Abstract
The kinase Aurora-A (Aur-A), which is enriched at centrosomes, is required for centrosome maturation and accurate chromosome segregation, and recent work implicates centrosomes as sites where the earliest activation of cyclin B1-cdc2 occurs. Here, we have used Xenopus egg extracts to investigate Aur-A's contribution to cell cycle progression and spindle morphology in the presence or absence of centrosomes. We find that addition of active Aur-A accelerates cdc2 activation and mitotic entry. Depletion of endogenous Aur-A or addition of inactive Aur-A, which lead to monopolar spindles, delays but does not block mitotic entry. These effects on timing and spindle structure do not require the presence of centrosomes or chromosomes. The catalytic domain alone of Aur-A is sufficient to restore spindle bipolarity; additional N-terminal sequences function in mitotic timing.
Keywords: Aurora kinases, G2/M transition, mitosis
The Aurora kinases are required for mitotic progression (reviewed in refs. 1–5). Aurora B (Aur-B) participates in the formation of correct attachment of spindle microtubules to chromosomes and in cytokinesis. Aurora A (Aur-A) is required for the maintenance of spindle bipolarity and the accurate completion of chromosome segregation. A portion of Aur-A localizes to centrosomes and nearby spindle microtubules, where it rapidly exchanges with cytoplasmic pools. Mechanistically, Aur-A's best understood roles are in the recruitment and regulation of proteins at centrosomes. For example, Aur-A is required for the localization of centrosomin, the microtubule nucleating γ-tubulin ring complex, TACC, and the subsequent TACC-dependent localization of the microtubule stabilizing factor MAP215. Reducing the amount of Aur-A protein or introducing inactive Aur-A interferes with the separation of duplicated centrosomes, the maintenance of centrosome separation, and/or defects in chromosome alignment.
Aur-A also plays a role in the timing of entry into M phase. Overexpression of Aur-A in G2-arrested Xenopus oocytes, where it was originally called Eg2, accelerates mitogen-induced entry into M phase of meiosis I (6, 7). This acceleration is due at least in part to Aur-A's phosphorylation of CPEB, a protein associated with the 3′ UTRs of certain stored, untranslated cytoplasmic mRNAs. This results in the cytoplasmic polyadenylation and translation of mRNA encoding Mos (8–10), a mitogen-activated protein kinase (MAPK) kinase kinase that is required for the G2/Meiosis I transition in frog oocytes (11). In mouse oocytes, Aur-A also stimulates translation of cyclin B mRNA (12). Aur-A levels can also affect the timing of mitotic entry in embryonic and somatic cells. In Caenorhabditis elegans embryos, loss of Aur-A results in a 1- to 2-min delay in time of mitotic entry (13). In mammalian somatic cells, Aur-A RNA interference (RNAi) treatment or injection of Aur-A antibody is reported to delay (14, 15) or block (15, 16) mitotic entry, possibly through translation effects (17). Cdc25B, a member of the phosphatase family that activates cyclin-dependent kinases at various cell cycle points, is enriched at centrosomes and appears to be responsible for an initial activation of cdc2 at centrosomes during late G2 (18–22). Recent work suggests that the Aur-A might contribute to early, centrosomal activation of cyclin B1-cdc2 through phosphorylation of cdc25B (23).
Here, we have carried out a systematic analysis of the effects of Aur-A on the timing of mitotic entry during normal cell cycle progression in Xenopus egg cycling extracts. In these extracts, neither the DNA nor spindle integrity checkpoint pathways are active (24–27), making it possible to investigate the basic cell cycle roles of Aur-A free from complications encountered in somatic cells with active checkpoint pathways. We find that immunodepletion of Aur-A or addition of inactive Aur-A, which disrupts spindle bipolarity, delays but does not block cdc2 activation and mitotic entry. The catalytic domain alone of Aur-A is adequate to restore spindle bipolarity; additional N-terminal sequences are required for Aur-A's ability to affect the timing of mitotic entry. Most notably, Aur-A depletion or overexpression delays or accelerates, respectively, the timing of mitotic entry even in the absence of centrosomes.
