Abstract
The aureolic acid antibiotic mithramycin (MTM) binds selectively to GC-rich DNA sequences and blocks preferentially binding of proteins, like Sp1 transcription factors, to GC-rich elements in gene promoters. Genetic approaches can be applied to alter the MTM biosynthetic pathway in the producing microorganism and obtain new products with improved pharmacological properties. Here, we report on a new analog, MTM SDK, obtained by targeted gene inactivation of the ketoreductase MtmW catalyzing the last step in MTM biosynthesis. SDK exhibited greater activity as transcriptional inhibitor compared to MTM. SDK was a potent inhibitor of Sp1-dependent reporter activity and interfered minimally with reporters of other transcription factors, indicating that it retained a high degree of selectivity toward GC-rich DNA-binding transcription factors. RT–PCR and microarray analysis showed that SDK repressed transcription of multiple genes implicated in critical aspects of cancer development and progression, including cell cycle, apoptosis, migration, invasion and angiogenesis, consistent with the pleiotropic role of Sp1 family transcription factors. SDK inhibited proliferation and was a potent inducer of apoptosis in ovarian cancer cells while it had minimal effects on viability of normal cells. The new MTM derivative SDK could be an effective agent for treatment of cancer and other diseases with abnormal expression or activity of GC-rich DNA-binding transcription factors.
INTRODUCTION
Deregulation of transcription factor activity is an important event in the pathogenesis of many human diseases, including cancer, chronic inflammatory, cardiovascular and neurodegenerative disorders (1,2). Compounds able to block over-active transcription factors and modulate gene expression could be very attractive therapeutic agents. Aureolic acid antibiotics, like mithramycin (MTM) and chromomycin, are natural polycyclic aromatic polyketides produced by various Streptomyces species (3). These compounds contain an identical tricyclic chromophore with a unique hydrophilic side chain attached in 3-position and different saccharide chains attached in 2 and 6 positions (3). Compounds like MTM have the interesting property of binding to GC-rich DNA sequences selectively (4). Mg2+-coordinated dimers of MTM bind non-covalently to DNA in the minor groove with the chromophores parallel to the sugar-phosphate backbone and saccharide chains wrapping across the minor groove (4,5). The chromophores form specific hydrogen bonds with the NH2 of guanines determining the selectivity for GC-rich sequences (4,5). As consequence of this sequence selectivity, MTM blocks preferentially binding of proteins, like Sp1 family transcription factors, to GC-rich sequences in gene promoters and inhibit transcription of genes regulated by these factors (6–10). In this manner, MTM has the potential to inhibit expression of many genes involved in cancer pathogenesis and therapeutically relevant (6–11). MTM is active against a variety of human cancers in experimental models and has been used clinically for many years to treat testicular carcinoma as well as hypercalcemia in patients with metastatic bone lesions and Paget's disease (3,12,13). Its current clinical use is limited by its severe side effects that include gastrointestinal, hepatic, kidney and bone marrow toxicity. Nevertheless, MTM has recently attracted renewed attention as an experimental therapeutic agent in cancer and non-cancer-related disorders (14–22). The availability of new analogs with improved pharmacological and toxicological properties and a better understanding of their effects on gene expression may open new perspectives for therapeutic use of this class of compounds.
Metabolic engineering, in which a compound's biosynthetic pathway is altered by gene inactivation, recombination or mutation, has been used successfully to increase biodiversity and produce new ‘unnatural’ natural products with an improved pharmacological profile (23,24). The MTM biosynthetic pathway has been almost completely elucidated in recent years (25–31). Its biosynthesis proceeds through the condensation of multiple acyl-coA monomers catalyzed by type II polyketide synthases. The initial condensation phase leads to the formation of the tetracyclic intermediate premithramycinone, which is subsequently modified by the addition of saccharide chains leading to the formation of premithramycin B (Figure 1). The final steps, which are catalyzed by the oxygenase MtmOIV and ketoreductase MtmW, are the oxidative cleavage of the fourth ring of premithramycin B followed by the decarboxylation and ketoreduction of the pentyl side chain in C-3 (25,29). We have applied genetic approaches to modify MTM biosynthesis in the attempt to produce compounds that would share the same basic mechanism of action of the parent compound, but have increased potency and/or improved therapeutic index. Attempts made over the years have yielded compounds exhibiting distinct structural changes and have allowed us to gain information on the structure-activity relationships of this type of compounds (11,28–30). While modifications of the carbohydrate and aglycon moieties disrupted almost completely activity, changes in the 3-side chain were more promising (11,28,29).
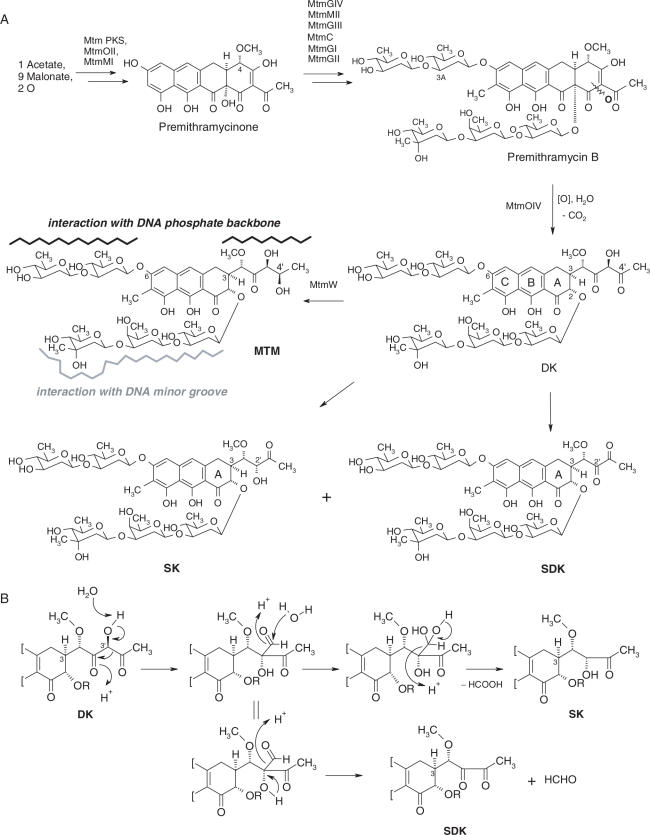
Figure 1.
Biosynthesis of the MTM analogs SDK and SK. (A) The oxygenase MtmOIV and ketoreductase MtmW catalyze the conversion of premithramycin B to mithramycin (MTM). Inactivation of MtmW prevents synthesis of MTM and results in accumulation of the intermediate product DK, which is then converted into SK and SDK. Regions of relevant MTM-DNA interaction are indicated. (B) Side chain rearrangements leading to formation of SK and SDK from the MtmOIV product DK.
In an effort to find new analogs with improved pharmacological properties, we have identified recently a new compound produced by targeted inactivation of the ketoreductase MtmW gene. The compound, named MTM SDK (SDK), had a fully glycosylated tricyclic aglycon and differed with respect to MTM only in the structure and length of the 3-side chain (Figure 1). Here, we show that SDK is a potent inhibitor of transcription, cancer cell proliferation and survival. Our data suggest that this new MTM analog could be a valid agent for treatment of cancer and other diseases with abnormal activity of GC-rich DNA-binding transcription factors and over-expression of their target genes.
MATERIALS AND METHODS
Isolation of SDK and SK
MTM and the secondary metabolite MTM SK (SK) (Figure 1) were initially isolated as described previously (29). The following procedure was later adopted to produce large amounts of SDK and SK. Seed cultures were first prepared by inoculating spores of Streptomyces argillaceus M7W1 in TSB (tryptone soya broth) medium and incubating in an orbital shaker at 30°C for 36 h (29). Seed cultures (3.5 ml/flask) were then used to inoculate 10 Erlenmeyer flasks with 100 ml of modified R5 medium. After three days, cultures were monitored by high-performance liquid chromatography (HPLC) to avoid decomposition of SDK into another secondary metabolite, MTM SA (29). The cultures were harvested as soon as SA was observed (Supplementary Data S1). Cells were centrifuged at 4000 r.p.m. (750 mL EC 2273 rotor; 4550 g) for 35 min, and the culture filtrate was passed through a 5 × 10 cm RP C18 column using vacuum (diaphragm pump, 1–5 mbar). The column was washed with 100% water and then eluted with a 5–30% acetonitrile gradient in water. Only fractions containing SK and SDK were collected. These fractions were combined and further purified on a 2 × 10 cm RP C18 column using a gradient of acetonitrile from 25 to 30% in water. SK was eluted first (yield ∼50 mg), followed by SDK (yield ∼30 mg). NMR spectroscopy and mass spectrometry data of SDK are presented as Supplementary Data S2. Stock solutions of MTM, SK and SDK were prepared in sterile DMSO at a concentration of 10 mM and stored at −20°C. For each experiment drug dilutions at the desired concentrations were made in tissue culture medium and used immediately.
Cell lines and plasmids
A2780, OVCAR3, OVCAR5, OVCAR8, OVCAR432, SKOV3 and IGROV1 ovarian cancer cell lines were maintained in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS). Primary cultures of normal human fibroblasts were maintained in DMEM supplemented with 10% heat-inactivated FBS. Luciferase reporter plasmids with the c-src, c-myc and ets2 promoters have been described previously (11,32,33). The TransLucent Sp1 reporter (Sp1-Luc) and control reporter (Control-Luc) vectors were purchased from Panomics (Redwood City, CA). The pCMV-Sp1 and PPREx3-tk-Luc were gifts of Dr D. Kardassis and Dr R. Evans, respectively. The AP1, SRE and NF-kB reporters were provided by Dr G. Natoli.
