA microarrays to study host-microbe interactions. Emerg. Infect. Dis. 6:513-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deghmane, A. E., S. Petit, A. Topilko, Y. Pereira, D. Giorgini, M. Larribe, and M. K. Taha. 2000. Intimate adhesion of Neisseria meningitidis to human epithelial cells is under the control of the crgA gene, a novel LysR-type transcriptional regulator. EMBO J. 19:1068-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Detweiler, C. S., D. B. Cunanan, and S. Falkow. 2001. Host microarray analysis reveals a role for the Salmonella response regulator phoP in human macrophage cell death. Proc. Natl. Acad. Sci. USA 98:5850-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Vries, F. P., E. A. van Der Ende, J. P. van Putten, and J. Dankert. 1996. Invasion of primary nasopharyngeal epithelial cells by Neisseria meningitidis is controlled by phase variation of multiple surface antigens. Infect. Immun. 64:2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards, U., A. Muller, S. Hammerschmidt, R. Gerardy-Schahn, and M. Frosch. 1994. Molecular analysis of the biosynthesis pathway of the alpha-2,8 polysialic acid capsule by Neisseria meningitidis serogroup B. Mol. Microbiol. 14:141-149. [DOI] [PubMed] [Google Scholar]
- 9.Frosch, M., and T. F. Meyer. 1992. Transformation-mediated exchange of virulence determinants by co-cultivation of pathogenic Neisseriae. FEMS Microbiol. Lett. 79:345-349. [DOI] [PubMed] [Google Scholar]
- 10.Frosch, M., C. Weisgerber, and T. F. Meyer. 1989. Molecular characterization and expression in Escherichia coli of the gene complex encoding the polysaccharide capsule of Neisseria meningitidis serogroup B. Proc. Natl. Acad. Sci. USA 86:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goetz, M., A. Bubert, G. Wang, I. Chico-Calero, J. A. Vazquez-Boland, M. Beck, J. Slaghuis, A. A. Szalay, and W. Goebel. 2001. Microinjection and growth of bacteria in the cytosol of mammalian cells. Proc. Natl. Acad. Sci. USA 98:12221-12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grifantini, R., E. Bartoloni, A. Muzzi, M. Draghi, E. Frigimelica, J. Berger, G. Ratti, R. Petracca, G. Galli, M. Agnusdei, M. M. Giuliani, L. Santini, B. Brunelli, H. Tettelin, R. Rappuoli, F. Randazzo, and G. Grandi. 2002. Previously unrecognized vaccine candidates against group B meningococcus identified by DNA microarrays. Nat. Biotechnol. 20:914-921. [DOI] [PubMed] [Google Scholar]
- 13.Guckenberger, M., S. Kurz, C. Aepinus, S. Theiss, S. Haller, T. Leimbach, U. Panzner, J. Weber, H. Paul, A. Unkmeir, M. Frosch, and G. Dietrich. 2002. Analysis of the heat shock response of Neisseria meningitidis with DNA microarrays. J. Bacteriol. 184:2546-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas, R., and T. F. Meyer. 1986. The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell 44:107-115. [DOI] [PubMed] [Google Scholar]
- 15.Hammerschmidt, S., R. Hilse., J. P. van Putten, R. Gerardy-Schahn, A. Unkmeir, and M. Frosch. 1996. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 15:192-198. [PMC free article] [PubMed] [Google Scholar]
- 16.Hammerschmidt, S., A. Muller, H. Sillmann, M. Muhlenhoff, R. Borrow, A. Fox, J. van Putten, W. D. Zollinger, R. Gerardy-Schahn, and M. Frosch. 1996. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol. Microbiol. 20:1211-1220. [DOI] [PubMed] [Google Scholar]
- 17.Hauck, C. R., and T. F. Meyer. 1997. The lysosomal/phagosomal membrane protein h-lamp-1 is a target of the IgA1 protease of Neisseria gonorrhoeae. FEBS Lett. 405:86-90. [DOI] [PubMed] [Google Scholar]
- 18.Hopper, S., B. Vasquez, A. Merz, S. Clary, J. S. Wilbur, and M. So. 2000. Effects of the immunoglobulin A1 protease on Neisseria gonorrhoeae trafficking across polarized T84 epithelial monolayers. Infect. Immun. 68:906-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolb-Mäurer, A., A. Unkmeir, U. Kämmerer, C. Hübner, T. Leimbach, A. Stade, E. Kaempgen, M. Frosch, and G. Dietrich. 2001. Interaction of Neisseria meningitidis with dendritic cells. Infect. Immun. 69:6912-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larson, J. A., D. L. Higashi, I. Stojiljkovic, and M. So. 2002. Replication of Neisseria meningitidis within epithelial cells requires TonB-dependent acquisition of host cell iron. Infect. Immun. 70:1461-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenzen, D. R., F. Düx, U. Wölk, A. Tsirpouchtsidis, G. Haas, and T. F. Meyer. 