Results
Aur-A and the Timing of Mitotic Entry.
To prepare cycling extracts, metaphase II-arrested eggs were activated by the addition of calcium ionophore to induce entry into interphase of the first mitotic cycle (28, 29). After 30 min, eggs were shifted to 4°C to stop cell cycle progression and a low speed supernatant was prepared. For immunodepletion experiments, only extracts in which >99% of the Aur-A protein was removed were used (Fig. 1A). Depletion of Aur-A did not reduce the level of Aur-B or reduce phosphorylation of histone H3 Ser-10, a target of Aur-B, indicating that the depletion was specific for Aur-A (not shown). Sperm nuclei were added, and extracts were warmed to 20°C to initiate resumption of the cell cycle.
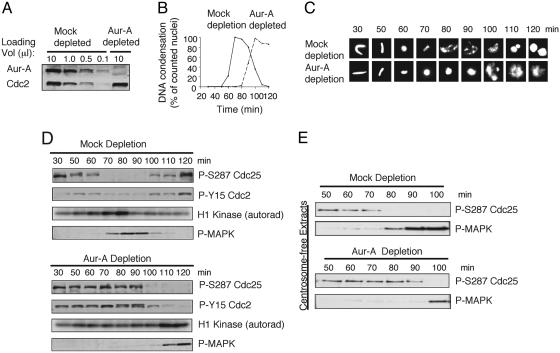
Fig. 1.
Depletion of Aur-A delays mitotic entry, but does not block it. (A) Cycling extracts were prepared 30 min after egg activation and immunodepleted with two rounds of IgG (mock) or affinity-purified Aur-A antibody. Samples were analyzed by SDS/PAGE followed by blotting with Aur-A and cdc2 antibodies. (B) Mock or Aur-A-depleted cycling extracts were supplemented with demembranated sperm nuclei (500 per μl). Samples were stained with Hoechst dye 33342 and visualized by fluorescence microscopy. At least 100 chromosome structures at each time point were quantified. (C) Representative chromatin structures from B. (D) Samples were analyzed by SDS/PAGE followed by blotting with indicated antibodies. Cdc2 activity toward histone H1 was assayed by incorporation of γ-32P, followed by SDS/PAGE and autoradiography. (E) Cycling extracts lacking sperm nuclei were mock- or Aur-A-depleted. Samples were blotted with the indicated antibodies.
Most nuclei in the mock-depleted extract entered mitosis between 60 and 70 min, as judged by chromosome condensation (Fig. 1 B and C). At the molecular level (Fig. 1D), mitotic entry was reflected by (i) removal of the inhibitory phosphorylation on Ser-287 on cdc25C (29, 30), the phosphatase that removes the inhibitory Tyr-15 phosphorylation from cdc2, (ii) dephosphorylation of cdc2 Tyr-15, and (iii) a rise in cdc2's histone H1 kinase activity. The mock-depleted extract continued through mitosis, as judged both morphologically and by the mid-mitotic activation of MAPK that is required to maintain chromosomes condensed after destruction of cyclin B and inactivation of cdc2 (31, 32). The extract then entered interphase of the second cell cycle around 100 min, as marked by rephosphorylation of cdc25C on Ser-287 and the formation of nuclei containing decondensed chromosomes.
In extracts depleted of Aur-A, cdc2 activation, chromosome condensation, and the subsequent activation of MAPK were all delayed by 20–30 min (Fig. 1 B–D). In eight separate experiments, the extent of the G2/M delay varied from 10 min to >30 min, with an average 18-min delay (not shown). Depletion also interfered with changes in chromosome morphology (Fig. 1C). Some chromosome condensation did occur, but individual chromosomes were hard to distinguish and partially decondensed chromatin arrays developed during the last time points taken, when both cdc2 and MAPK activity were still high. In other experiments where cell cycle kinetics were faster, it was seen that Aur-A depletion did not block inactivation of either cdc2 or MAPK but did interfere with chromosome decondensation and spindle disassembly (Fig. 5, which is published as supporting information on the PNAS web site).
