Abstract
The p53 tumor suppressor protein is a homotetrameric transcription factor whose gene is mutated in nearly half of all human cancers. In an unrelated screen of RNA/protein interactions using the yeast three-hybrid system, we inadvertently detected p53 interactions with several different RNAs. A literature review revealed previous reports of both sequence-specific and -non-specific interactions between p53 and RNA. Using yeast three-hybrid selections to identify preferred RNA partners for p53, we failed to identify primary RNA sequences or obvious secondary structures required for p53 binding. The cationic p53 C-terminus was shown to be required for RNA binding in yeast. We show that while p53 strongly discriminates between certain RNAs in the yeast three-hybrid assay, the same RNAs are bound equally by p53 in vitro. We further show that the p53 RNA-binding preferences in yeast are mirrored almost exactly by a recombinant tetrameric form of the HIV-1 nucleocapsid (NC) protein thought to be a sequence-nonspecific RNA-binding protein. However, the possibility of specific RNA binding by p53 could not be ruled out because p53 and HIV-1 NC displayed certain differences in RNA-binding preference. We conclude that (1) p53 binds RNA in vivo, (2) RNA binding by p53 is largely sequence-nonspecific in the yeast nucleus, (3) some structure-specific RNA binding by p53 cannot be ruled out, and (4) caution is required when interpreting results of RNA screens in the yeast three-hybrid system because sequence-dependent differences in RNA folding and display can masquerade as sequence-dependent differences in protein recognition.
Keywords: p53, transcription factor, yeast genetic selection, RNA secondary structure, three-hybrid system
INTRODUCTION
The p53 tumor suppressor protein plays multiple roles in facilitating growth arrest and/or apoptosis in response to DNA damage. Its vital function in growth control is supported by the observation that p53 mutations are among the most prevalent genetic abnormalities in human cancer (Vousden and Prives 2005). Perhaps because p53 is one of the most intensely studied proteins of recent years, reports continue to surface proposing previously unknown functions and binding partners for this transcription factor. Much remains to be learned of p53 function beyond its conventional role as a sequence-specific DNA-binding protein.
p53 exists as a homotetramer and is unusual among transcription factors in apparently containing two distinct binding domains for nucleic acids, each with a different specificity (Ahn and Prives 2001). A sequence-specific DNA-binding domain comprises the central core of p53 (residues 100–300) (Baptiste and Prives 2004). This domain is responsible for recognition of the p53 consensus sequence in promoters of target genes. Most tumor-derived missense mutations occur in this core domain of the protein (Ahn and Prives 2001). A second nucleic-acid-binding domain resides near the C-terminus of p53 (residues 363–393) (Baptiste and Prives 2004). This highly basic domain has been described as exhibiting little sequence preference and is reported to bind with high affinity to single-stranded DNA and RNA, irradiated DNA, and four-way DNA junctions (Oberosler et al. 1993; Reed et al. 1995; Ahn and Prives 2001). The ability of the p53 C-terminal domain to bind to unusual nucleic acid structures suggested a direct role in DNA damage recognition. Other p53 domains include an acidic N-terminal transcription activation domain, a proline-rich domain, and a tetramerization domain (Clore et al. 1994, 1995a,Clore et al. b; Baptiste and Prives 2004). Alternatively spliced forms of p53 have been reported in mice and humans (Miner and Kulesz-Martin 1997; Rehberger et al. 1997; Rohaly et al. 2005).
A latency model for the p53 protein suggests that, in addition to being kept at low levels by MDM2-dependent proteolysis, p53 does not function as a transcription factor until activated during times of cellular stress, including DNA damage (Yakovleva et al. 2001). Activation of p53 transcriptional activity was provoked in vitro by C-terminal deletion of p53; mild proteolysis of full-length p53, Escherichia coli dnaK (which disrupts protein complexes), or casein kinase II (and coincident phosphorylation of a C-terminal site on p53); or by the addition of PAb421, a monoclonal antibody that binds in the C-terminal domain (Hupp et al. 1992, 1993, 1995). The addition of C-terminal peptides to full-length p53 was also reported to activate the protein in vitro (Selivanova et al. 1997, 1999). These results suggested an allosteric model for p53 latency in which the C-terminus of p53 interacted with and influenced the conformation of the other domains (Yakovleva et al. 2001). However, NMR analysis suggested that C-terminally truncated (active) and full-length (latent) p53 dimers had the same overall conformation, and that the C-terminal domain did not interact with the core domain (Ayed et al. 2001).
Another hypothesis for the mechanism of p53 latency proposed that the p53 C-terminus binds nonspecifically to DNA in a manner that blocks sequence-specific DNA binding through steric hindrance (Anderson et al. 1997). Complicating this hypothesis, C-terminal p53 binding to short single-stranded DNA appeared to activate sequence-specific binding in vitro (Jayaraman and Prives 1995; Anderson et al. 1997).
Most latency models for p53 activation reflect reports that the C-terminal basic domain modulates sequence-specific DNA binding. The C-terminus contains six critical lysine residues involved in many post-translational modifications (Gu and Roeder 1997; Rodriguez et al. 2000; Brooks and Gu 2003; Chuikov et al. 2004; Muller et al. 2004; Xirodimas et al. 2004). The C-terminal residues that interact nonspecifically with DNA have been identified, and they are known to be targets of acetylation (Friedler et al. 2005). In mouse thymocytes, a mutant form of p53 in which all of the six C-terminal lysines were mutated to arginines was activated more easily after DNA damage than was wild-type p53 (Krummel et al. 2005). Other work with comparable p53 arginine substitutions demonstrated normal p53 stabilization before and after DNA damage but concluded that post-translational modifications of the p53 C-terminus enhance p53 activity after DNA damage (Feng et al. 2005). Although the p53 C-terminus is often invoked, no consensus has yet emerged regarding the correct model for p53 activation (Kaeser and Iggo 2002; Yakovleva et al. 2002).
Beyond its well-characterized activity as a transcription factor, new potential roles for p53 are frequently proposed (Cassiday and Maher 2002). In 1993, Oberosler et al. argued that p53 acts as a general “anti-helicase” in vitro, inhibiting several unrelated DNA and RNA helicases (Oberosler et al. 1993). Intriguingly, p53 was shown to have a much higher affinity for single-stranded rather than double-stranded nucleic acids, and RNA was found to serve as a more efficient competitor for p53 binding than single-stranded DNA (Oberosler et al. 1993). The antihelicase activity of p53 was reported to increase the rate of RNA–RNA renaturation by >200-fold, and this activity was not dependent on base composition or RNA length (Oberosler et al. 1993). It was also reported that p53 altered RNA structure and/or disrupted dynamic RNA/RNA interactions (Oberosler et al. 1993). This proposed activity would be analogous to that of RNA chaperones such as the E. coli ribosomal protein S12 (Coetzee et al. 1994) and the HIV-1-nucleocapsid protein (Hargittai et al. 2004).