Luciferase reporter assays
Cells were plated at a density of 2 × 104 cells/well in 48-well plates and transfected with 200 ng of reporter plasmid using Lipofectamine2000 (Invitrogen). After 4 h, the medium was removed and replaced with fresh medium containing the desired concentrations of each compound. Cells were incubated for additional 18 h and luciferase activity was measured in cell extracts using the Promega Luciferase assay system. Reporter activity was normalized to protein content in cell extracts measured with the BioRad assay.
DNA-binding assays
Fluorescence spectroscopy experiments were carried at 20°C in 20 mM Tris–HCl buffer, pH 8.0. Drugs were pre-incubated in the same buffer containing 10 mM MgCl2 for 1 h at room temperature to ensure complete drug–Mg2+ complex formation. Salmon sperm DNA was pre-incubated in the same buffer containing 10 mM MgCl2 to avoid cation-induced changes in conformation during its association with the drug. DNA was added in small aliquots to the samples before taking fluorescence measurements. DNA concentration during the titration experiments ranged from 0 to 200 µM while total drug concentration was essentially kept constant at 20 µM. Fluorescence emission spectra were collected between 500 and 640 nm upon excitation at 470 nm (bandwidths of 5 and 10 nm for the excitation and emission, respectively) using a PerkinElmer LS 55 Luminescence spectrometer equipped with a 100 µl quartz cuvette. A total of six scans per experiment were recorded and the error in the fluorescence intensity between measurements was <5%. The change in fluorescence emission intensity (ΔF; λex = 470 nm and λemi = 515 nm) after each addition of DNA and its maximum value (ΔFmax) were calculated for each titration experiment. Dissociation constants of the drug–DNA complexes were determined by means of non-linear curve fitting analysis of fluorescence titration data from three independent experiments. DNase I footprinting experiments were done on a 223 bp c-src promoter fragment as described previously (11). Samples were analyzed by electrophoresis on denaturating polyacrylamide gels.
Electrophoretic mobility shift assays
Nuclear extracts were prepared using NE-PERR Nuclear and Cytoplasmic extraction kit (Pierce Biotechnology) according to manufacturer's instructions. Protein concentration in nuclear extracts was determined using the BCA protein assay kit (Pierce Biotechnology). HPLC-purified single-stranded oligonucleotide containing a canonical Sp1 binding sequence (5′-ATTCGATCGGGGCGGGGCGAGC-3′) and the complementary oligonucleotide were purchased from Sigma Genosys (Steinheim, Germany). Both oligonucleotides were labeled by adding biotinylated ribonucleotides at the 3′-OH end using the Biotin 3′ end DNA labeling kit (Pierce Biotechnology). Duplex DNA was formed by incubating the labeled oligonucleotides in 10 mM Tris–HCl, pH 8.0, 50 mM NaCl, 1 mM EDTA and 10 mM MgCl2 for 15 min at 95°C followed by slow cooling to room temperature. Duplex DNA (50–75 fmol) was then incubated with the compounds in a buffer containing 10 mM Tris–HCl, pH 7.5 and 5 mM MgCl2 for 1 h at 20°C. Nuclear extract was pre-incubated on ice in binding buffer containing 10 mM Tris–HCl, pH 7.5, 50 mM KCl, 1 mM DTT, 2.5% glycerol, 2.5 mM MgCl2, 25 ng Poly(dI–dC), 0.5 mM EDTA and 0.05% NP-40. Aliquots of nuclear extract (6 µg of protein/sample) were combined with drug–DNA complexes and incubated for additional 20 min at 20°C. Gel mobility shift assays were performed using the LightShift Chemiluminescent EMSA kit (Pierce Biotechnology) following the manufacturer's instructions. Non-denaturing 6% polyacrylamide gels were pre-run for 1.5 h at 4°C and electrophoresis was carried out at 150 V for 45 min using 0.5× TBE as running buffer. Gels were transferred to positive-charged BiodyneR B Nylon membranes (Pierce Biotechnology) and cross-linked in BioRad GS Gene Linker. Detection of biotin-labeled DNA was done using Streptavidin–horseradish peroxidase conjugate and Chemilumiscent Substrate (Pierce Biotechnology).
Cellular uptake
A2780 cells were incubated with 100 µM of each compound for 2 or 4 h. At the end of the drug incubation, cells were washed with ice-cold phosphate-buffered saline (PBS) repeatedly, harvested and immediately analyzed by flow cytometry. To isolate intact nuclei, cells were suspended in ice-cold nuclei extraction buffer (320 mM sucrose, 5 mM MgCl2, 10 mM HEPES and 1% Triton X-100 at pH 7.4) and incubated for 10 min on ice. Nuclei were recovered by centrifugation, washed twice and then analyzed by flow cytometry. Nuclei yield and integrity were confirmed by trypan blue staining and microscopic examination.
RNA isolation, RT–PCR and real-time PCR
Cells were seeded at a density of 1 × 105 cells/flask and treated with drugs for the indicated times. Total RNA was isolated using Trizol (Invitrogen) and further purified with RNeasy MiniKit (Qiagen). RNA concentration was determined using a NanoDrop spectrophotometer (Witec AG, CH). RT–PCR was performed under non-saturating conditions using the SuperScript One-Step RT–PCR system (Invitrogen) and gene-specific primers (for primer sequences and PCR conditions see Supplementary Data S3). PCR products were separated on 2% agarose gels, stained with ethidium bromide and visualized using the AlphaImager 3400 (AlphaInnotech). Band intensity was determined using the AlphaEase Software. For quantitative real-time PCR, 1 µg of total RNA was reverse-transcribed using SuperScript First-Strand Synthesis System (Invitrogen). Real-time PCR was performed in triplicate samples on an ABI Prism 7000 Sequence Analyzer using primer sets for VEGF and β-2-microglobulin (Assays-on-Demand, ABI) and TaqMan Universal PCR Mastermix (ABI). Standard curves were generated for each primer set and β-2-microglobulin RNA was used as control to quantify VEGF RNA.
Immunoblotting
Cell lysates were prepared from control and drug-treated cells and proteins separated on SDS–polyacrylamide gels as described (32). Blots were incubated with an antibody able to detect the cleaved 85 kDa form of PARP (BD Pharmingen), while an antibody against α-tubulin (Santa Cruz) was used to control sample loading. Sp1 protein level was determined using an anti-Sp1 antibody (clone sc59, SantaCruz). Blots were developed using peroxidase-conjugated secondary antibodies and the enhanced chemiluminescence system (ECL, Amersham).
Microarray analysis
Cells were plated in tissue culture flasks and incubated without or with 100 nM SDK for 6 h. Three independent replicates were done for each experimental group. Total RNA was isolated using Trizol and RNeasy columns (Qiagen). RNA quality was verified using the 2100 BioAnalyzer (Agilent Technologies). Of total RNA 10 µg was labeled and hybridized to HG-U133A and B Affymetrix GeneChips according to the manufacturer's protocol. After overnight incubation, the arrays were washed on the Fluidics Station 400 and images collected on GeneChip Scanner 3000 using the GeneChip Operating System (GCOS) software. Raw signals were extracted with MAS 5.0 software (Affymetrix) and further analysis was performed using the Bioconductor statistical package (www.bioconductor.org). Expression values were calculated from the raw CEL files using RMA (34). To remove non-varying genes, a variational filter was applied to the data to retain genes with a SD of at least 0.2 across all samples. Transcripts with statistically significant changes in expression between the two treatment groups were identified from this set using SAM (35). A cut-off value of Δ = 0.5 was used giving an estimated FDR of 0.027 and q-values for individual transcripts smaller than 0.031. Approximately 3400 transcripts were found to be differentially expressed with most transcripts (n = 3355) down-regulated and only 49 transcripts up-regulated. A large number of transcripts (∼2400) were reduced <2-fold, while ∼900 transcripts had a ≥2-fold change in expression. Only genes (n = 460) with ≥2.5-fold change in expression were considered for further analysis and functional classification (Supplementary Data S2). The Panther Classification System was used to classify affected genes in biological process, molecular function and pathway classes (36). Presence of GC-rich sequence and Sp1-binding sites in gene promoters was searched using GeneSpring 7.2 software (Silicon Genetics), DBTSS (http://dbtss.hgc.jp/index.html) or AliBaba 2.1 (http://darwin.nmsu.edu).
Cell proliferation, cell cycle and apoptosis assays
Cells were seeded in 96-well plates at a density of 1.5–2 × 103 cells/well and incubated with drugs for 72 h. Each treatment was performed in triplicate and experiments were repeated at least three times. The number of viable cells at the end of the drug incubation was determined using a colorimetric MTT assay (37). For cell cycle analysis, cells (1 × 105cells/flask) were treated for 24 or 48 h and then harvested, washed with PBS, and fixed in 80% ETOH. After staining with propidium iodide, samples were analyzed by flow cytometry (FACSCalibur, Becton Dickinson) as described previously (37). Cell cycle distribution and percentage of apoptotic cells were determined using ModFit (Verity). For detection of apoptotic cells, control and drug-treated cells were harvested, washed with PBS, and incubated with Annexin V-FITC and propidium iodide before analysis by flow cytometry.