Addition of kinase-dead Aur-A D281A (33) also delayed but did not block cdc2 activation and mitotic entry (Fig. 2 A and B). Thus, in addition to previously known dominant negative effects of Aur-A kinase mutants on spindle morphology (34) (Fig. 2 C and D), kinase-dead Aur-A also interferes with the timing the G2/M transition. Supplementing extracts with 5-fold more active Aur-A during mid-interphase (at 50 min) accelerated cdc2 activation and mitotic entry. Thus, not only are normal levels of Aur-A required for timely mitotic entry, but elevated levels can advance the time of cdc2 activation and the G2/M transition.
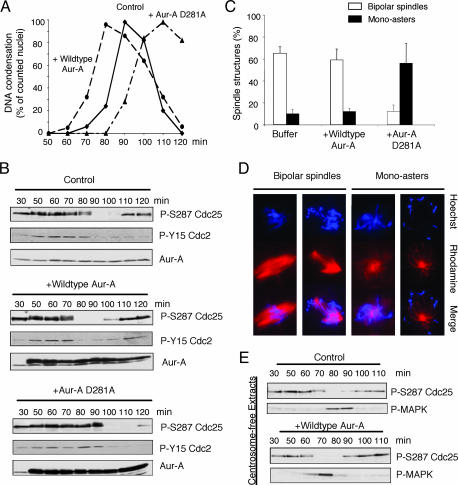
Fig. 2.
Overexpression of Aur-A accelerates the G2/M transition. Kinase-dead Aur-A delays mitotic entry and leads to monopolar spindles. (A) Recombinant wild-type or kinase-dead D281A Aur-A (100 ng/μl) or buffer was added to cycling extracts at 50 min. Samples were stained with Hoechst dye 33342. At least 100 nuclei were scored for each condition. (B) Samples in A were blotted with the indicated antibodies. (C) Cycling extracts were supplemented with nuclei and rhodamine-labeled tubulin. Wild-type or D281A Aur-A (100 ng/μl) or buffer was added at 50 min. Samples were stained with Hoechst dye 33342. The percentage of spindle structures was averaged from three independent experiments. At each time point, at least 100 spindle structures were counted. (D) Representative spindle structures are shown. (E) Cycling extracts lacking sperm nuclei were supplemented with recombinant wild-type Aur-A (100 ng/μl) or buffer at 50 min. Samples were blotted as above.
Aur-A's Role in the Timing of Mitotic Entry Does Not Require the Presence of Centrosomes.
Because Xenopus eggs lack centrosomes, it is possible to use egg extracts that have not been supplemented with sperm nuclei (the source of centrosomes in fertilized eggs) to ask whether Aur-A's ability to affect the timing of mitotic entry depends on its association with, or effects on, centrosomes. Immunodepletion of Aur-A from extracts lacking centrosomes clearly delayed activation of cdc2, as judged by dephosphorylation of cdc25C and the subsequent mid-mitotic activation of MAPK (Fig. 1E). Overexpression of Aur-A in centrosome-free extracts accelerated cdc25C dephosphorylation and activation of MAPK (Fig. 2E). Thus, in the early embryonic cell cycles, the effects of Aur-A on the timing of mitotic entry are through a centrosome-independent pathway. Because nuclei are also the source of chromatin, these results also indicate that the effects of Aur-A or interacting proteins on timing of cdc2 activation does not depend on the presence of chromatin.
Replacement of Endogenous Aur-A with Active Recombinant Aur-A Corrects the Timing of Mitotic Entry and Increases the Frequency of Bipolar Spindle Formation.