Besides its apparent sequence-independent RNA/DNA annealing activity in vitro, p53 has been proposed as a participant in the translational control of certain mRNA transcripts. An unexpected link to translation was first suggested by the early report that 5.8S ribosomal RNA is covalently bound to phosphoserine-389 of purified mouse p53 (Fontoura et al. 1992). Furthermore, Mosner et al. argued that murine p53 regulates the translation of its own mRNA (Mosner et al. 1995). Murine p53 was reported to preferentially bind the murine p53 mRNA 5′-untranslated region (5′-UTR) to inhibit translation of the p53 transcript in vitro. These authors proposed that stable RNA stem–loop structures may form in the 5′-UTR of p53 mRNA. The disruption of such secondary structures during translation initiation might require an RNA helicase, for example, eukaryotic translation initiation factor 4A (eIF-4A) (Jaramillo et al. 1991). It was therefore suggested that the in vitro RNA antihelicase activity of p53 (Oberosler et al. 1993) might be functionally relevant to the proposed translational inhibition of certain mRNA transcripts by p53. Translational regulation by p53 has also been proposed for the Cdk4 mRNA and human fibroblast growth factor 2 mRNA (Ewen et al. 1995; Miller et al. 2000; Galy et al. 2001).
In addition to reports of specific RNA interactions, p53 has been coprecipitated with heterogeneous RNA from MCF7 cells, and this RNA coprecipitation is dependent on the p53 C-terminal domain (Yoshida et al. 2004).
Despite evidence suggesting that p53 might be capable of sequence- or structure-specific RNA binding, precise RNA sequences and/or secondary structure targets have not been identified. The yeast three-hybrid system is a sensitive in vivo assay for detecting RNA–protein interactions (Fig. 1A ▶; Sen-Gupta et al. 1996, 1999; Zhang et al. 1999; Kraemer et al. 2000; Zhang et al. 2000; Hook et al. 2005). Transcription of HIS3 and LacZ reporter genes depends on a trimolecular interaction within the yeast nucleus (SenGupta et al. 1996). The present study was initiated when we unexpectedly detected RNA binding by p53 in the yeast three-hybrid system. Our results suggest that p53 binds RNA with little sequence or structure specificity, but the possibility of specific, high-affinity RNA targets cannot be ruled out. Our data suggest that in the yeast three-hybrid system, sequence-dependent differences in RNA folding and display can masquerade as sequence-specific differences in protein recognition.
FIGURE 1.
RNA recognition by p53. (A) The yeast three-hybrid system. Transcription of LacZ and HIS3 reporter genes integrated at independent sites in the yeast genome depends on a trimolecular interaction between hybrid 1 (LexA/MS2 coat protein fusion); hybrid 2, an RNA comprised of one or more MS2 recognition sequences (gray) and a test RNA sequence; and hybrid 3 (GAL4AD/p53 protein fusion). (B) Predicted secondary structures of hybrid RNAs tested in a preliminary yeast three-hybrid system assay. The RNA designated –/(MS2)2 encodes two copies of the MS2 recognition sequence (gray), as well as RNase P RPR1 leader and terminator sequences (SenGupta et al. 1996). In addition to the sequences present in the –/(MS2)2 RNA, α-p50/(MS2)2 encodes an in vitro-selected RNA aptamer targeted to transcription factor NF-κB (Cassiday and Maher 2001), and IRE/(MS2)2 encodes the iron response element RNA (Sen-Gupta et al. 1996). (C) Colony filter-lift assay for LacZ reporter gene expression in the yeast three-hybrid system. Two independent transformants of each yeast strain are shown.
RESULTS
Promiscuous RNA binding by p53 in the yeast three-hybrid system
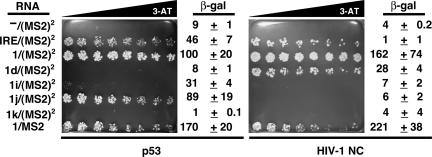
The experiments described in this work arose from our serendipitous observation that the p53 tumor suppressor protein binds RNA in the yeast three-hybrid system. In an unrelated study, we were demonstrating the specificity of an RNA aptamer against transcription factor NF-κB (α-p50) (Fig. 1B ▶; Cassiday and Maher 2001) and intended to show that the aptamer did not bind to irrelevant proteins (e.g., the iron regulatory protein [IRP] or mouse p53) in the yeast three-hybrid system. To our surprise, we found that the p53 protein not only bound the 317-nt α-p50/(MS2)2 hybrid RNA (Fig. 1B ▶), but also all other RNAs tested in the three-hybrid system, including a 271-nt RNA containing MS2 phage coat recognition sequence [–/(MS2)2] (Fig. 1B ▶) and a 303-nt RNA containing the iron response element hybrid RNA [IRE/(MS2)2] (Fig. 1B,C ▶). Interestingly, p53 appeared to exhibit some degree of sequence preference in RNA recognition, with LacZ reporter expression greatest for the yeast three-hybrid strain expressing IRE/(MS2)2 and weakest for the strain expressing –/(MS2)2 (Fig. 1C ▶).
To characterize factors driving preferential binding of p53 to certain RNA sequences, we performed a yeast three-hybrid selection to identify RNA sequences that bind strongly to p53. A 60-nt random DNA library was cloned upstream of two MS2 recognition sites in the yeast three-hybrid system RNA vector –/(MS2)2. The resulting hybrid RNA-encoding plasmid library was transformed into the yeast three-hybrid system host strain L40-coat (SenGupta et al. 1996) expressing GAL4AD/p53. RNA sequences were selected for improved in vivo binding to p53 by plating the transformation mixture on media containing a higher 3-AT concentration than that tolerated by the yeast strain expressing GAL4AD/p53 with IRE/(MS2)2. This procedure screened ~107 hybrid RNAs for those that induced higher levels of HIS3 reporter gene expression than previously observed for the strongest of the protein/RNA interactions tested in Figure 1C ▶.
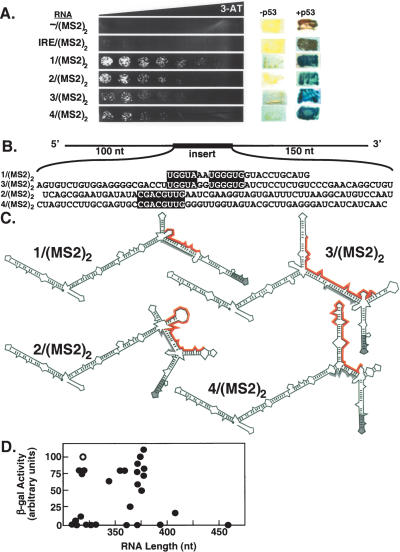
We present four selected RNA sequences that showed markedly improved binding to p53 compared to IRE/(MS2)2 [RNAs 1–4/(MS2)2] (Fig. 2 ▶). Activation of LacZ by RNAs 1–4/(MS2)2 required p53 (Fig. 2A ▶, right), indicating that these RNAs did not autoactivate or bind to the GAL4 activation domain. Interestingly, blocks of nucleotide identity were observed within the randomized regions of the selected RNA sequences (Fig. 2B ▶, boxes). The hybrid RNA sequence that induced the highest level of HIS3 reporter gene expression actually contained the smallest region of randomized sequence [RNA 1/(MS2)2] (Fig. 2B ▶). Although the majority of hybrid RNAs within the library contained 60 nt of random sequence, the randomized region of RNA 1/(MS2)2 was only 24 nt in length.
FIGURE 2.