RESULTS
Biosynthesis of SDK
The last steps in MTM biosynthesis are catalyzed by the oxygenase MtmOIV and ketoreductase MtmW (25,29). Modifications of the isolation procedure of MTM metabolites from the S.argillaceus M7W1 mutant strain showed that targeted inactivation of the MtmW gene resulted in the accumulation of a new product, SDK, in addition to the previously described SK (Figure 1A). Detection of SDK, which is acid sensitive and poorly soluble in ethylacetate, had been missed in earlier work because an ethylacetate extraction step and HPLC acidic elution solvent were used in the isolation procedure (29). The newly discovered analog SDK (for short side chain, diketo) had the fully glycosylated tricyclic aglycon and 3-butyl side chain similar to SK, but with two keto groups instead the keto- and secondary alcohol group of SK. Figure 1B shows the proposed pathway leading to the formation of SK and SDK via a rearrangement process triggered by the inactivation of MtmW. This mechanism is supported by 13C incorporation studies, which proved that C-3′ is excised during the formation of SK (29). In the absence of the ketoreductase, the product of the MtmOIV reaction, MTM DK (DK), undergoes a spontaneous rearrangement in its 3-β-diketo side chain, triggered by a β-shift and followed by elimination of either formic acid or formaldehyde to form SK and SDK, respectively. This view has been confirmed recently through studies with the isolated enzyme MtmOIV, showing that SDK is a major end product of the conversion of premithramycin B at pH 8.25 (38). A similar compound with a short α-diketo-butyl side chain was also found as a product of a mutant of the chromomycin producer Streptomyces griseus ssp. Griseus (39).
Transcription inhibition by MTM analogs
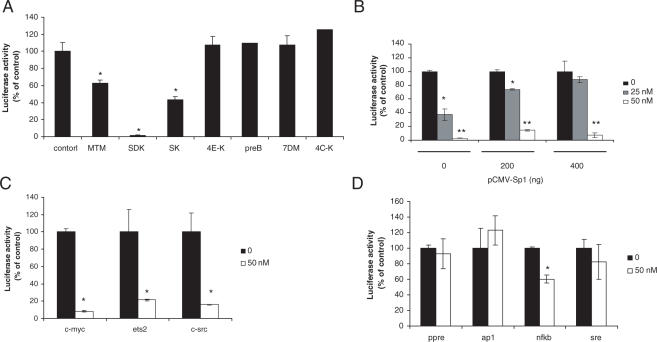
We used a luciferase reporter assay to compare the ability of MTM derivatives to block transcription driven by GC-rich DNA-binding transcription factors, like Sp1. Transfection of A2780 cells with the Sp1 reporter gave high levels of activity with ≥30-fold induction compared with the control vector lacking Sp1-binding sites, consistent with the high Sp1 protein level seen in western blots (data not shown). Next, cells were transfected with the Sp1 reporter and incubated with 100 nM of each MTM derivative for 18 h before measuring luciferase activity. The new analog SDK was the most potent compound in the reporter assays inducing >90% inhibition at 100 nM, to a level comparable with that seen with the control vector lacking Sp1 binding sites (Figure 2A). At this dose, MTM and SK inhibited the Sp1 reporter only 40 and 60%, respectively. All other MTM derivatives were inactive at this dose. SDK was active also at lower concentrations with ∼60 and 90% inhibition at 25 and 50 nM, respectively (Figure 2B). Transfection of increasing amounts of a Sp1 expression vector prior to SDK treatment prevented inhibition of the reporter completely at 25 nM and partially at 50 nM, confirming that the compound acted by competing with Sp1 for binding to GC-rich consensus sequence (Figure 2B). Next, we extended the analysis to promoter reporters of genes, like c-myc, ets2 and c-src, containing GC-rich elements and Sp1-binding sites. Activity of these reporters was inhibited by SDK with 80–90% inhibition at 50 nM (Figure 2C). On the other hand, reporter constructs containing responsive elements for other transcription factors (i.e. AP1, SRE, PPRE and NF-kB) without GC-rich elements were not or much less affected by SDK compared with the Sp1 reporter (Figure 2D).
Figure 2.
Effects of MTM analogs on promoter reporter activity. (A) A2780 cells were transfected with Sp1 reporter vector for 4 h and incubated for 18 h in medium with or without 100 nM of the indicated compounds before measuring luciferase activity. Data are mean ± SD of triplicate samples. *P < 0.01 as compared with control cells. MTM, mithramycin; SDK, mithramycin SDK; SK, mithramycin SK; 4E-K, 4E-keto-mithramycin; PreB, premithramycin B; 7DM, 7-demethylmithramycin; 4C-K, 4C-keto-demycarosylmithramycin. (B) Sp1 reporter activity in A2780 cells transfected with increasing amounts of Sp1 expression vector or empty vector and treated with 25 and 50 nM of SDK. *P < 0.01and **P < 0.001 compared with control cells. (C) Luciferase activity in A2780 cells transfected with c-myc, ets2 and c-src promoter reporters and treated with 50 nM of SDK. *P < 0.01 compared with control cells. (D) Luciferase activity in cells transfected with PPRE, AP1, NF-kB and SRE reporters and treated with 50 nM of SDK.
DNA-binding properties and cellular uptake of MTM analogs
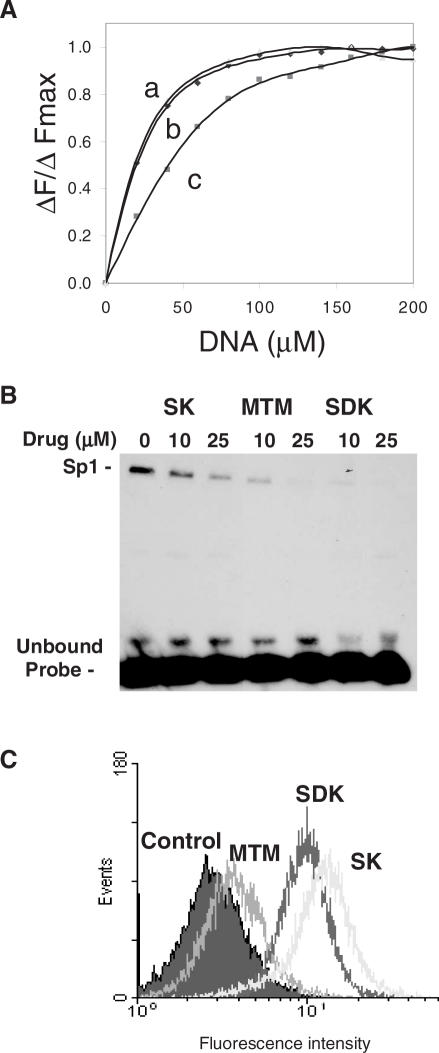
To determine the factors that might be responsible for the increased potency of the new analog, we compared DNA-binding properties, ability to block Sp1 binding and cellular uptake of MTM, SK and SDK. DNA-binding properties of the three compounds were studied by monitoring changes in fluorescence emission spectra at increasing DNA concentrations. Binding to DNA induces dequenching of the drug's fluorochromes with an increase in fluorescence intensity. Figure 3A shows the changes of fluorescence intensity normalized to the maximum fluorescence intensity (ΔF/ΔFmax) for each compound as a function of the DNA concentration in the binding reactions. SDK and MTM bound to DNA with similar affinity, while SK had lower affinity. Dissociation constants (Kd) calculated from the fluorescence titration experiments were 19, 20 and 40 µM for MTM, SDK and SK, respectively. Footprinting experiments done on a 223 bp fragment of the c-src promoter (11) confirmed that SDK had similar preference for GC-rich sequences as MTM and SK (data not shown).
Figure 3.
DNA binding, inhibition of Sp1 binding and cellular uptake of MTM analogs. (A) Fluorescence spectroscopy. Normalized changes in fluorescence intensity of MTM (curve a), SDK (curve b), SK (curve c) are plotted as a function of increasing concentration of salmon sperm DNA. (B) Gel mobility shift assay. The biotin-labeled duplex oligonucleotide with an Sp1 binding site was incubated with MTM, SK and SDK at concentrations of 0, 10 and 20 µM for 1 h prior to the addition of nuclear extract. Samples were incubated for 20 min and analyzed on a non-denaturating polyacrylamide gel. The position of Sp1–DNA complex and unbound probe are indicated. (C) Flow cytometry analysis of drug uptake. Cells were incubated with 100 µM of MTM, SK and SDK. After 4 h, cells were harvested, repeatedly washed with ice-cold PBS and analyzed by FACS to determine the amount of compound accumulated in cells.
Next, we examined the ability of the compounds to interfere with GC-rich DNA-binding proteins, like Sp1, in gel mobility shift assays. A duplex oligonucleotide probe containing a canonical Sp1-binding sequence was incubated with the compounds before the addition of nuclear extract. Presence of Sp1 and binding to the probe had been demonstrated in separate experiments by Western blotting and competition assays (data not shown). SDK at 10 µM inhibited almost completely formation of the Sp1–DNA complex (Figure 3B). MTM was less effective in blocking Sp1 binding despite the similar binding affinity shown in fluorescence titration experiments. SK, consistent with its reduced binding to DNA, was less effective than both MTM and SDK in blocking Sp1 binding.
The ability to inhibit transcription in cells could be greatly affected by differences in cellular uptake. Therefore, flow cytometry analysis was carried out to measure drug accumulation following incubation of cells with the compounds. SDK was taken up by cells very rapidly and accumulated in much larger amounts than MTM (Figure 3C). In addition to enter cells rapidly, SDK accumulated in cell nuclei as shown by fluorescence microscopy and flow cytometry of isolated nuclei (data not shown). The amount of SK entering cells was also higher than MTM, a fact that could compensate at least in part for its reduced DNA-binding properties.