Mock- or Aur-A-depleted cycling extracts were supplemented with either recombinant active wild-type Aur-A or the D281A mutant to give a final concentration that was slightly higher (40 ng/μl) than endogenous Aur-A. Adding back wild-type Aur-A, but not the D281A mutant, abrogated the delay in the G2/M transition (Fig. 3A). This result argues that Aur-A, rather than a coprecipitating protein, is required for the proper kinetics of the G2/M transition, and that, more specifically, Aur-A's kinase activity is required for the correct timing of mitotic entry. Adding back wild-type Aur-A also rescued the bipolar spindle defects. A quantitative summary of four independent experiments are shown in Fig. 3B. Representative spindle structures seen in mitosis are shown in Fig. 3C.
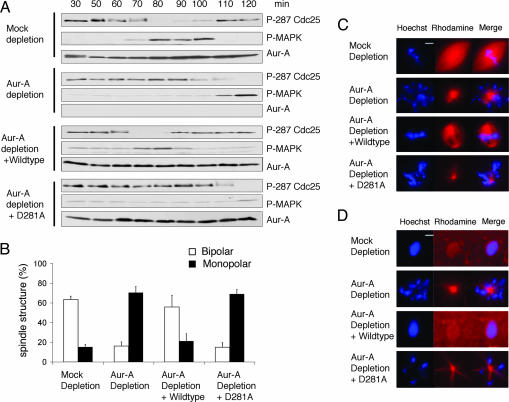
Fig. 3.
Adding back wild-type, but not kinase-dead form of Aur-A, to Aur-A-depleted cycling extracts corrects the timing of G2/M transition and rescues the bipolar spindle formation. (A) Mock or Aur-A-depleted extracts were supplemented with nuclei (500 per μl) and rhodamine-labeled tubulin. Wild-type Aur-A, kinase-dead D281A Aur-A, or buffer control was added to Aur-A-depleted cycling extracts at 30 min. Samples were analyzed as in Fig. 2. (B) Samples were taken during mitosis and stained with Hoechst dye 33342. The percentage of distinct spindle structures in mitosis was averaged from four independent experiments. At each time point, >100 spindle structures were counted. (C) Representative spindle structures are shown. (D) Sixty minutes after mitosis, samples were again taken and examined as above.
The Catalytic Domain Alone Is Sufficient to Enhance Spindle Bipolarity, but Additional N-Terminal Sequences Are Required for Proper Chromosome Alignment and the Timing of Mitotic Entry.
Whereas catalytically active full-length Aur-A was able to correct the timing of mitotic entry, the C-terminal catalytic domain itself (Aur-A 137–407) was not, indicating the requirement for additional N-terminal sequences (Fig. 4 A and B). Aur-A 19–407 completely rescued mitotic timing, but Aur-A 69–407 did not. Thus, in addition to Aur-A's catalytic activity, sequences within the N-terminal region 19–68 are in some way required for setting the timing of the G2/M transition.
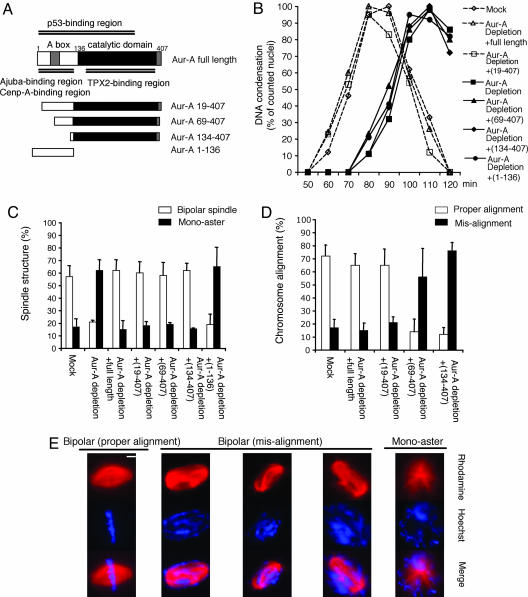
Fig. 4.