Selection of hybrid RNAs that bind strongly to p53 in the yeast three-hybrid system. (A) Yeast three-hybrid screen of a hybrid RNA library containing ~60 nt of random sequence yielded four clones [hybrid RNAs 1–4/(MS2)2] with improved p53 binding compared to –/(MS2)2 or IRE/(MS2)2. (Left) Growth assay for HIS3 reporter gene expression on selective medium containing a gradient of 0–40 mM 3-AT. (Right) RNAs –/(MS2)2, IRE/(MS2)2, and 1– 4/(MS2)2 do not activate reporter genes in the absence of p53. (B) Sequences of the selected random inserts (shown 5′ to 3′) within hybrid RNAs 1–4. Blocks of nucleotide identity between aligned sequences (1 and 3) and (2 and 4) are highlighted in black. (C) Predicted secondary structures of hybrid RNAs 1–4. MS2 recognition sequences are shown in gray. Unique sequences in each RNA are highlighted in red. (D) LacZ activation is independent of RNA length. Data points reflect the mean of at least three independent transformants for each yeast strain.
To confirm that p53 did not bind RNA based on the length of the yeast three-hybrid RNA construct, we plotted relative β-galactosidase activity as a function of RNA length for several RNAs we have tested for p53 interaction (Fig. 2D ▶). RNA 1/(MS2)2 is shown as an open circle. There was no detectable relationship between reporter gene activation and RNA length.
When predicted secondary structures of RNAs 1–4/(MS2)2 were compared (Jaeger et al. 1989a,b; Zuker 1989), a common and unanticipated secondary-structural feature was observed. These RNAs were predicted to have only one properly folded MS2 recognition site (gray in Fig. 2C ▶). Interestingly, the second block of nucleotide identity between RNAs 1/(MS2)2 and 3/(MS2)2 (5′-UG3UG) (Fig. 2B ▶) was predicted in both structures to form base pairs with complementary nucleotides within one MS2 recognition site, causing it to mis-fold (Fig. 2C ▶). Likewise, although RNA 2/(MS2)2 did not share this block of primary sequence, the predicted secondary structure suggested that a similar (5′-G3UG) motif might be created by bulging of the intervening (5′-AU) sequence (Fig. 2C ▶). The predicted secondary structure of RNA 4/(MS2)2 suggested that it also disrupted proper folding of one MS2 recognition site, but by a different mechanism (Fig. 2C ▶). These observations suggested that some aspect of global RNA folding or display drove the selection of RNAs 1–4/(MS2)2. However, we caution that RNA secondary structural predictions derived from folding algorithms are not definitive.
Analysis of yeast-three-hybrid-selected RNA 1/(MS2)2
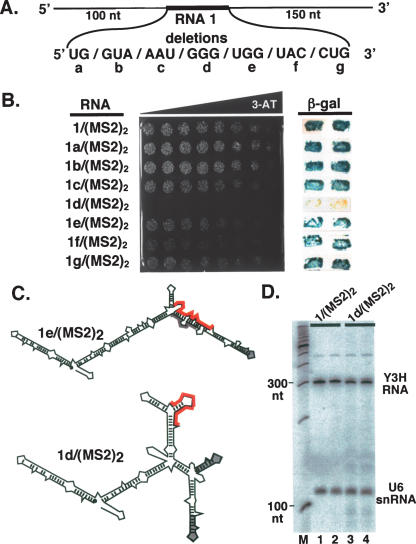
Because RNA 1/(MS2)2 appeared to be the strongest p53-binding RNA among the selected sequences, we further characterized this sequence using the yeast three-hybrid system. Blocks of two or three nucleotides within the randomized region of RNA 1/(MS2)2 were consecutively deleted (Fig. 3A ▶), and the resulting hybrid RNAs were tested for binding to p53 in HIS3 and LacZ reporter gene assays (Fig. 3B ▶). This deletion analysis was surprisingly definitive, showing that only one RNA trinucleotide block was absolutely essential for strong p53 binding. Deletion of this G3 sequence [RNA 1d/(MS2)2] (Fig. 3B ▶) completely abolished strong HIS3 (Fig. 3B ▶, left) and LacZ (Fig. 3B ▶, right) reporter gene activity. Replacing the three G residues with three A residues had the same detrimental effect on p53 binding (data not shown). Upon examining the predicted secondary structures of hybrid RNAs 1a–g/(MS2)2, it was observed that, with the exception of nonfunctional RNA 1d/(MS2)2, all of the other functional mutant RNAs were predicted to fold similarly to RNA 1/(MS2)2 [e.g., RNA 1e/(MS2)2] (Fig. 3C ▶). RNA 1d/(MS2)2 was predicted to fold much differently than RNA 1/(MS2)2 (Fig. 3C ▶), allowing both MS2 sequences to fold into native conformations (Fig. 3C ▶). This prediction raised the possibility that disruption of one MS2 secondary structure was somehow directly or indirectly essential for the strong binding of p53 to RNA 1/(MS2)2.
FIGURE 3.
Analysis of high-affinity RNA 1/(MS2)2. (A) Sequences within the randomized region of RNA 1/(MS2)2 were individually deleted to obtain hybrid RNAs 1a–g/(MS2)2. (B, left) Growth assay for HIS3 expression on selective medium containing a gradient of 0–35 mM 3-AT. (Right) Colony filter-lift assay for LacZ reporter gene expression. Two independent transformants of each strain are shown. (C) Predicted secondary structures of hybrid RNAs 1d/(MS2)2 and 1e/ (MS2)2. MS2 recognition sequences are shown in gray, and unique sequences are highlighted in red. (D) RNAs 1/(MS2)2 and 1d/(MS2)2 accumulate to the same level. Yeast total RNA was extracted and assayed by Northern blotting, probing with radiolabeled oligonucleotides complementary to the RNase P RPR1 leader and to the U6 snRNA. Mobilities of DNA size markers are indicated.
To determine if differences in hybrid RNA accumulation contributed to differences in HIS3 and LacZ reporter gene activation by GAL4AD/p53, levels of RNA 1/(MS2)2 and RNA 1d/(MS2)2 were compared by Northern blotting. As shown in Figure 3D ▶, the hybrid RNAs accumulated to comparable levels.
Importance of the p53 C-terminus for RNA recognition in the yeast three-hybrid system
A cationic domain comprising the 30 C-terminal amino acids of p53 has been implicated in nonspecific RNA binding (Oberosler et al. 1993; Wu et al. 1995; Nedbal et al. 1997). We wondered whether RNA binding by p53 in the yeast three-hybrid system required the p53 C-terminal domain. We therefore prepared a GAL4 activation domain fusion protein incorporating a version of mouse p53 lacking the final 31 amino acids. When tested for specific DNA binding and transcriptional activation in a yeast one-hybrid system (Fig. 4A ▶, top), we found that this p53 C-terminal truncation (residues 70–359) functioned comparably to the p53 protein with an intact C-terminus (residues 70–389) in HIS3 and LacZ reporter gene assays (Fig. 4A ▶, bottom). This result is consistent with previous studies showing that the p53 C-terminus is dispensable for specific DNA binding (Hupp et al. 1992, 1993, 1995). In contrast to p53(70–389), p53(70–359) was completely incapable of recognizing RNA, indicating a requirement for the 31 C-terminal residues (Fig. 4B ▶).
FIGURE 4.