Effects of MTM analogs on endogenous gene transcription
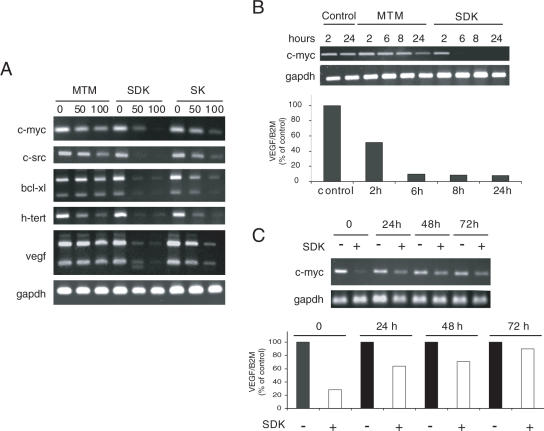
To assess the effects on transcription of endogenous genes, A2780 cells were incubated with MTM, SK and SDK for 24 h and expression of genes, like c-myc, c-src, hTERT, VEGF and bcl-xL, with known Sp1-binding sites was monitored by RT–PCR. All three compounds inhibited transcription, although there were clear differences in their relative potency (Figure 4A). SDK inhibited transcription of all the genes by at least 90% at 50 nM. SK was less potent than SDK, reaching comparable levels of inhibition at 100 nM. MTM reduced c-myc, c-src and hTERT transcript levels by 50–70% at 100 nM, while it inhibited bcl-xL and VEGF <50%. Thus, both SDK and SK inhibited transcription at lower concentrations and to a higher degree than MTM.
Figure 4.
Inhibition of transcription of Sp1-regulated genes by MTM analogs. (A) A2780 cells were incubated with 50 or 100 nM of MTM, SK and SDK for 24 h. Total RNA was isolated from control and drug-treated cells and individual transcripts assessed by RT–PCR. (B) A2780 cells were treated with 100 nM of MTM or SDK and total RNA was isolated at the indicated times. Individual transcripts were measured by RT–PCR (c-myc and GAPDH) or real-time RT–PCR (VEGF and β2-microglobulin). (C) A2780 cells were treated with 50 nM SDK for 6 h and then incubated for 24, 48 or 72 h in drug-free medium. Total RNA was isolated and transcript levels were determined by RT–PCR or real-time RT–PCR as above.
The kinetics of SDK-induced transcription inhibition was determined by measuring c-myc and VEGF RNA levels at different times from the start of the treatment. Reduction of c-myc RNA to ≤20% of control was seen within 6 h of treatment with SDK (Figure 4B). At this time, MTM induced only marginal effects on c-myc RNA, while a more consistent decrease was seen only after 24 h. A similar kinetics of inhibition was observed measuring VEGF RNA by real-time RT–PCR with ≥80% reduction in transcript levels within 6 h of treatment with SDK (Figure 4B, lower panel). We assessed also the reversibility of the SDK effects on transcription. Both c-myc and VEGF RNA levels were reduced ≥80% after 6 h with 50 nM of SDK (Figure 4C). Following drug removal, transcription of both genes was slowly restored over time. RNA levels were ∼50% of the control levels at 24 and 48 h after drug removal, while an almost complete recovery was seen after 72 h.
Gene expression profiling
GC-rich DNA-binding compounds, like SDK, could affect transcription of several genes. To better understand the extent of the transcriptional response induced by SDK and the mechanism underlying its anti-proliferative and pro-apoptotic activity, we examined gene expression of control and drug-treated cells using the HG-U133 Affymetrix GeneChip set, which contained probes for ∼45 000 transcripts. Approximately 460 genes were down-regulated ≥2.5-fold upon SDK treatment (Supplementary Data S4). Table 1 shows the top 50 genes with the highest level of down-regulation (≥4-fold). Many of the top down-regulated genes (e.g. MYC, VEGF, DKK1, MCL1, ID1, ID3 and ZNF217) are known to be up-regulated in cancer cells and have a defined role in cancer pathogenesis. Furthermore, most of these genes have GC-rich elements and Sp1 binding sites in their regulatory regions.
Table 1.
SDK down-regulated genesa
| Gene symbol | Gene title | Fold change | Expression level |
|---|---|---|---|
| ZNF217 | Zinc finger protein 217 | 7.6 | 8.9 |
| NA | CDNA FLJ11397 fis, clone HEMBA1000622 | 6.7 | 6.6 |
| SPRY2 | Sprouty homolog 2 (Drosophila) | 6.5 | 10.9 |
| ADNP | Activity-dependent neuroprotector | 6.2 | 9.9 |
| DKK1 | Dickkopf homolog 1 (Xenopus laevis) | 6.2 | 11.6 |
| MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | 6.1 | 10.6 |
| SPRY1 | Sprouty homolog 1, antagonist of FGF signaling (Drosophila) | 6.0 | 11.1 |
| PUM1 | Pumilio homolog 1 (Drosophila) | 5.8 | 10 |
| PDE4D | Phosphodiesterase 4D, cAMP-specific | 5.7 | 9.6 |
| CTCF | CCCTC-binding factor (zinc finger protein) | 5.7 | 9.7 |
| ARID5B | AT rich interactive domain 5B (MRF1-like) | 5.6 | 9.5 |
| NA | CDNA FLJ11397 fis, clone HEMBA1000622 | 5.5 | 8.4 |
| NUP153 | Nucleoporin 153kDa | 5.4 | 9.6 |
| DYRK1A | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | 5.4 | 9.7 |
| RAI17 | Retinoic acid induced 17 | 5.4 | 10.4 |
| PHLDA1 | Pleckstrin homology-like domain, family A, member 1 | 5.4 | 8.3 |
| NEDD9 | Neural precursor cell expressed, developmentally down-regulated 9 | 5.2 | 7.6 |
| MCL1 | Myeloid cell leukemia sequence 1 (BCL2-related) | 5.2 | 10.7 |
| EIF2AK3 | Eukaryotic translation initiation factor 2-alpha kinase 3 | 5.2 | 7.6 |
| SPRED2 | Sprouty-related, EVH1 domain containing 2 | 5.1 | 10.5 |
| TSC22D2 | TSC22 domain family 2 | 5.1 | 8.2 |
| RNF184 | Ring finger protein 184 | 5.1 | 8.4 |
| NA | Solute carrier family 38, member 2 | 5.0 | 11.9 |
| ID3 | Inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 5.0 | 12 |
| PPP1R15B | Protein phosphatase 1, regulatory (inhibitor) subunit 15B | 5.0 | 10.3 |
| CRSP6 | Cofactor required for Sp1 transcriptional activation, subunit 6, 77kDa | 4.9 | 8.7 |
| C19orf7 | Chromosome 19 open reading frame 7 | 4.9 | 9.4 |
| TGIF | TGFB-induced factor (TALE family homeobox) | 4.9 | 9 |
| PUM2 | Pumilio homolog 2 (Drosophila) | 4.9 | 9.5 |
| E2F6 | E2F transcription factor 6 | 4.8 | 9.5 |
| WAC | WW domain containing adaptor with coiled-coil | 4.8 | 10 |
| CHD1 | Chromodomain helicase DNA binding protein 1 | 4.7 | 6.8 |
| SPRY4 | Sprouty homolog 4 (Drosophila) | 4.7 | 12 |
| KLF10 | Kruppel-like factor 10 | 4.7 | 10.6 |
| ARID4B | AT rich interactive domain 4B (RBP1- like) | 4.7 | 6.8 |
| KIAA0232 | KIAA0232 gene product | 4.6 | 7.9 |
| KIAA0261 | KIAA0261 | 4.6 | 7.9 |
| ZCCHC8 | Zinc finger, CCHC domain containing 8 | 4.6 | 8.2 |
| RBM16 | RNA-binding motif protein 16 | 4.5 | 8.4 |
| ZNF278 | Zinc finger protein 278 | 4.5 | 8.7 |
| RSBN1 | Round spermatid basic protein 1 | 4.5 | 7.8 |
| NA | CDNA clone IMAGE:5263531, partial cds | 4.5 | 8.9 |
| SLC38A2 | Solute carrier family 38, member 2 | 4.4 | 11.8 |
| MAT2A | Methionine adenosyltransferase II, alpha | 4.4 | 10.8 |
| C20orf158 | Chromosome 20 open reading frame 158 | 4.4 | 8.5 |
| DDX20 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 | 4.4 | 7.8 |
| SERTAD2 | SERTA domain containing 2 | 4.4 | 9 |
| LOC58486 | Transposon-derived Buster1 transposase-like protein gene | 4.3 | 7.9 |
| ZNRF3 | Zinc and ring finger 3 | 4.3 | 8.6 |
| IFRD1 | Interferon-related developmental regulator 1 | 4.3 | 7.4 |
aDifferentially expressed genes after treatment of ovarian cancer cells with 100 nM SDK for 6 h were identified using SAM. The top 50 genes with the highest degree of down-regulation (fold change) are shown. The expression level in control cells is also indicated.