The Aur-A catalytic domain alone is sufficient to allow formation of bipolar spindle, but additional N-terminal sequences are required to restore the correct timing of mitotic entry and proper chromosome alignment. (A) Schematic of Aur-A proteins used in add-back experiments. (B) Mock- or Aur-A-depleted extracts were supplemented with nuclei (500 per μl) and rhodamine-labeled tubulin. Aur-A proteins or buffer were added to the Aur-A-depleted extract. Samples were analyzed as in Fig. 2. (C and D) The percentage of spindle structures and chromosome alignment was averaged from three independent experiments. At each time point, >100 spindle structures were counted. (E) Representative spindle structures. (Scale bar, 10 μm.)
In a separate experiment, the Aur-A truncations were tested for their ability to restore spindle bipolarity to Aur-A-depleted extracts (Fig. 4 C and E). The N-terminal fragment showed no obvious rescue effects. Full-length Aur-A, the C-terminal catalytic domain alone, and the constructs 134–407 and 69–407 each promoted an increase in the number of bipolar structures. However, the spindles that assembled either in the presence of the catalytic domain or a larger (69–407) fragment had misaligned chromosomes (Fig. 4 D and E). Thus, the catalytic domain of Aur-A by itself appears to be sufficient for establishing bipolarity, but additional N-terminal sequences are in some way important for proper chromosome alignment.
Aur-A and Mitotic Exit.
In most cell types, spindle defects lead to activation of the spindle integrity checkpoint, which then arrests the cell cycle in mitosis, both biochemically and morphologically. As this checkpoint does not operate in Xenopus early embryos, it is possible to ask whether Aur-A is directly required for the inactivation of cdc2 and/or MAPK during mitotic exit or for the return of chromosomes and microtubules to the interphase state. Depletion of Aur-A did not prevent the inactivation of either cdc2 or MAPK (Fig. 5). However, depletion interfered with morphological events of mitotic exit (Fig. 3D): partially decondensed chromosomes and focused arrays of microtubules remained for extended periods. Thus, in addition to its well known roles in centrosome maturation and mitotic spindle morphology, Aur-A is required to couple inactivation of cdc2 and MAPK to the morphological events of mitotic exit.
Discussion
Aur-A and Timing of the G2/M Transition.
A main finding of this study is that increasing the level of Aur-A protein in Xenopus cycling egg extracts advances the timing of cdc2 activation and mitotic entry, and removing Aur-A delays, but does not block cdc2 activation and mitotic entry. Moreover, these effects on mitotic timing are independent of the presence of both centrosomes and chromatin. This finding has bearing on a proposed role for Aur-A in the activation of cyclin B1-cdc2. In somatic cells, a phosphorylated version of cyclin B1 that may be specific for active cyclin B1-cdc2 complexes localizes first at centrosomes (22) and Aur-A appears to be required for this localization (15). Moreover, some of the phosphatase cdc25B that catalyzes this early activation is also found at centrosomes (18, 19, 21), and there is some evidence that cdc25B might be a substrate of Aur-A (23). However, our finding that Aur-A affects the timing of cdc2 activation in egg extracts even in the absence of centrosomes suggest that there is an additional level of regulation.
Previously identified translational targets of Aur-A in G2-arrested oocytes are mos and cyclin B mRNAs (8, 12). Mos is a MAPK kinase kinase that is required for breaking the G2/Meiosis I transition in Xenopus oocytes (35). However, an Aur-A-mediated increase in Mos protein seems an unlikely explanation for the early mitotic cell cycles. First, addition of Aur-A protein during interphase does not lead to a detectable increase in Mos (H. Wu and J.V.R., unpublished data). Second, addition of Mos or MAP kinase blocks mitotic entry rather than accelerating it (36, 37). CPEB-dependent translation of cyclin B1 is required for mitotic entry in egg extracts (38) and overexpression of Aur-A enhances polyadenylation of cyclin mRNA in somatic cells (17). Although we were unable to detect any obvious change in cyclin B levels, we cannot rule out small effects at this time.