The p53 C-terminus is necessary for RNA recognition of IRE and RNA 1/(MS2)2 in the yeast three-hybrid system. (A) The p53 C-terminus is not necessary for DNA binding. (Top) Binding of GAL4AD/p53 to p53 recognition sites in DNA drives transcription of HIS3 and LacZ reporter genes. (Bottom left) Growth assay for HIS3 expression on selective medium containing a gradient of 0–5 mM 3-AT. (Bottom right) β-Galactosidase specific activities, normalized to the yeast strain expressing p53 amino acids 70–359 (signal set at 100 arbitrary units). Data reflect four independent transformants for each yeast strain. (B) Yeast three-hybrid system HIS3 and LacZ reporter gene assays for strains expressing (top) full-length p53 (70–389) or (bottom) the p53 C-terminal truncation (70–359). (Left) Growth assay for HIS3 expression on selective medium containing a gradient of 0–30 mM 3-AT. (Right) β-Galactosidase specific activities, normalized to the yeast strain expressing full-length p53 (amino acids 70–389) with RNA 1/(MS2)2. Data reflect four independent transformants for each yeast strain.
Mutational analysis of p53 binding to RNA 1/(MS2)2
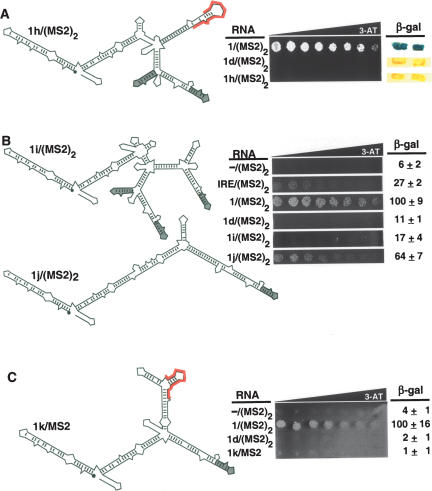
Our observations suggested that RNA secondary structure might be important for strong p53 recognition of RNA 1/(MS2)2. We tested a hybrid RNA that contained the primary sequence of the functional RNA 1 insert, but in a different RNA context with a different predicted secondary structure in which both MS2 recognition sites are properly folded [RNA 1h/(MS2)2] (Fig. 5A ▶, left). Hybrid RNA 1h/(MS2)2 failed to bind p53 in HIS3 and LacZ reporter assays (Fig. 5A ▶, right). This result underscores the importance of RNA secondary structure for strong p53 recognition in the yeast three-hybrid system.
FIGURE 5.
Probing features of RNA 1/(MS2)2 important for p53 recognition. (A) The primary insert sequence of RNA 1/(MS2)2 is not recognized by p53 in a different structural context (1h/(MS2)2). (Left) Predicted secondary structure of hybrid RNA 1h/(MS2)2 with MS2 recognition sequences shown in gray. The unique sequence from RNA 1/(MS2)2 is highlighted in red. (Right) Yeast three-hybrid growth assay for HIS3 expression on selective medium containing a gradient of 0–35 mM 3-AT and β-galactosidase assay. Two independent transformants of each strain are shown. (B) Role for a predicted three-way RNA junction in p53 recognition depends on context. (C) Presentation of a single MS2 sequence cannot increase p53 recognition of the primary sequence for RNA 1d/(MS2)2.
For a further comparison, we prepared a version of RNA 1/(MS2)2 that was predicted to present a truncated three-way junction, but with three MS2 recognition sites folded in the native conformation [RNA 1i/(MS2)2] (Fig. 5B ▶, top left). This RNA was not recognized by p53, suggesting that subtle RNA features may be important for yeast display and/or p53 recognition.
Perhaps p53 preferentially recognizes any hybrid RNA with only one properly folded MS2 recognition site. This hypothesis implies that RNAs 1–4/(MS2)2 were selected by virtue of their ability to disrupt the native conformation of one of the two MS2 recognition sites. To test this concept, we cloned a version of RNA 1/(MS2)2 having an insert region that is perfectly complementary to the MS2 site [RNA 1j/(MS2)2] (Fig. 5B ▶, bottom). RNA 1j/(MS2)2 retains the second region of nucleotide identity with RNAs 1/(MS2)2 and 3/(MS2)2, but deletes the first region, removes nucleotide bulges, and replaces a U-G wobble base pair with a U-A Watson-Crick base pair (Fig. 5B ▶, bottom). Thus, the insert region of RNA 1j/ (MS2)2 would be predicted to increase the strength of the base-pairing interaction between the random insert region of RNA 1/(MS2)2 and the MS2 recognition site. Interestingly, RNA 1j/(MS2)2 binds p53 but somewhat less strongly than RNA 1/ (MS2)2 (Fig. 5B ▶, right). This result suggests that although base-pairing with an MS2 recognition site appears to contribute to strong recognition of RNA 1/(MS2)2 by p53, other regions of the random insert are important for strong p53 binding.
To further explore the hypothesis that disruption of proper folding of one MS2 recognition site is sufficient for the strong p53 binding activity of RNA 1/(MS2)2, we cloned the primary sequence from nonfunctional RNA 1d/(MS2)2 into a vector containing a single MS2 recognition sequence (RNA 1k/MS2) (Fig. 5C ▶, left). This RNA remained incompetent to bind p53, implying that the presence of a single MS2 recognition hairpin is not sufficient for p53 recognition.
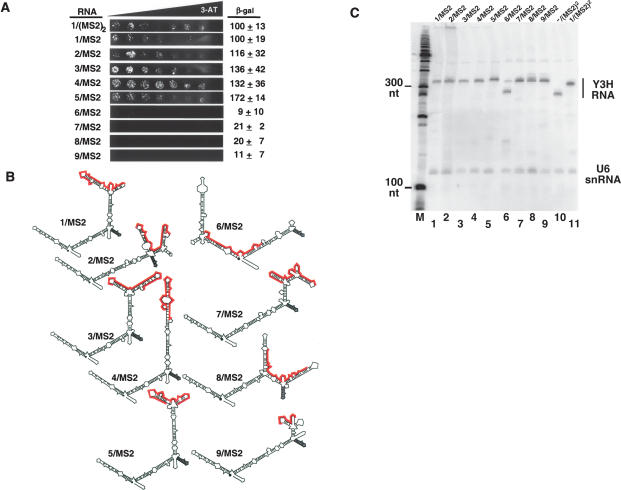
To eliminate the number of MS2 sequences as a variable in p53 affinity measurements, we performed an additional yeast three-hybrid selection for RNA sequences that bind strongly to p53 in the context of only a single MS2 recognition site in the yeast three-hybrid system RNA vector –/MS2. As before, the library encoded 60 random nucleotides. Random RNA sequences were again selected for improved in vivo binding to p53 by plating the three-hybrid strain transformation mixture on selective medium containing 15 mM 3-AT. This procedure identified hybrid RNAs that induced high levels of HIS3 reporter gene expression, comparable to those found by the initial selection. Examples of selected RNAs are shown in Figure 6 ▶. Activation of LacZ by RNAs 1–5/MS2 required p53 (data not shown), again indicating that these RNAs did not autoactivate or bind to the GAL4AD. We also conducted a separate screen for RNAs that failed to induce HIS3 or LacZ reporter gene expression (Fig. 6A ▶). The predicted secondary structures of RNAs 1–9/MS2 are shown in Figure 6B ▶ with the MS2 recognition sequence noted in gray and unique insert sequences in red. Our results indicate that RNAs with a full range of apparent p53 affinities can be selected from a library containing only a single MS2 sequence.
FIGURE 6.