Functional classification of the genes down-regulated by SDK was done using the Panther Classification tool, which gives also a statistical estimate of the degree of over-representation of a given functional class. Table 2 shows biological process, molecular function and pathway classes that were over-represented among SDK down-regulated genes with the highest statistical significance. SDK affected a considerable number of biological processes highly relevant to cancer pathogenesis, including developmental processes, cell cycle control, signal transduction pathways, cell proliferation and differentiation, oncogenesis and angiogenesis, consistent with the pleiotropic role of Sp1 family transcription factors. Analysis of molecular function and biological process classes indicated a predominance of genes involved in transcription regulation and nucleic acid metabolism, suggesting that SDK treatment could affect expression of multiple transcriptional regulatory and co-regulatory factors. This is likely a secondary effect due to the presence of GC-regulatory elements and Sp1-binding sites in the promoter of many of these genes. Critical cellular pathways, like the Wnt, TGF-β, Hedgehog and apoptosis signaling pathway, were also over-represented among the SDK down-regulated genes, although only few genes could be assigned to the distinct pathways with limited statistical power.
Table 2.
Gene Ontology classes differentially affected by SDK as assessed by Panthera
| U133 (REF) (Number of genes) | SDK down (Number of genes) | Expected (Number of genes) | P-value | |
|---|---|---|---|---|
| Biological process | ||||
| mRNA transcription | 1462 | 92 | 32.21 | 2.33E−20 |
| mRNA transcription regulation | 1205 | 80 | 26.55 | 6.16E−19 |
| Nucleoside, nucleotide and nucleic acid metabolism | 2607 | 122 | 57.44 | 6.12E−17 |
| Developmental processes | 1776 | 62 | 39.13 | 1.94E−04 |
| Cell cycle control | 354 | 19 | 7.8 | 4.08E−04 |
| MAPKKK cascade | 194 | 13 | 4.27 | 4.58E−04 |
| Stress response | 164 | 11 | 3.61 | 1.22E−03 |
| Other developmental process | 94 | 8 | 2.07 | 1.31E−03 |
| Angiogenesis | 31 | 4 | 0.68 | 5.23E−03 |
| Cell proliferation and differentiation | 621 | 24 | 13.68 | 6.20E−03 |
| Oncogenesis | 499 | 20 | 10.99 | 8.25E−03 |
| Molecular function | ||||
| Transcription factor | 1602 | 102 | 35.3 | 4.04E−23 |
| Nucleic acid binding | 2121 | 90 | 46.73 | 5.85E−10 |
| Zinc finger transcription factor | 619 | 38 | 13.64 | 1.97E−08 |
| Transcription cofactor | 137 | 16 | 3.02 | 1.08E−07 |
| Other transcription factor | 341 | 24 | 7.51 | 9.02E−07 |
| Nuclease | 220 | 14 | 4.85 | 4.66E−04 |
| KRAB box transcription factor | 423 | 20 | 9.32 | 1.35E−03 |
| Chromatin/chromatin-binding protein | 106 | 8 | 2.34 | 2.73E−03 |
| Homeobox transcription factor | 166 | 10 | 3.66 | 4.26E−03 |
| Other zinc finger transcription factor | 100 | 7 | 2.2 | 7.32E−03 |
| Other signaling molecule | 154 | 9 | 3.39 | 7.88E−03 |
| HMG box transcription factor | 36 | 4 | 0.79 | 8.73E−03 |
| Pathway | ||||
| Transcription regulation by bZIP transcription factor | 42 | 5 | 0.93 | 2.59E−03 |
| Interferon-gamma signaling pathway | 32 | 4 | 0.71 | 5.83E−03 |
| Wnt signaling pathway | 289 | 13 | 6.37 | 1.30E−02 |
| TGF-beta signaling pathway | 105 | 6 | 2.31 | 3.02E−02 |
| Hedgehog signaling pathway | 33 | 3 | 0.73 | 3.74E−02 |
| Apoptosis signaling pathway | 123 | 6 | 2.71 | 5.69E−02 |
aPanther analysis was done on the 460 transcripts with ≥2.5-fold decrease in expression after SDK treatment. Only classes found to be over-represented among SDK down-regulated genes and with lowest P-values are shown.
Notably, a small number of genes (n = 49) were up-regulated upon treatment with SDK, with few transcripts having a ≥2-fold change in expression (Supplementary Data S5). Among the up-regulated genes were the gene encoding spermidine/spermine N1-acetyltransferase (SSAT) and the gene encoding the BCL2-interacting protein BNIP3L. Both proteins have a pro-apoptotic function.
Effects of MTM analogs on proliferation of human ovarian cancer cell lines
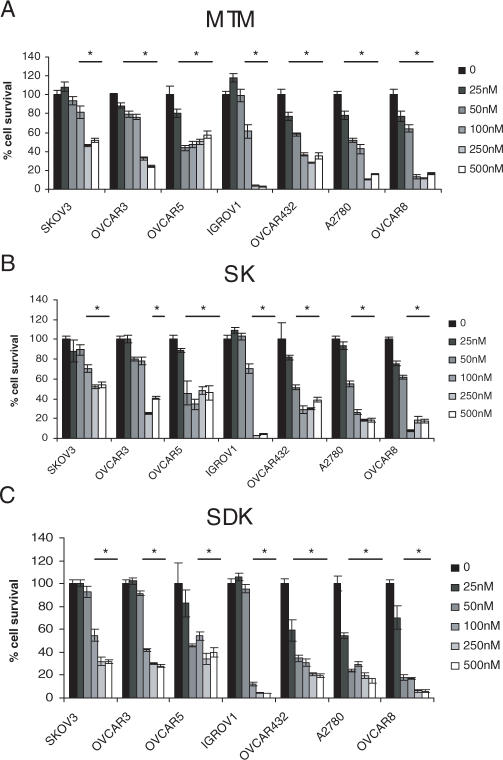
SDK was an effective inhibitor of transcription of genes critical to cell growth and survival and thus could represent a useful cancer therapeutic agent. Therefore, we examined the effects of MTM, SK and SDK on proliferation of ovarian cancer cells. Upon a 72 h continuous exposure, all three compounds inhibited growth of ovarian cancer cells at nanomolar concentrations (Figure 5). SDK was the most potent compound with IC50 up to 2-fold lower than MTM and SK in the majority of cases (Table 3). Short incubations with SDK were equally effective in inhibiting cell growth. Incubation of A2780 cells with 100 nM of SDK for 4 or 6 h followed by incubation in drug-free medium for up to 72 h produced an effect identical to the 72 h continuous exposure (i.e. ≥80% inhibition), indicating that the drug's effects were not reversible upon its removal (data not shown). Thus, even brief exposures to relatively low doses of SDK-induced persistent effects on cell growth.
Figure 5.
Anti-proliferative effects of MTM analogs in ovarian cancer cells. Ovarian cancer cells were incubated with increasing concentrations of MTM (A), SK (B) and SDK (C). After 72 h cell survival was measured by MTT assay. Data represent mean ± SD of triplicate samples. *P < 0.001 compared with control cells.
Table 3.
Effects of MTM and MTM analogs SK and SDK on growth of ovarian cancer cell linesa
| SKOV3 | OVCAR3 | OVCAR5 | OVCAR432 | OVCAR8 | IGROV1 | A2780 | |
|---|---|---|---|---|---|---|---|
| MTM (nM) | 392.3 ± 100 | 184.8 ± 29 | 145 ± 73.8 | 83.7 ± 19.7 | 53.4 ± 15.4 | 136.2 ± 76.8 | 64.1 ± 9 |
| SK (nM) | 359.7 ± 67 | 215.6 ± 63.3 | 111.9 ± 50.6 | 80.2 ± 26.7 | 55.4 ± 18 | 147.3 ± 81.6 | 72.5 ± 20.4 |
| SDK (nM) | 182.5 ± 45.5 | 149.9 ± 46.4 | 108.4 ± 34.9 | 38.5 ± 7.1 | 26.6 ± 7.9 | 82.1 ± 49.2 | 28.9 ± 6.8 |
aMTT assays were performed after 72 h of incubation of ovarian cancer cell lines with increasing concentrations of the compounds. Concentrations that inhibited cell growth by 50% (IC50) compared to control cells are reported. Data are mean ± SD of triplicate experiments.