Domains of Aur-A Required for Timing of Mitotic Entry, Spindle Bipolarity, and Chromosome Alignment.
Most sequence differences among the Aurora kinases lie in their noncatalytic N termini and, as shown here, the N terminus of Aur-A is needed for both correct timing of mitotic entry and proper alignment of chromosomes. Ajuba, a protein that may enhance Aur-A activity during G2, promotes translation of both cyclin B and cdc2 and advance entry into M phase. Moreover, Ajuba binds to the N terminus of Aur-A (17, 39). Regarding chromosome alignment, one obvious target is CENP-A, a variant of histone H3 that resides in nucleosomes of the centromeric chromatin at the inner plate of the kinetochore (40). CENP-A, which binds to the N terminus of Aur-A, appears to be phosphorylated on Ser-7, first by Aur-A and then by Aur-B; both kinases are required for the CENP-A-dependent restriction of Aur-B to the inner centromere (41). When Ser-7 phosphorylation is prevented, mitotic cells show defects in the ability of kinetochores to attach to microtubules, providing a possible explanation for the requirement for Aur-A phosphorylation in chromosome alignment.
By contrast, the catalytic domain alone is sufficient to restore spindle bipolarity. TPX2, a spindle assembly factor that enhances Aur-A activity during mitosis, can bind and activate Aur-A lacking its N terminus (42). TPX2 is required for targeting Aur-A to spindle microtubules, which would concentrate it near potential spindle substrates such as Eg5, a microtubule motor required for spindle bipolarity (reviewed in ref. 43). This discrimination between pathways that require Aur-A's N terminus vs. those that do not may help to organize known and future Aur-A targets into a more cohesive picture.
Materials and Methods
Cycling extracts were prepared from Xenopus eggs (28, 29). Demembranated sperm nuclei (final concentration 500 nuclei per μl) and rhodamine-labeled tubulin (Cytoskeleton, final concentration 50 ng/μl) were added. Typically, 50 μl of cycling extract was used for each individual time course. Cdc2 activity was assayed as described (29)
Bacterially expressed N-terminally His-tagged Aur-A proteins were prepared were prepared as described (44). Kinase-dead Aur-A D281A (33) was created by site-directed mutagenesis (Hua Wu, personal communication). For immunoblots, 2-μl extract samples were separated by SDS/PAGE, blotted and reacted with antibodies against Aur-A (45), active MAPK (Cell Signaling Technology, Beverly, MA; catalog no. 9101S), phospho-cdc2(Tyr-15) (Cell Signaling Technology, catalog no. 911L), phospho-cdc25(Ser-287) (46) as described.
Supplementary Material
Acknowledgments
We are grateful to Bedrick Gadea, Ryoma Puck Ohi, Aaron Groen, and Tim Mitchison (Harvard Medical School) for invaluable advice and discussion and to Jennifer Waters of the Nikon Imaging Center at Harvard Medical School for assistance. This work was supported by National Institutes of Health Grant HD23696 (to J.V.R.) and a National Research Service Award (to Q.L.).