The number of MS2 sites does not determine p53 affinity for RNA. (A) Example RNAs isolated from a yeast three-hybrid screen of a hybrid RNA library containing ~60 nt of random sequence in an expression cassette containing a single MS2 sequence. Hybrid RNAs 1–5/MS2 bind p53 well, while hybrid RNAs 6–9/MS2 do not bind p53. (Left) Growth assay for HIS3 reporter gene expression on selective medium containing a gradient of 0–40 mM 3-AT. (Right) β-Galactosidase specific activities, normalized to the yeast strain expressing RNA 1/(MS2)2. Data reflect at least three independent transformants for each yeast strain. (B) Predicted secondary structures of RNAs that bind p53 (left) and fail to bind p53 (right). MS2 recognition sequences are shown in gray, and unique sequences are highlighted in red. (C) p53 binding does not reflect target RNA abundance. For strains expressing various RNAs (above), yeast total RNA was extracted and assayed by Northern blotting using radiolabeled oligonucleotides complementary to the RNaseP RPR1 leader and to the U6 snRNA as probes. Mobilities of DNA size markers are indicated (left).
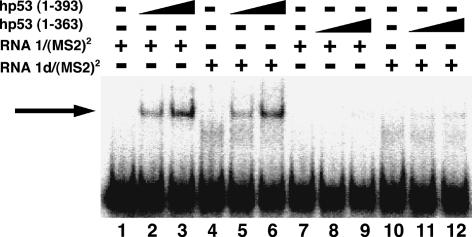
Human p53 binds RNA 1/(MS2)2 and 1d/(MS2)2 equally in vitro
RNA sequences are evidently distinguished by murine p53 in the yeast three-hybrid assay. To test the specificity of p53 binding to RNA in vitro, we performed electrophoretic mobility shift assays using either full-length (amino acids 1–393) or C-terminally truncated (amino acids 1–363) human p53. To our surprise, full-length p53 bound both RNAs 1/(MS2)2 and 1d/(MS2)2 with comparable affinity (Fig. 7 ▶, cf. lanes 2,3 and 5,6). This observation was unexpected because these RNAs are strongly distinguished by p53 in the yeast three-hybrid system. Similar data were obtained for RNAs 6–9/MS2 (data not shown). C-terminally truncated p53 did not bind either RNA in vitro (Fig. 7 ▶, lanes 7–12), as predicted by yeast three-hybrid experiments (Fig. 4 ▶). Similar results were obtained with or without gradual RNA cooling to promote folding (data not shown). These data confirm that p53 binds RNA, but suggest that the RNA specificity of p53 recognition in the yeast nucleus is not retained under the tested in vitro conditions.
FIGURE 7.
RNA binding by full-length human p53 in vitro. Full-length human p53(1–393) (lanes 2,3,5,6) or truncated human p53(1–363) (lanes 8,9,11,12) were incubated with 32P-labeled RNA 1/(MS2)2 (lanes 1–3,7–9) or 1d/(MS2)2 (lanes 4–6,10–12), and reaction mixtures were analyzed by EMSA. The arrow indicates the protein–RNA complex.
p53 and a nonspecific RNA-binding protein show similar preference for a panel of RNAs
Our in vitro and in vivo data could be reconciled if p53 were actually a nonspecific RNA-binding protein, and if RNAs selected for p53 interaction in the yeast three-hybrid system were simply better displayed on the surface of the MS2 coat protein. To test this hypothesis, we performed a yeast three-hybrid experiment that compared the pattern of p53 RNA affinity to that of an artificial tetrameric form the sequence nonspecific RNA-binding protein HIV-1-nucleo-capsid protein (HIV-1 NC) (Levin et al. 2005). The results of both liquid β-galactosidase and 3-AT gradient plate assays as seen in Figure 8 ▶ (p53 and HIV-1 NC) revealed strikingly similar apparent RNA specificities: both proteins interacted poorly with –/(MS2)2, 1i/(MS2)2, and 1k/MS2, while both proteins interacted strongly with IRE/(MS2)2, 1/(MS2)2, and 1/MS2. This result suggests that both proteins are sequence-nonspecific RNA binders, and it is the quality of RNA display that is being reported in the yeast three-hybrid assays. However, it is important to note that p53 and HIV-1 NC strongly differed in their abilities to bind RNA 1d/(MS2)2 and 1j/(MS2)2. This latter result rules out the possibility that both tetrameric HIV-1 NC and p53 are completely nonspecific RNA-binding proteins.
FIGURE 8.
p53 and HIV-NC bind a panel of RNAs in a similar pattern. Various RNAs (far left) were tested for ability to interact with either p53 (left) or tetramerized HIV-1 NC (right). The growth assay for HIS3 reporter gene expression is on selective medium containing a gradient of 0–40 mM 3-AT, and normalized β-galactosidase specific activities are included. Data reflect at least three independent transformants for each yeast strain.
DISCUSSION
p53 binds RNA in yeast
Our inadvertent observation of RNA binding by p53 led us to design experiments to better understand the RNA specificity of this well-studied tumor suppressor protein. Interesting reports of RNA interactions with p53 have been published (Fontoura et al. 1992; Oberosler et al. 1993; Ewen et al. 1995; Nedbal et al. 1997; Miller et al. 2000; Galy et al. 2001; Yoshida et al. 2004). However, this previous work made it difficult to judge whether the p53 protein is truly capable of sequence-or structure-specific RNA recognition (Ewen et al. 1995; Miller et al. 2000; Galy et al. 2001), or whether the protein has a general sequence-independent affinity for nucleic acids (Oberosler et al. 1993; Nedbal et al. 1997; Yoshida et al. 2004). We show that RNA binding by murine p53 in the yeast three-hybrid system absolutely requires the 30 C-terminal amino acids of the protein previously implicated in sequence-nonspecific nucleic acid interactions (Yoshida et al. 2004). In this work, we acknowledge that post-translational modifications of p53 undoubtedly play key roles in the natural regulation of this protein in mammalian cells (Rodriguez et al. 2000; Brooks and Gu 2003; Chuikov et al. 2004; Muller et al. 2004; Xirodimas et al. 2004; Feng et al. 2005; Friedler et al. 2005; Krummel et al. 2005), and that such modifications are probably not recapitulated in yeast.
Does p53 recognize sequence-specific RNA structure or quality of RNA display in yeast?
Our results initially suggested that p53 recognition of RNA involves subtle features of tertiary structure that are not revealed in the analysis of primary sequence or predicted secondary structure. We emphasize the alternative possibility that p53 lacks RNA sequence specificity, and that selection in the yeast three-hybrid system could be at the level of effective or favorable display of the test RNA, rather than its interaction with p53. This view is supported by the observation that purified p53 protein could not discriminate between RNAs in vitro even when the RNAs displayed dramatically different apparent affinities for p53 in yeast.
p53 and a nonspecific RNA-binding protein have similar apparent RNA specificities
Three sequence-nonspecific RNA-binding proteins were compared with p53: the E. coli S12 ribosomal protein (Coetzee et al. 1994), the double-stranded RNA-binding domain of the double-stranded RNA-dependent protein kinase (Carlson et al. 2003), and the zinc finger domain of the HIV-1 nucleocapsid protein (Kanevsky et al. 2005). Both S12 and HIV-1 NC have been discussed as “RNA chaperones,” an activity that has also been mentioned for p53. We found that an engineered tetrameric form of HIV-1 NC shared many RNA preferences with p53. This result suggests that favorable RNA display, not specific RNA sequence or structural features, was selected in our search for the best RNA partners for p53. However, comparison of the RNA preferences of tetrameric HIV-1 NC and p53 revealed some differences that cannot be explained by invoking only nonspecific RNA affinity.