1999. Immunoglobulin A1 protease, an exoenzyme of pathogenic Neisseriae, is a potent inducer of proinflammatory cytokines. J. Exp. Med. 190:1049-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 23.Pizza, M., V. Scarlato, V. Masignani, M. M. Giuliani, B. Arico, M. Comanducci, G. T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, C. L. Galeotti, E. Luzzi, R. Manetti, E. Marchetti, M. Mora, S. Nuti, G. Ratti, L. Santini, S. Savino, M. Scarselli, E. Storni, P. Zuo, M. Broeker, E. Hundt, B. Knapp, E. Blair, T. Mason, H. Tettelin, D. W. Hood, A. C. Jeffries, N. J. Saunders, D. M. Granoff, J. C. Venter, E. R. Moxon, G. Grandi, and R. Rappuoli. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816-1820. [DOI] [PubMed] [Google Scholar]
- 24.Pujol, C., E. Eugene, M. de Saint, and X. Nassif. 1997. Interaction of Neisseria meningitidis with a polarized monolayer of epithelial cells. Infect. Immun. 65:4836-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rappuoli, R. 2000. Pushing the limits of cellular microbiology: microarrays to study bacteria-host cell intimate contacts. Proc. Natl. Acad. Sci. USA 97:13467-13469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sampson, B. A., and E. C. Gotschlich. 1992. Neisseria meningitidis encodes an FK506-inhibitable rotamase. Proc. Natl. Acad. Sci. USA 89:1164-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saunders, N. J., A. C. Jeffries, J. F. Peden, D. W. Hood, H. Tettelin, R. Rappuoli, and E. R. Moxon. 2000. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol. Microbiol. 37:207-215. [DOI] [PubMed] [Google Scholar]
- 28.Scheuerpflug, I., T. Rudel, R. Ryll, J. Pandit, and T. F. Meyer. 1999. Roles of PilC and PilE proteins in pilus-mediated adherence of Neisseria gonorrhoeae and Neisseria meningitidis to human erythrocytes and endothelial and epithelial cells. Infect. Immun. 67:834-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens, D. S., and M. M. Farley. 1991. Pathogenic events during infection of the human nasopharynx with Neisseria meningitidis and Haemophilus influenzae. Rev. Infect. Dis. 13:22-33. [DOI] [PubMed] [Google Scholar]
- 30.Stins, M. F., F. Gilles, and K. S. Kim. 1997. Selective expression of adhesion molecules on human brain microvascular endothelial cells. J. Neuroimmunol. 76:81-90. [DOI] [PubMed] [Google Scholar]
- 31.Sun, Y. H., S. Bakshi, R. Chalmers, and C. M. Tang. 2000. Functional genomics of Neisseria meningitidis pathogenesis. Nat. Med. 6:1269-1273. [DOI] [PubMed] [Google Scholar]
- 32.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 33.Virji, M., H. Kayhty, D. J. P. Ferguson, C. Alexandrescu, J. E. Heckels, and E. R. Moxon. 1991. The role of pili in the interactions of pathogenic Neisseria with cultured human endothelial cells. Mol. Microbiol. 5:1831-1841. [DOI] [PubMed] [Google Scholar]
- 34.Virji, M., K. Makepeace, D. J. Ferguson, M. Achtmann, and E. R. Moxon. 1993. Meningococcal Opa and Opc proteins: their role in colonization and invasion of human epithelial and endothelial cells. Mol. Microbiol. 10:499-510. [DOI] [PubMed] [Google Scholar]
- 35.Virji, M., K. Makepeace, I. R. Peak, D. J. Ferguson, M. P. Jennings, and E. R. Moxon. 1995. Opc- and pilus-dependent interactions of meningococci with human endothelial cells: molecular mechanisms and modulation by surface polysaccharides. Mol. Microbiol. 18:741-754. [DOI] [PubMed] [Google Scholar]
- 36.Vogel, U., and M. Frosch. 1999. Mechanisms of neisserial serum resistance. Mol. Microbiol. 32:1133-1139. [DOI] [PubMed] [Google Scholar]
- 37.von Loewenich, F. D., E. Wintermeyer, M. Dumig, and M. Frosch. 2001. Analysis of transcriptional control mechanisms of capsule expression in Neisseria meningitidis. Int. J. Med. Microbiol. 291:361-369. [DOI] [PubMed] [Google Scholar]
- 38.Wells, D. B., P. J. Tighe, K. G. Wooldridge, K. Robinson, and D. A. Ala'Aldeen. 2001. Differential gene expression during meningeal-meningococcal interaction: evidence for self-defense and early release of cytokines and chemokines. Infect. Immun. 69:2718-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woods, J. P., and J. G. Cannon. 1990. Variation in expression of class 1 and class 5 outer membrane proteins during nasopharyngeal carriage of Neisseria meningitidis. Infect. Immun. 58:569-572. [DOI] [PMC free article] [PubMed] [Google Scholar]