Abbreviations
- Aur-A
Aurora A
- Aur-B
Aurora B
- MAP
mitogen-activated protein
- MAPK
MAP kinase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Blagden S. P., Glover D. M. Nat. Cell Biol. 2003;5:505–511. doi: 10.1038/ncb0603-505. [DOI] [PubMed] [Google Scholar]
- 2.Ducat D., Zheng Y. Exp. Cell Res. 2004;301:60–67. doi: 10.1016/j.yexcr.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Meraldi P., Honda R., Nigg E. A. Curr. Opin. Genet. Dev. 2004;14:29–36. doi: 10.1016/j.gde.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Varmark H. J. Cell Biochem. 2004;91:904–914. doi: 10.1002/jcb.20013. [DOI] [PubMed] [Google Scholar]
- 5.Brittle A. L., Ohkura H. Curr. Biol. 2005;15:R880–R882. doi: 10.1016/j.cub.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Andresson T., Ruderman J. V. EMBO J. 1998;17:5627–5637. doi: 10.1093/emboj/17.19.5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma C., Cummings C., Liu X. J. Mol. Cell. Biol. 2003;23:1703–1716. doi: 10.1128/MCB.23.5.1703-1716.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendez R., Hake L. E., Andresson T., Littlepage L. E., Ruderman J. V., Richter J. D. Nature. 2000;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- 9.Mendez R., Murthy K. G., Ryan K., Manley J. L., Richter J. D. Mol. Cell. 2000;6:1253–1259. doi: 10.1016/s1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- 10.Hodgman R., Tay J., Mendez R., Richter J. D. Development (Cambridge, U.K.) 2001;128:2815–2822. doi: 10.1242/dev.128.14.2815. [DOI] [PubMed] [Google Scholar]
- 11.Sagata N. BioEssays. 1997;19:13–21. doi: 10.1002/bies.950190105. [DOI] [PubMed] [Google Scholar]
- 12.Tay J., Hodgman R., Richter J. D. Dev. Biol. 2000;221:1–9. doi: 10.1006/dbio.2000.9669. [DOI] [PubMed] [Google Scholar]
- 13.Hannak E., Kirkham M., Hyman A. A., Oegema K. J. Cell Biol. 2001;155:1109–1116. doi: 10.1083/jcb.200108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marumoto T., Hirota T., Morisaki T., Kunitoku N., Zhang D., Ichikawa Y., Sasayama T., Kuninaka S., Mimori T., Tamaki N., et al. Genes Cells. 2002;7:1173–1182. doi: 10.1046/j.1365-2443.2002.00592.x. [DOI] [PubMed] [Google Scholar]
- 15.Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., Saya H. Cell. 2003;114:585–598. doi: 10.1016/s0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- 16.Du J., Hannon G. J. Proc. Natl. Acad. Sci. USA. 2004;101:8975–8980. doi: 10.1073/pnas.0308484101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasayama T., Marumoto T., Kunitoku N., Zhang D., Tamaki N., Kohmura E., Saya H., Hirota T. Genes Cells. 2005;10:627–638. doi: 10.1111/j.1365-2443.2005.00870.x. [DOI] [PubMed] [Google Scholar]
- 18.Gabrielli B. G., De Souza C. P., Tonks I. D., Clark J. M., Hayward N. K., Ellem K. A. J. Cell Sci. 1996;109:1081–1093. doi: 10.1242/jcs.109.5.1081. [DOI] [PubMed] [Google Scholar]
- 19.Lammer C., Wagerer S., Saffrich R., Mertens D., Ansorge W., Hoffmann I. J. Cell Sci. 1998;111:2445–2453. doi: 10.1242/jcs.111.16.2445. [DOI] [PubMed] [Google Scholar]
- 20.Karlsson C., Katich S., Hagting A., Hoffmann I., Pines J. J. Cell Biol. 1999;146:573–584. doi: 10.1083/jcb.146.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Souza C. P., Ellem K. A., Gabrielli B. G. Exp. Cell Res. 2000;257:11–21. doi: 10.1006/excr.2000.4872. [DOI] [PubMed] [Google Scholar]
- 22.Jackman M., Lindon C., Nigg E. A., Pines J. Nat. Cell Biol. 2003;5:143–148. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- 23.Dutertre S., Cazales M., Quaranta M., Froment C., Trabut V., Dozier C., Mirey G., Bouche J. P., Theis-Febvre N., Schmitt E., et al. J. Cell Sci. 2004;117:2523–2531. doi: 10.1242/jcs.01108. [DOI] [PubMed] [Google Scholar]