Implications
This work highlights the challenge of distinguishing sequence- or structure-specific recognition from general nucleic acid affinity. We show that results obtained by screening random RNA libraries in the yeast three-hybrid system must be interpreted with caution. Because the processes of binding and displaying test RNAs at the chromosome are both sensitive to RNA sequence and structure, a strong signal does not simply imply a specific RNA/protein interaction. Our results underscore the need to confirm yeast three-hybrid interactions with in vitro binding assays.
The p53 protein has been described as natively denatured under physiological conditions in vitro (Bell et al. 2002). The isolated C-terminal domain appears to be unstructured in biophysical experiments, and apparently does not reorganize in the presence of DNA (Friedler et al. 2005). It is challenging to imagine how this unstructured protein domain can be responsible for sequence- or structure-specific RNA recognition, unless particular RNAs can drive the p53 C-terminus into a specific stable structure.
The plethora of modifications reported for the p53 C-terminus are important given the activity of this domain in nucleic acid binding. Without modifications to reduce the cationic character of this domain (Marston et al. 1998; Chuikov et al. 2004; Feng et al. 2005; Friedler et al. 2005; Krummel et al. 2005), p53 would be expected to exist perpetually bound to RNA. Our results highlight the possibility that RNA may be a participant in the elegant regulation of p53. Particularly intriguing is the question of whether p53 has specific RNA partners in vivo. If so, perturbation of such partnerships could contribute to pathologies involving this powerful protein.
MATERIALS AND METHODS
Hybrid RNA and protein expression constructs
–/MS2, α-p50/MS2, and IRE/MS2 RNAs were expressed from the yeast three-hybrid system vector pIIIA/MS2-2 (SenGupta et al. 1996), encoding MS2 hybrid RNAs under the control of the RNase P RPR1 promoter. RNAs 1–4, 1a–g, 1i, 1j, and 1k were expressed from pJ713, a version of pIIIA/MS2-2 with additional cloning sites. RNA 1h was expressed from pJ714, a version of pIIIA/MS2-2 encoding MS2 hybrid RNAs with a G/C “clamp” flanking the inserted sequences. The GAL4AD/p53 hybrid protein expression construct (pGAD53m; Clontech) encodes the yeast GAL4 transcriptional activation domain fused to mouse p53 residues 70–389. The C-terminal truncated GAL4AD/p53 hybrid protein construct was generated by PCR amplification of p53 amino acid residues 70–359 from pGAD53m (Clontech), appending a BamHI restriction site and using a downstream primer that inserts a stop codon in place of mouse p53 amino acid residue 360.
Yeast one- and three-hybrid system reporter gene assays
For HIS3 reporter gene assays, yeast strains were grown to an optical density of ~1.0 at 600 nm and diluted for plating of 1000-cell spots on selective medium containing a 3-amino-1,2,4-triazole (3-AT; Sigma) gradient over the concentration range specified in the figure legends. LacZ reporter gene expression was assayed qualitatively by a colony filter-lift assay (Invitrogen) and quantitatively using the substrate o-nitrophenyl-β-D-galactopyranoside as described (Zhang et al. 2000).
Synthesis and cloning of random hybrid RNA libraries
A 60-nt random DNA oligonucleotide library was synthesized by phosphoramidite methodology. The random oligonucleotide library was PCR-amplified, appending 5′ XhoI and 3′ SphI restriction sites for cloning. Following gel purification and digestion, the random library was cloned between XhoI and SphI sites of pJ713, a version of pIIIA/MS2-2 (SenGupta et al. 1996), or pJ1194, a version of pJ713 cut with SalI and re-ligated to eliminate one of the two MS2 sequences.
Yeast three-hybrid screens for RNAs that bind strongly to p53
The host yeast strain L40-coat (SenGupta et al. 1996) expressing the GAL4AD/p53 hybrid protein was transformed using Clontech’s lithium acetate-mediated transformation with a hybrid RNA plasmid library encoding ~60 nt of random sequence upstream of one or two copies of the MS2 recognition sequence. The transformations were plated on synthetic dropout medium lacking histidine, leucine, and uracil and containing 25 mM 3-AT. An estimated 2 × 106 plasmid sequences were screened for strong binding to p53. After incubating for 6 d at 30°C, 140 transformants were patched onto fresh medium lacking histidine, leucine, and uracil and containing 30 mM 3-AT. After incubating for 3 d at 30°C, a β-galactosidase filter assay was performed. RNA-encoding plasmids were isolated from the strongest and weakest HIS3- and LacZ-positive transformants and re-transformed into fresh L40-coat expressing GAL4AD/ p53 to confirm phenotypes. True positive clones were then sequenced to analyze selected hybrid RNAs.
Northern blot analysis
Total RNA was extracted from yeast three-hybrid strains as described (Caponigro et al. 1993). Northern blot analysis was conducted as described (Cassiday and Maher 2001), simultaneously hybridizing to radiolabeled oligonucleotide probes complementary to the RNase P RPR1 leader (5′-AGCAC2ACAGCGTAC2ATGT) and to the U6 snRNA (5′-TC2T2ATGCAG4A2CTGC). Radioactivity was detected and analyzed by storage phosphor technology.
Electrophoretic gel mobility shift assays
Recombinant, purified human full-length (1–393) and C-terminally truncated (1–363) p53 were graciously provided by C. Prives (Columbia University) (Jayaraman and Prives 1995). RNAs 1/(MS2)2 and 1d/(MS2)2 were transcribed in vitro using T7 RNA polymerase (Epicentre). Transcribed RNAs were purified using 6% denaturing polyacrylamide gels and were quantitated by UV spectrophotometry. The RNA probes were labeled by T4 RNA ligase (New England Biolabs) using 32P-pCp (Amersham). Reaction mixtures contained 20 mM HEPES (pH 7.9), 25 mM KCl, 0.1 mM EDTA, 10% glycerol, 2 mM MgCl2, 0.1 mg/mL bovine serum albumin, 0.025% NP-40, 2 mM spermidine, 0.5 mM DTT, 8.5 ng/μL salmon sperm DNA (Invitrogen), ~3 nM probe RNA, proteins as indicated, and water to a total volume of 20 μL. Reaction mixtures were incubated at room temperature for 15 min before electrophoresis on native 4% polyacrylamide (30:1 acrylamide:bisacrylamide) gels containing 0.5× TBE buffer and electrophoresed at room temperature at 165 V for 1.5 h. Radioactivity was detected and analyzed by storage phosphor technology.
HIV-1 NC interaction with RNA
HIV-1 NC was cloned by PCR-amplifying the HIV-1 NC-coding region residues 1–55 from a construct provided by Robert J. Gorelick (National Cancer Institute-Frederick Cancer Research and Development Center, Frederick, MD), appending a 5′ EcoRI site and a 3′ BglII restriction site. The PCR product was cloned between EcoRI and BamHI sites of pGAD424 (Clontech), to create pJ1295. This plasmid was used to construct a GAL4 activation domain–p53 tetramerization domain–HIV-1 NC fusion protein expression plasmid. A DNA sequence encoding the p53 tetramerization domain (amino acids 316–357 of mouse p53) was amplified by PCR from pGAD53m (Clontech), and inserted by in vivo homologous recombination (Oldenburg et al. 1997) into pJ1295 to produce pJ1316.