Cell cycle alterations and apoptosis induced by MTM analogs
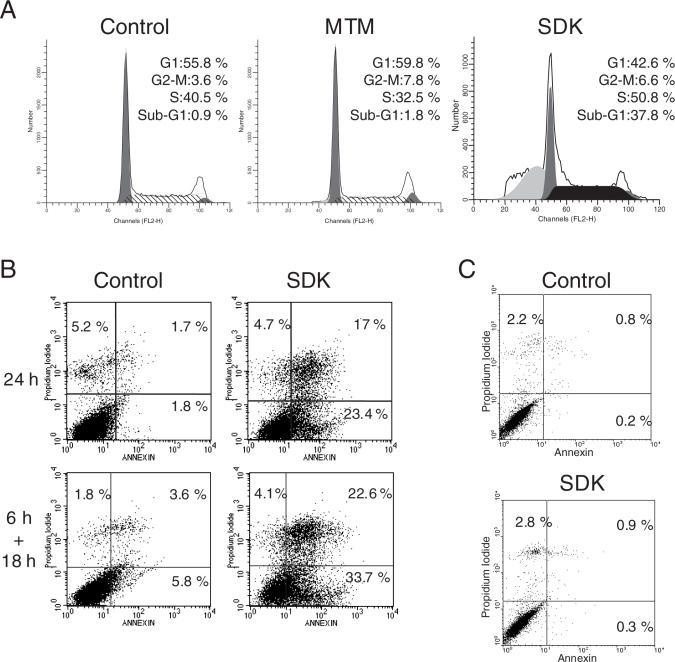
SDK inhibited growth of cancer cells, an effect that could be related to arrest of cell cycle progression and/or induction of cell death. To examine the processes leading to growth inhibition, A2780 cells were treated with the three compounds for 24 and 48 h and cell cycle distribution monitored by flow cytometry. Cells incubated with 100 nM of MTM for 24 h showed a decrease of S phase cells (42% versus 32%), indicative of G1/S phase arrest, while minimal induction of cell death (i.e. cells with sub-G1 DNA content) was evident at this time (Figure 6A). Induction of cell death became more evident only after 48 h of treatment with MTM (∼20% of sub-G1 cells). Unlike MTM, the major effect of SDK in these cells was induction of cell death. The fraction of cells with sub-G1 DNA content was 38% already at 24 h and increased to ∼50% at 48 h (Figure 6A). With respect to the induction of apoptosis, SK behavior was intermediate between that of SDK and MTM, with a higher percentage of apoptotic cells than MTM both at 24 and 48 h (10 and 40%, respectively). Induction of cell death by SDK was confirmed by immunoblotting showing PARP cleavage, which indicates caspase activation, within 24 h of treatment with SDK (data not shown). Furthermore, annexin V-PI staining and flow cytometry showed ∼50% of apoptotic cells following 24 h incubation with SDK (Figure 6B, upper panels). Treatment of cells with SDK for 6 h followed by incubation in drug-free medium for 18 h resulted in a similar level of apoptosis as the 24 h continuous treatment (Figure 6C, lower panels). Overall, the induction of apoptosis appeared as a major event underlying the anticancer effect of SDK, which acted at lower concentrations and more rapidly than MTM.
Figure 6.
Induction of apoptotic cell death in SDK-treated ovarian cancer cells. (A) A2780 cells were incubated with 100 nM of MTM or SDK for 24 h. Cells were fixed, stained with propidium iodide and analyzed by flow cytometry. (B) Cells were incubated without or with 50 nM of SDK for 24 h (upper panels) or for 6 h followed by 18 h in drug-free medium (lower panels). Control and drug-treated cells were harvested and incubated with Annexin V-FITC and propidium iodide before being analyzed by flow cytometry. Percentages of AnnexinV- and PI- positive cells are indicated. (C) Normal human fibroblasts were incubated with or without 50 nM of SDK for 24 h. Presence of AnnexinV- and PI- positive cells was determined by flow cytometry.
SDK had a profound effect on proliferation and survival of cancer cells. We wondered whether SDK would have similar cytotoxic effects in normal cells, like normal human fibroblasts. Annexin V-PI staining showed the absence of cell death in normal fibroblasts incubated for 24 h with 50 nM SDK (Figure 6C). The percentage of Annexin V and PI positive cells was essentially identical in untreated and SDK-treated NHF, while >50% of A2780 cells were apoptotic under the same conditions. Thus, the pro-apoptotic activity of SDK in ovarian cancer cells was not associated with comparable cytoxicity in normal cells.
DISCUSSION
Polyketides comprise a large family of structurally diverse natural products that include widely used pharmaceuticals with potent anticancer, antibacterial and antifungal activity (23,24). MTM is a polycyclic aromatic polyketide and, like other compounds of this class, has interesting pharmacological properties. Clinical use of MTM and similar compounds, however, is limited at the present by the induction of severe side effects. The availability of new MTM analogs with an improved pharmacological and toxicological profile may open new possibilities for exploiting the unique properties of this class of compounds for therapeutic applications.
The MTM biosynthetic pathway has been fully characterized in recent years, making it possible to apply metabolic engineering methods to produce new analogs (25–31). Using this approach, we have identified new compounds, SDK and SK, which have improved properties compared with MTM. SDK was discovered after reevaluating the isolation procedures of the secondary metabolites of the mutant strain M7W1, in which the last step in MTM biosynthesis had been genetically altered. This new compound was a much more potent inhibitor of transcription than MTM. SDK inhibited the activity of a Sp1 reporter by ≥90% at 50 nM while it interfered minimally with other transcription factors, indicating that the compound retained a high degree of selectivity for GC-rich DNA-binding transcription factors, like Sp1. Furthermore, expression of increasing amounts of Sp1 reduced the effects of SDK on Sp1 reporter activity, indicating a direct competition between the drug and the transcription factor. The potency of SDK as transcriptional inhibitor was confirmed by determining its effects on endogenous gene expression by RT–PCR and microarrays. The ability of SDK to modulate expression of multiple genes involved in a variety of cellular functions at relatively low doses is an important feature that could be exploited for therapeutic applications in cancer and other diseases.
The modified 3-side chain apparently confers to SDK unique properties that result in improved biological activity compared to MTM. The pentyl side chain attached at C-3 is a structural element highly conserved among aureolic acid antibiotics exhibiting anticancer and antibacterial activity (3). However, unlike the chromophore and saccharide chains, the role of the 3-side chain is much less characterized. Indeed, the availability of compounds like SK and SDK can offer for the first time the opportunity to investigate the relevance of the 3-side chain. The 3′-OH and 4′-OH groups in the 3-side chain of MTM form H-bonds with the DNA phosphate backbone with the 4′-OH serving as H-bond donor and perhaps stabilize DNA binding (4). Both the 3′- and 4′-OH groups are missing or are in different positions in the 3-side chains of SDK and SK. The 3-side chain of SDK without OH groups can act only as H-bond acceptor and does not provide any H-bond donor function. The 3-side chain of SK has both H-bond donor and acceptor function, but its potential donor group is in a position where both MTM and SDK have a keto group, a potential H-bond acceptor. Considering the absence and repositioning of these functional groups in the 3-side chains of SDK and SK compared to MTM, one can imagine differences in the DNA-binding mode of the three compounds. Thus, the increased activity of SDK could be due to improvements in the ability to bind DNA and/or interfere with DNA binding proteins. Fluorescence titration experiments indicated that SDK and MTM had similar binding affinity, while footprinting studies showed that the new analog retained the GC-rich sequence selectivity of the parent compound. Despite similar DNA-binding properties, however, SDK had a greater ability to prevent Sp1 binding in gel mobility shift assays, suggesting that SDK might bind in ways to block more effectively protein binding to DNA than MTM perhaps as a result of slight differences in its binding mode. The greater activity of SDK as transcriptional inhibitor in cells was also related to improved cellular uptake. In fact, SDK accumulated in cells more rapidly and to a greater extent than MTM. Thus, the combination of tight DNA binding, efficient inhibition of protein binding and rapid accumulation in cells seems at the basis of the increased potency of SDK compared to MTM. In addition to bind tightly to DNA in vitro, SDK accumulated in large amounts in cell nuclei, suggesting that the new analog might have easier access and bind more efficiently to nuclear and chromatin targets compared with MTM. Similarly, the large amount of SK taken up by cells could compensate for the reduced DNA binding in vitro and explain the greater activity of this compound as transcriptional inhibitor compared with MTM.
The ability of SDK to inhibit expression of relevant genes at relatively low concentrations is an important property. To better characterize the effects of this compound on transcription, we compared gene expression of control and drug-treated cancer cells using microarrays. SDK inhibited expression of many genes confirming its potent anti-transcriptional activity. As expected, genes that were actively transcribed and possessed GC-rich promoter elements appeared to be more susceptible to drug-induced inhibition. Overall, SDK had a profound effect on the transcriptional profile and appeared able to reverse multiple consequences of the deregulated transcription program intrinsic of cancer cells. Many genes affected by SDK were involved in various aspects of cancer development and progression and represent per se interesting targets for therapeutic intervention. In addition to genes involved in cell cycle, cell proliferation and survival, SDK inhibited expression of genes implicated in cell migration, invasion and angiogenesis. Thus, the compound might undermine the ability of cancer cells to invade tissues, migrate, produce metastasis and stimulate angiogenesis, in addition to exert a direct effect on cancer cell proliferation. By inhibiting expression of anti-apoptotic and drug resistance genes, SDK might also be able to modulate the sensitivity of cancer cells to chemo- and radio-therapy as proposed recently for MTM (21,22).
The main targets of GC-rich DNA-binding drugs, like SDK, are likely to be transcription factors of the Sp1 family and most of the effects of this compound on cell proliferation and survival are probably mediated, directly or indirectly, by interference with the activity of these transcription factors. Sp1 is the prototype of the family and was the first transcription factor to be purified, cloned and characterized in mammalian cells (40). Sp1 is a ubiquitous factor controlling expression of several cellular genes (40,41). Recent evidence implicates Sp1 in control of cell growth, survival and differentiation and point to its involvement in cancer pathogenesis (40,42). Over-expression of Sp1 has been observed in cancers, including pancreatic, breast, gastric and thyroid cancers, and is a negative prognostic factor (43–46). Sp1 binding and transcriptional activity can increase also as result of post-translational modifications or altered interactions with oncogenic or tumor suppressor proteins (40). Increased transcription of certain cancer-related genes has been found recently to be a consequence of genetic polymorphisms at the level of Sp1-binding sites in the genes promoters (21,47,48). Because of the large number of Sp1-regulated genes, abnormal Sp1 activity can contribute to various aspects of cancer, promoting growth, survival, invasion, metastasis and angiogenesis. As other transcription factors, Sp1 can be a valid therapeutic target (1,2,42). Several strategies have been investigated to inhibit Sp1 activity in experimental settings. Inhibition of Sp1 by decoy oligonucleotides and peptide nucleic acids, small-interfering RNAs and ribozymes results in growth inhibition and induction of apoptosis in various in vitro and in vivo cancer models (49–51). Thus, GC-rich DNA-binding compounds like SDK might represent valid agents for treatment of cancers with aberrant Sp1 activity. The new MTM analog was very effective in inhibiting growth and survival of ovarian cancer cells that exhibited high levels of Sp1 protein and binding activity. Even a short exposure to low doses of SDK committed ovarian cancer cells to death and growth arrest. Importantly, the potent anti-proliferative and pro-apoptotic activity of SDK in cancer cells was not associated with significant cytotoxicity in normal cells. This aspect is particularly relevant in light of the concerns regarding selectivity and toxicity of DNA-binding agents like SDK and will need to be further studied in appropriate in vitro and in vivo animal models.