- 24.Dasso M., Newport J. W. Cell. 1990;61:811–823. doi: 10.1016/0092-8674(90)90191-g. [DOI] [PubMed] [Google Scholar]
- 25.Minshull J., Sun H., Tonks N. K., Murray A. W. Cell. 1994;79:475–486. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- 26.Anderson J. A., Lewellyn A. L., Maller J. L. Mol. Biol. Cell. 1997;8:1195–1206. doi: 10.1091/mbc.8.7.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumagai A., Yakowec P. S., Dunphy W. G. Mol. Biol. Cell. 1998;9:345–354. doi: 10.1091/mbc.9.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray A. W. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 29.Gadea B. B., Ruderman J. V. Mol. Biol. Cell. 2005;16:1305–1318. doi: 10.1091/mbc.E04-10-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanford J. S., Ruderman J. V. Mol. Biol. Cell. 2005;16:5749–5760. doi: 10.1091/mbc.E05-06-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chau A. S., Shibuya E. K. Biol. Cell. 1998;90:565–572. [PubMed] [Google Scholar]
- 32.Guadagno T. M., Ferrell J. E., Jr. Science. 1998;282:1312–1315. doi: 10.1126/science.282.5392.1312. [DOI] [PubMed] [Google Scholar]
- 33.Eyers P. A., Erikson E., Chen L. G., Maller J. L. Curr. Biol. 2003;13:691–697. doi: 10.1016/s0960-9822(03)00166-0. [DOI] [PubMed] [Google Scholar]
- 34.Roghi C., Giet R., Uzbekov R., Morin N., Chartrain I., Le Guellec R., Couturier A., Doree M., Philippe M., Prigent C. J. Cell Sci. 1998;111:557–572. doi: 10.1242/jcs.111.5.557. [DOI] [PubMed] [Google Scholar]
- 35.Schmitt A., Nebreda A. R. J. Cell Sci. 2002;115:2457–2459. doi: 10.1242/jcs.115.12.2457. [DOI] [PubMed] [Google Scholar]
- 36.Walter S. A., Guadagno T. M., Ferrell J. E., Jr. Mol. Biol. Cell. 1997;8:2157–2169. doi: 10.1091/mbc.8.11.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bitangcol J. C., Chau A. S., Stadnick E., Lohka M. J., Dicken B., Shibuya E. K. Mol. Biol. Cell. 1998;9:451–467. doi: 10.1091/mbc.9.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Groisman I., Huang Y. S., Mendez R., Cao Q., Richter J. D. Cold Spring Harbor Symp. Quant. Biol. 2001;66:345–351. doi: 10.1101/sqb.2001.66.345. [DOI] [PubMed] [Google Scholar]
- 39.Goyal R. K., Lin P., Kanungo J., Payne A. S., Muslin A. J., Longmore G. D. Mol. Cell. Biol. 1999;19:4379–4389. doi: 10.1128/mcb.19.6.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henikoff S., Dalal Y. Curr. Opin. Genet. Dev. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Kunitoku N., Sasayama T., Marumoto T., Zhang D., Honda S., Kobayashi O., Hatakeyama K., Ushio Y., Saya H., Hirota T. Dev. Cell. 2003;5:853–864. doi: 10.1016/s1534-5807(03)00364-2. [DOI] [PubMed] [Google Scholar]
- 42.Bayliss R., Sardon T., Vernos I., Conti E. Mol. Cell. 2003;12:851–862. doi: 10.1016/s1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- 43.Gadde S., Heald R. Curr. Biol. 2004;14:R797–R805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 44.Tsai M. Y., Wiese C., Cao K., Martin O., Donovan P., Ruderman J., Prigent C., Zheng Y. Nat. Cell Biol. 2003;5:242–248. doi: 10.1038/ncb936. [DOI] [PubMed] [Google Scholar]
- 45.Littlepage L. E., Ruderman J. V. Genes Dev. 2002;16:2274–2285. doi: 10.1101/gad.1007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duckworth B. C., Weaver J. S., Ruderman J. V. Proc. Natl. Acad. Sci. USA. 2002;99:16794–16799. doi: 10.1073/pnas.222661299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.