Acknowledgments
We thank Marvin Wickens, Robert Gorelick, Karin Musier-Forsyth, and Carol Prives for materials and advice. We acknowledge the Mayo Clinic College of Medicine DNA Sequencing and Synthesis Facility and appreciate the technical suggestions of W. Scott Moye-Rowley, Joseph Gera, Wendy Olivas, Roy Parker, and Grazia Isaya.
This work was supported by the Mayo Foundation, American Cancer Society Grant GMC-98585, and NIH Grant GM68128. L.A.C. was supported by a Howard Hughes Medical Institute Predoctoral Fellowship. A.K. was supported by a Dr. Scholl Summer Undergraduate Research Fellowship.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2286706.
REFERENCES
- Ahn, J. and Prives, C. 2001. The C-terminus of p53: The more you learn the less you know. Nat. Struct. Biol. 8: 730–732. [DOI] [PubMed] [Google Scholar]
- Anderson, M.E., Woelker, B., Reed, M., Wang, P., and Tegtmeyer, P. 1997. Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53: Implications for regulation. Mol. Cell. Biol. 17: 6255–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayed, A., Mulder, F.A., Yi, G.S., Lu, Y., Kay, L.E., and Arrowsmith, C.H. 2001. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 8: 756–760. [DOI] [PubMed] [Google Scholar]
- Baptiste, N. and Prives, C. 2004. p53 in the cytoplasm: A question of overkill? Cell 116: 487–489. [DOI] [PubMed] [Google Scholar]
- Bell, S., Klein, C., Muller, L., Hansen, S., and Buchner, J. 2002. p53 contains large unstructured regions in its native state. J. Mol. Biol. 322: 917–927. [DOI] [PubMed] [Google Scholar]
- Brooks, C.L. and Gu, W. 2003. Ubiquitination, phosphorylation and acetylation: The molecular basis for p53 regulation. Curr. Opin. Cell Biol. 15: 164–171. [DOI] [PubMed] [Google Scholar]
- Caponigro, G., Muhlrad, D., and Parker, R. 1993. A small segment of the MAT α1 transcript promotes mRNA decay in Saccharomyces cerevisiae: A stimulatory role for rare codons. Mol. Cell. Biol. 13: 5141–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, C.B., Stephens, O.M., and Beal, P.A. 2003. Recognition of double-stranded RNA by proteins and small molecules. Biopolymers 70: 86–102. [DOI] [PubMed] [Google Scholar]
- Cassiday, L.A. and Maher III, L.J. 2001. In vivo recognition of an RNA aptamer by its transcription factor target. Biochemistry 40: 2433–2438. [DOI] [PubMed] [Google Scholar]
- ———. 2002. Having it both ways: Transcription factors that bind DNA and RNA. Nucleic Acids Res. 30: 4118–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov, S., Kurash, J.K., Wilson, J.R., Xiao, B., Justin, N., Ivanov, G.S., McKinney, K., Tempst, P., Prives, C., Gamblin, S.J., et al. 2004. Regulation of p53 activity through lysine methylation. Nature 432: 353–360. [DOI] [PubMed] [Google Scholar]
- Clore, G.M., Omichinski, J.G., Sakaguchi, K., Zambrano, N., Sakamoto, H., Appella, E., and Gronenborn, A.M. 1994. High-resolution structure of the oligomerization domain of p53 by multidimensional NMR. Science 265: 386–391. [DOI] [PubMed] [Google Scholar]
- Clore, G.M., Ernst, J., Clubb, R., Omichinski, J.G., Kennedy, W.M., Sakaguchi, K., Appella, E., and Gronenborn, A.M. 1995a. Refined solution structure of the oligomerization domain of the tumour suppressor p53. Nat. Struct. Biol. 2: 321–333. [DOI] [PubMed] [Google Scholar]
- Clore, G.M., Omichinski, J.G., Sakaguchi, K., Zambrano, N., Sakamoto, H., Appella, E., and Gronenborn, A.M. 1995b. Interhelical angles in the solution structure of the oligomerization domain of p53: Correction. Science 267: 1515–1516. [DOI] [PubMed] [Google Scholar]
- Coetzee, T., Herschlag, D., and Belfort, M. 1994. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes & Dev. 8: 1575–1588. [DOI] [PubMed] [Google Scholar]
- Ewen, M.E., Oliver, C.J., Sluss, H.K., Miller, S.J., and Peeper, D.S. 1995. p53-dependent repression of CDK4 translation in TGF-β-induced G1 cell-cycle arrest. Genes & Dev. 9: 204–217. [DOI] [PubMed] [Google Scholar]
- Feng, L., Lin, T., Uranishi, H., Gu, W., and Xu, Y. 2005. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol. Cell. Biol. 25: 5389–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura, B.M., Sorokina, E.A., David, E., and Carroll, R.B. 1992. p53 is covalently linked to 5.8S rRNA. Mol. Cell. Biol. 12: 5145–5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedler, A., Veprintsev, D.B., Freund, S.M., von Glos, K.I., and Fersht, A.R. 2005. Modulation of binding of DNA to the C-terminal domain of p53 by acetylation. Structure 13: 629–636. [DOI] [PubMed] [Google Scholar]
- Galy, B., Creancier, L., Prado-Lourenco, L., Prats, A.C., and Prats, H. 2001. p53 directs conformational change and translation initiation blockade of human fibroblast growth factor 2 mRNA. Oncogene 20: 4613–4620. [DOI] [PubMed] [Google Scholar]
- Gu, W. and Roeder, R.G. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90: 595–606. [DOI] [PubMed] [Google Scholar]
- Hargittai, M.R., Gorelick, R.J., Rouzina, I., and Musier-Forsyth, K. 2004. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J. Mol. Biol. 337: 951–968. [DOI] [PubMed] [Google Scholar]
- Hook, B., Bernstein, D., Zhang, B., and Wickens, M. 2005. RNA–protein interactions in the yeast three-hybrid system: Affinity, sensitivity, and enhanced library screening. RNA 11: 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupp, T.R., Meek, D.W., Midgley, C.A., and Lane, D.P. 1992. Regulation of the specific DNA binding function of p53. Cell 71: 875–886. [DOI] [PubMed] [Google Scholar]
- ———. 1993. Activation of the cryptic DNA binding function of mutant forms of p53. Nucleic Acids Res. 21: 3167–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupp, T.R., Sparks, A., and Lane, D.P. 1995. Small peptides activate the latent sequence-specific DNA binding function of p53. Cell 83: 237–245. [DOI] [PubMed] [Google Scholar]
- Jaeger, J.A., Turner, D.H., and Zuker, M. 1989a. Improved predictions of secondary structures for RNA. Proc. Natl. Acad. Sci. 86: 7706–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1989b. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 183: 281–306. [DOI] [PubMed] [Google Scholar]
- Jaramillo, M., Dever, T.E., Merrick, W.C., and Sonenberg, N. 1991. RNA unwinding in translation: Assembly of helicase complex intermediates comprising eukaryotic initiation factors eIF-4F and eIF-4B. Mol. Cell. Biol. 11: 5992–5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman, J. and Prives, C. 1995. Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell 81: 1021–1029. [DOI] [PubMed] [Google Scholar]