Abnormal Sp1 activity is involved also in diseases other than cancer, including neurological and chronic degenerative diseases (19,20,52–56). MTM has been investigated recently as an experimental agent in neurological, hematological, cardiovascular, chronic inflammatory and viral diseases (14–20). MTM was found to protect cortical neurons from oxidative stress-induced apoptosis (14) and to improve symptoms in a Huntington's disease mouse model (18). The drug's activity in these conditions could be attributed at least in part to its ability to interact with GC-rich DNA sequences and interfere with Sp1 or other DNA-binding proteins (14,16,19,20). New MTM analogs, like SDK, should also be investigated in these disease models, where they might exhibit improved activity and lower toxicity compared to MTM.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Dr D. Kardassis (Institute of Molecular Biology and Biotechnology, Heraklion, Greece), Dr R. Evans (The Salk Institute for Biological Studies, La Jolla, CA) and Dr G. Natoli (Institute for Research in Biomedicine, Bellinzona, Switzerland) for the gift of reporter and expression vectors. We acknowledge the support of the mass spectrometry and NMR core facilities of the University of Kentucky for the physicochemical characterization of SDK. This study was supported by Grant CA091901 from National Institutes of Health (to J.R.) and a grant from the Fondazione Ticinese per la Ricerca sul Cancro (to C.V.C.). The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Darnell J.E., Jr Transcription factors as targets for cancer therapy. Nature Rev. Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh D., Papavassiliou A.G. Transcription factor therapeutics: long-shot or lodestone. Curr. Med. Chem. 2005;12:691–701. doi: 10.2174/0929867053202197. [DOI] [PubMed] [Google Scholar]
- 3.Gause G.F. Olivomycin, mithramycin, chromomycin: three related cancerostatic antibiotics. Adv. Chemother. 1965;2:179–195. doi: 10.1016/b978-1-4831-9930-6.50009-5. [DOI] [PubMed] [Google Scholar]
- 4.Sastry M., Patel D.J. Solution structure of the mithramycin dimer–DNA complex. Biochemistry. 1993;32:6588–6604. doi: 10.1021/bi00077a012. [DOI] [PubMed] [Google Scholar]
- 5.Sastry M., Fiala R., Patel D.J. Solution structure of mithramycin dimers bound to partially overlapping sites on DNA. J. Mol. Biol. 1995;251:674–689. doi: 10.1006/jmbi.1995.0464. [DOI] [PubMed] [Google Scholar]
- 6.Miller D.M., Polansky D.A., Thomas S.D., Ray R., Campbell V.W., Sanchez J., Koller C.A. Mithramycin selectively inhibits transcription of G-C containing DNA. Am. J. Med. Sci. 1987;294:388–394. doi: 10.1097/00000441-198711000-00015. [DOI] [PubMed] [Google Scholar]
- 7.Ray R., Snyder R.C., Thomas S., Koller C.A., Miller D.M. Mithramycin blocks protein binding and function of the SV40 early promoter. J. Clin. Invest. 1989;83:2003–2007. doi: 10.1172/JCI114110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ray R., Thomas S., Miller D.M. Mithramycin selectively inhibits the transcriptional activity of a transfected human c-myc gene. Am. J. Med. Sci. 1990;300:203–208. doi: 10.1097/00000441-199010000-00001. [DOI] [PubMed] [Google Scholar]
- 9.Snyder R.C., Ray R., Blume S., Miller D.M. Mithramycin blocks transcriptional initiation of the c-myc P1 and P2 promoters. Biochemistry. 1991;30:4290–4297. doi: 10.1021/bi00231a027. [DOI] [PubMed] [Google Scholar]
- 10.Blume S.W., Snyder R.C., Ray R., Thomas S., Koller C.A., Miller D.M. Mithramycin inhibits SP1 binding and selectively inhibits transcriptional activity of the dihydrofolate reductase gene in vitro and in vivo. J. Clin. Invest. 1991;88:1613–1621. doi: 10.1172/JCI115474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Remsing L.L., Bahadori H.R., Carbone G.M., McGuffie E.M., Catapano C.V., Rohr J. Inhibition of c-src transcription by mithramycin: structure-activity relationships of biosynthetically produced mithramycin analogues using the c-src promoter as target. Biochemistry. 2003;42:8313–8324. doi: 10.1021/bi034091z. [DOI] [PubMed] [Google Scholar]
- 12.Brown J.H., Kennedy B.J. Mithramycin in the treatment of disseminated testicular neoplasms. N. Engl. J. Med. 1965;272:111–118. doi: 10.1056/NEJM196501212720301. [DOI] [PubMed] [Google Scholar]
- 13.Koller C.A., Miller D.M. Preliminary observations on the therapy of the myeloid blast phase of chronic granulocytic leukemia with plicamycin and hydroxyurea. N. Engl. J. Med. 1986;315:1433–1438. doi: 10.1056/NEJM198612043152301. [DOI] [PubMed] [Google Scholar]
- 14.Chatterjee S., Zaman K., Ryu H., Conforto A., Ratan R.R. Sequence-selective DNA binding drugs mithramycin A and chromomycin A3 are potent inhibitors of neuronal apoptosis induced by oxidative stress and DNA damage in cortical neurons. Ann. Neurol. 2001;49:345–354. [PubMed] [Google Scholar]
- 15.Fibach E., Bianchi N., Borgatti M., Prus E., Gambari R. Mithramycin induces fetal hemoglobin production in normal and thalassemic human erythroid precursor cells. Blood. 2003;102:1276–1281. doi: 10.1182/blood-2002-10-3096. [DOI] [PubMed] [Google Scholar]
- 16.Chen S.J., Chen Y.F., Miller D.M., Li H., Oparil S. Mithramycin inhibits myointimal proliferation after balloon injury of the rat carotid artery in vivo. Circulation. 1994;90:2468–2473. doi: 10.1161/01.cir.90.5.2468. [DOI] [PubMed] [Google Scholar]
- 17.Bianchi N., Passadore M., Rutigliano C., Feriotto G., Mischiati C., Gambari R. Targeting of the Sp1 binding sites of HIV-1 long terminal repeat with chromomycin. Disruption of nuclear factor.DNA complexes and inhibition of in vitro transcription. Biochem. Pharmacol. 1996;52:1489–1498. doi: 10.1016/s0006-2952(96)00510-2. [DOI] [PubMed] [Google Scholar]
- 18.Ferrante R.J., Ryu H., Kubilus J.K., D'Mello S., Sugars K.L., Lee J., Lu P., Smith K., Browne S., Beal M.F., et al. Chemotherapy for the brain: the antitumor antibiotic mithramycin prolongs survival in a mouse model of Huntington's disease. J. Neurosci. 2004;24:10335–10342. doi: 10.1523/JNEUROSCI.2599-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen M.A., Zhou W., Qing H., Lehman A., Philipsen S., Song W. Transcriptional regulation of BACE1, the beta-amyloid precursor protein beta-secretase, by Sp1. Mol. Cell. Biol. 2004;24:865–874. doi: 10.1128/MCB.24.2.865-874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes A., Abraham D.J., Chen Y., Denton C., Shi-wen X., Black C.M., Leask A. Constitutive connective tissue growth factor expression in scleroderma fibroblasts is dependent on Sp1. J. Biol. Chem. 2003;278:41728–41733. doi: 10.1074/jbc.M305019200. [DOI] [PubMed] [Google Scholar]
- 21.Bond G.L., Hu W., Bond E.E., Robins H., Lutzker S.G., Arva N.C., Bargonetti J., Bartel F., Taubert H., Wuerl P., et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 22.Duverger V., Murphy A.M., Sheehan D., England K., Cotter T.G., Hayes I., Murphy F.J. The anticancer drug mithramycin A sensitises tumour cells to apoptosis induced by tumour necrosis factor (TNF) Br. J. Cancer. 2004;90:2025–2031. doi: 10.1038/sj.bjc.6601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cane D.E., Walsh C.T., Khosla C. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 24.Khosla C., Keasling J.D. Metabolic engineering for drug discovery and development. Nature Rev. Drug Discov. 2003;2:1019–1025. doi: 10.1038/nrd1256. [DOI] [PubMed] [Google Scholar]
- 25.Prado L., Fernandez E., Weissbach U., Blanco G., Quiros L.M., Brana A.F., Mendez C., Rohr J., Salas J.A. Oxidative cleavage of premithramycin B is one of the last steps in the biosynthesis of the antitumor drug mithramycin. Chem. Biol. 1999;6:19–30. doi: 10.1016/s1074-5521(99)80017-9. [DOI] [PubMed] [Google Scholar]
- 26.Lozano M.J., Remsing L.L., Quiros L.M., Brana A.F., Fernandez E., Sanchez C., Mendez C., Rohr J., Salas J.A. Characterization of two polyketide methyltransferases involved in the biosynthesis of the antitumor drug mithramycin by Streptomyces argillaceus. J. Biol. Chem. 2000;275:3065–3074. doi: 10.1074/jbc.275.5.3065. [DOI] [PubMed] [Google Scholar]
- 27.Nur-e-Alam M., Mendez C., Salas J.A., Rohr J. Elucidation of the glycosylation sequence of mithramycin biosynthesis: isolation of 3A-deolivosylpremithramycin B and its conversion to premithramycin B by glycosyltransferase MtmGII. Chembiochem. 2005;6:632–636. doi: 10.1002/cbic.200400309. [DOI] [PubMed] [Google Scholar]