- Kaeser, M.D. and Iggo, R.D. 2002. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc. Natl. Acad. Sci. 99: 95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanevsky, I., Chaminade, F., Ficheux, D., Moumen, A., Gorelick, R., Negroni, M., Darlix, J.L., and Fosse, P. 2005. Specific interactions between HIV-1 nucleocapsid protein and the TAR element. J. Mol. Biol. 348: 1059–1077. [DOI] [PubMed] [Google Scholar]
- Kraemer, B., Zhang, B., SenGupta, D., Fields, S., and Wickens, M. 2000. Using the yeast three-hybrid system to detect and analyze RNA–protein interactions. Methods Enzymol. 328: 297–321. [DOI] [PubMed] [Google Scholar]
- Krummel, K.A., Lee, C.J., Toledo, F., and Wahl, G.M. 2005. The C-terminal lysines fine-tune p53 stress responses in a mouse model but are not required for stability control or transactivation. Proc. Natl. Acad. Sci. 102: 10188–10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, J.G., Guo, J., Rouzina, I., and Musier-Forsyth, K. 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 80: 217–286. [DOI] [PubMed] [Google Scholar]
- Marston, N.J., Ludwig, R.L., and Vousden, K.H. 1998. Activation of p53 DNA binding activity by point mutation. Oncogene 16: 3123–3131. [DOI] [PubMed] [Google Scholar]
- Miller, S.J., Suthiphongchai, T., Zambetti, G.P., and Ewen, M.E. 2000. p53 binds selectively to the 5′ untranslated region of cdk4, an RNA element necessary and sufficient for transforming growth factor βand p53-mediated translational inhibition of cdk4. Mol. Cell. Biol. 20: 8420–8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner, Z. and Kulesz-Martin, M. 1997. DNA binding specificity of proteins derived from alternatively spliced mouse p53 mRNAs. Nucleic Acids Res. 25: 1319–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosner, J., Mummenbrauer, T., Bauer, C., Sczakiel, G., Grosse, F., and Deppert, W. 1995. Negative feedback regulation of wild-type p53 biosynthesis. EMBO J. 14: 4442–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, S., Ledl, A., and Schmidt, D. 2004. SUMO: A regulator of gene expression and genome integrity. Oncogene 23: 1998–2008. [DOI] [PubMed] [Google Scholar]
- Nedbal, W., Frey, M., Willemann, B., Zentgraf, H., and Sczakiel, G. 1997. Mechanistic insights into p53-promoted RNA–RNA annealing. J. Mol. Biol. 266: 677–687. [DOI] [PubMed] [Google Scholar]
- Oberosler, P., Hloch, P., Ramsperger, U., and Stahl, H. 1993. p53-catalyzed annealing of complementary single-stranded nucleic acids. EMBO J. 12: 2389–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg, K.R., Vo, K.T., Michaelis, S., and Paddon, C. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25: 451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, M., Woelker, B., Wang, P., Wang, Y., Anderson, M.E., and Tegtmeyer, P. 1995. The C-terminal domain of p53 recognizes DNA damaged by ionizing radiation. Proc. Natl. Acad. Sci. 92: 9455–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehberger, P.A., Richter, K.H., Schwartz, D., Goldfinger, N., Oskato, R., Almog, N., Marks, F., and Rotter, V. 1997. Differential expression of the regularly spliced wild-type p53 and its COOH-terminal alternatively spliced form during epidermal differentiation. Cell Growth Differ. 8: 851–860. [PubMed] [Google Scholar]
- Rodriguez, M.S., Desterro, J.M., Lain, S., Lane, D.P., and Hay, R.T. 2000. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell. Biol. 20: 8458–8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohaly, G., Chemnitz, J., Dehde, S., Nunez, A.M., Heukeshoven, J., Deppert, W., and Dornreiter, I. 2005. A novel human p53 isoform is an essential element of the ATR-intra-S phase checkpoint. Cell 122: 21–32. [DOI] [PubMed] [Google Scholar]
- Selivanova, G., Iotsova, V., Okan, I., Fritsche, M., Strom, M., Groner, B., Grafstrom, R.C., and Wiman, K.G. 1997. Restoration of the growth suppression function of mutant p53 by a synthetic peptide derived from the p53 C-terminal domain. Nat. Med. 3: 632–638. [DOI] [PubMed] [Google Scholar]
- Selivanova, G., Ryabchenko, L., Jansson, E., Iotsova, V., and Wiman, K.G. 1999. Reactivation of mutant p53 through interaction of a C-terminal peptide with the core domain. Mol. Cell. Biol. 19: 3395–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta, D.J., Zhang, B., Kraemer, B., Pochart, P., Fields, S., and Wickens, M. 1996. A three-hybrid system to detect RNA–protein interactions in vivo. Proc. Natl. Acad. Sci. 93: 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta, D.J., Wickens, M., and Fields, S. 1999. Identification of RNAs that bind to a specific protein using the yeast three-hybrid system. RNA 5: 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden, K.H. and Prives, C. 2005. p53 and prognosis: New insights and further complexity. Cell 120: 7–10. [DOI] [PubMed] [Google Scholar]
- Wu, L., Bayle, J.H., Elenbaas, B., Pavletich, N.P., and Levine, A.J. 1995. Alternatively spliced forms in the carboxy-terminal domain of the p53 protein regulate its ability to promote annealing of complementary single strands of nucleic acids. Mol. Cell. Biol. 15: 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xirodimas, D.P., Saville, M.K., Bourdon, J.C., Hay, R.T., and Lane, D.P. 2004. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118: 83–97. [DOI] [PubMed] [Google Scholar]
- Yakovleva, T., Pramanik, A., Kawasaki, T., Tan-No, K., Gileva, I., Lindegren, H., Langel, U., Ekstrom, T.J., Rigler, R., Terenius, L., et al. 2001. p53 Latency. C-terminal domain prevents binding of p53 core to target but not to nonspecific DNA sequences. J. Biol. Chem. 276: 15650–15658. [DOI] [PubMed] [Google Scholar]
- Yakovleva, T., Pramanik, A., Terenius, L., Ekstrom, T.J., and Bakalkin, G. 2002. p53 latency—Out of the blind alley. Trends Biochem. Sci. 27: 612–618. [DOI] [PubMed] [Google Scholar]
- Yoshida, Y., Izumi, H., Torigoe, T., Ishiguchi, H., Yoshida, T., Itoh, H., and Kohno, K. 2004. Binding of RNA to p53 regulates its oligomerization and DNA-binding activity. Oncogene 23: 4371–4379. [DOI] [PubMed] [Google Scholar]
- Zhang, T. and Prives, C. 2001. Cyclin A-CDK phosphorylation regulates MDM2 protein interactions. J. Biol. Chem. 276: 29702–29710. [DOI] [PubMed] [Google Scholar]
- Zhang, B., Kraemer, B., SenGupta, D., Fields, S., and Wickens, M. 1999. Yeast three-hybrid system to detect and analyze interactions between RNA and protein. Methods Enzymol. 306: 93–113. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Yeast three-hybrid system to detect and analyze RNA–protein interactions. Methods Enzymol. 318: 399–419. [DOI] [PubMed] [Google Scholar]
- Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244: 48–52. [DOI] [PubMed] [Google Scholar]