- 28.Remsing L.L., Garcia-Bernardo J., Gonzalez A., Kunzel E., Rix U., Brana A.F., Bearden D.W., Mendez C., Salas J.A., Rohr J. Ketopremithramycins and ketomithramycins, four new aureolic acid-type compounds obtained upon inactivation of two genes involved in the biosynthesis of the deoxysugar moieties of the antitumor drug mithramycin by Streptomyces argillaceus, reveal novel insights into post-PKS tailoring steps of the mithramycin biosynthetic pathway. J. Am. Chem. Soc. 2002;124:1606–1614. doi: 10.1021/ja0105156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Remsing L.L., Gonzalez A.M., Nur-e-Alam M., Fernandez-Lozano M.J., Brana A.F., Rix U., Oliveira M.A., Mendez C., Salas J.A., Rohr J. Mithramycin SK, a novel antitumor drug with improved therapeutic index, mithramycin SA, and demycarosyl-mithramycin SK: three new products generated in the mithramycin producer Streptomyces argillaceus through combinatorial biosynthesis. J. Am. Chem. Soc. 2003;125:5745–5753. doi: 10.1021/ja034162h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trefzer A., Blanco G., Remsing L., Kunzel E., Rix U., Lipata F., Brana A.F., Mendez C., Rohr J., Bechthold A., et al. Rationally designed glycosylated premithramycins: hybrid aromatic polyketides using genes from three different biosynthetic pathways. J. Am. Chem. Soc. 2002;124:6056–6062. doi: 10.1021/ja017385l. [DOI] [PubMed] [Google Scholar]
- 31.Blanco G., Fernandez E., Fernandez M.J., Brana A.F., Weissbach U., Kunzel E., Rohr J., Mendez C., Salas J.A. Characterization of two glycosyltransferases involved in early glycosylation steps during biosynthesis of the antitumor polyketide mithramycin by Streptomyces argillaceus. Mol. Gen. Genet. 2000;262:991–1000. doi: 10.1007/pl00008667. [DOI] [PubMed] [Google Scholar]
- 32.Carbone G.M., McGuffie E.M., Collier A., Catapano C.V. Selective inhibition of transcription of the Ets2 gene in prostate cancer cells by a triplex-forming oligonucleotide. Nucleic Acids Res. 2003;31:833–843. doi: 10.1093/nar/gkg198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carbone G.M., McGuffie E., Napoli S., Flanagan C.E., Dembech C., Negri U., Arcamone F., Capobianco M.L., Catapano C.V. DNA binding and antigene activity of a daunomycin-conjugated triplex-forming oligonucleotide targeting the P2 promoter of the human c-myc gene. Nucleic Acids Res. 2004;32:2396–2410. doi: 10.1093/nar/gkh527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 35.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carbone G.M., Napoli S., Valentini A., Cavalli F., Watson D.K., Catapano C.V. Triplex DNA-mediated downregulation of Ets2 expression results in growth inhibition and apoptosis in human prostate cancer cells. Nucleic Acids Res. 2004;32:4358–4367. doi: 10.1093/nar/gkh744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson M., Nur-e-alam M., Lipata F., Oliveira M.A., Rohr J. Characterization of kinetics and products of the Baeyer–Villiger oxygenase MtmOIV, the key enzyme of the biosynthetic pathway toward the natural product anticancer drug mithramycin from Streptomyces argillaceus. J. Am. Chem. Soc. 2005;127:17594–17595. doi: 10.1021/ja055750t. [DOI] [PubMed] [Google Scholar]
- 39.Menendez N., Nur-e-Alam M., Brana A.F., Rohr J., Salas J.A., Mendez C. Biosynthesis of the antitumor chromomycin A3 in Streptomyces griseus: analysis of the gene cluster and rational design of novel chromomycin analogs. Chem. Biol. 2004;11:21–32. doi: 10.1016/j.chembiol.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 40.Black A.R., Black J.D., Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell. Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 41.Philipsen S., Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Safe S., Abdelrahim M. Sp transcription factor family and its role in cancer. Eur. J. Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Shi Q., Le X., Abbruzzese J.L., Peng Z., Qian C.N., Tang H., Xiong Q., Wang B., Li X.C., Xie K. Constitutive Sp1 activity is essential for differential constitutive expression of vascular endothelial growth factor in human pancreatic adenocarcinoma. Cancer Res. 2001;61:4143–4154. [PubMed] [Google Scholar]
- 44.Zannetti A., Del Vecchio S., Carriero M.V., Fonti R., Franco P., Botti G., D'Aiuto G., Stoppelli M.P., Salvatore M. Coordinate up-regulation of Sp1 DNA-binding activity and urokinase receptor expression in breast carcinoma. Cancer Res. 2000;60:1546–1551. [PubMed] [Google Scholar]
- 45.Kitadai Y., Yasui W., Yokozaki H., Kuniyasu H., Haruma K., Kajiyama G., Tahara E. The level of a transcription factor Sp1 is correlated with the expression of EGF receptor in human gastric carcinomas. Biochem. Biophys. Res. Commun. 1992;189:1342–1348. doi: 10.1016/0006-291x(92)90221-6. [DOI] [PubMed] [Google Scholar]
- 46.Chiefari E., Brunetti A., Arturi F., Bidart J.M., Russo D., Schlumberger M., Filetti S. Increased expression of AP2 and Sp1 transcription factors in human thyroid tumors: a role in NIS expression regulation? BMC Cancer. 2002;2:35. doi: 10.1186/1471-2407-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu W., Innocenti F., Wu M.H., Desai A.A., Dolan M.E., Cook E.H., Jr, Ratain M.J. A functional common polymorphism in a Sp1 recognition site of the epidermal growth factor receptor gene promoter. Cancer Res. 2005;65:46–53. [PubMed] [Google Scholar]
- 48.Yu C., Zhou Y., Miao X., Xiong P., Tan W., Lin D. Functional haplotypes in the promoter of matrix metalloproteinase-2 predict risk of the occurrence and metastasis of esophageal cancer. Cancer Res. 2004;64:7622–7628. doi: 10.1158/0008-5472.CAN-04-1521. [DOI] [PubMed] [Google Scholar]
- 49.Abdelrahim M., Samudio I., Smith R., III, Burghardt R., Safe S. Small inhibitory RNA duplexes for Sp1 mRNA block basal and estrogen-induced gene expression and cell cycle progression in MCF-7 breast cancer cells. J. Biol. Chem. 2002;277:28815–28822. doi: 10.1074/jbc.M203828200. [DOI] [PubMed] [Google Scholar]
- 50.Ishibashi H., Nakagawa K., Onimaru M., Castellanous E.J., Kaneda Y., Nakashima Y., Shirasuna K., Sueishi K. Sp1 decoy transfected to carcinoma cells suppresses the expression of vascular endothelial growth factor, transforming growth factor beta1, and tissue factor and also cell growth and invasion activities. Cancer Res. 2000;60:6531–6536. [PubMed] [Google Scholar]
- 51.Lou Z., O'Reilly S., Liang H., Maher V.M., Sleight S.D., McCormick J.J. Down-regulation of overexpressed sp1 protein in human fibrosarcoma cell lines inhibits tumor formation. Cancer Res. 2005;65:1007–1017. [PubMed] [Google Scholar]
- 52.Dunah A.W., Jeong H., Griffin A., Kim Y.M., Standaert D.G., Hersch S.M., Mouradian M.M., Young A.B., Tanese N., Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 53.Li S.H., Cheng A.L., Zhou H., Lam S., Rao M., Li H., Li X.J. Interaction of Huntington disease protein with transcriptional activator Sp1. Mol. Cell. Biol. 2002;22:1277–1287. doi: 10.1128/mcb.22.5.1277-1287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Osawa H., Yamada K., Onuma H., Murakami A., Ochi M., Kawata H., Nishimiya T., Niiya T., Shimizu I., Nishida W., et al. The G/G genotype of a resistin single-nucleotide polymorphism at −420 increases type 2 diabetes mellitus susceptibility by inducing promoter activity through specific binding of Sp1/3. Am. J. Hum. Genet. 2004;75:678–686. doi: 10.1086/424761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryu H., Lee J., Zaman K., Kubilis J., Ferrante R.J., Ross B.D., Neve R., Ratan R.R. Sp1 and Sp3 are oxidative stress-inducible, antideath transcription factors in cortical neurons. J. Neurosci. 2003;23:3597–3606. doi: 10.1523/JNEUROSCI.23-09-03597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson J.F., Lloyd D.B., Lira M.E., Milos P.M. Cholesteryl ester transfer protein promoter single-nucleotide polymorphisms in Sp1-binding sites affect transcription and are associated with high-density lipoprotein cholesterol. Clin. Genet. 2004;66:223–228. doi: 10.1111/j.1399-0004.2004.00289.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.