Abstract
Shared transcription factor binding sites that are conserved in distance and orientation help control the expression of gene products that act together in the same biological context. New bioinformatics approaches allow the rapid characterization of shared promoter structures and can be used to find novel interacting molecules. Here, these principles are demonstrated by using molecules linked to the unique functional unit of the glomerular slit diaphragm. An evolutionarily conserved promoter model was generated by comparative genomics in the proximal promoter regions of the slit diaphragm-associated molecule nephrin. Phylogenetic promoter fingerprints of known elements of the slit diaphragm complex identified the nephrin model in the promoter region of zonula occludens-1 (ZO-1). Genome-wide scans using this promoter model effectively predicted a previously unrecognized slit diaphragm molecule, cadherin-5. Nephrin, ZO-1, and cadherin-5 mRNA showed stringent coexpression across a diverse set of human glomerular diseases. Comparative promoter analysis can identify regulatory pathways at work in tissue homeostasis and disease processes.
Keywords: bioinformatics, gene regulation, podocyte, slit diaphragm
The prediction and analysis of the regulatory networks underlying gene expression is a major challenge in functional genomics (1, 2). The cell- and signal-specific regulation of genes is controlled to a significant degree by functional elements within their proximal promoter regions. Proximal promoters are represented by the nucleotide sequence immediately upstream from the site of transcriptional initiation, often overlapping with the respective transcribed sequence. The relative order and spacing of regulatory elements in promoters, such as transcription factor (TF) binding sites (TFBSs), are often highly conserved through evolution, highlighting their importance in regulation. Recent methods offer rapid identification of these sets of elements through comparative genomics, an approach similar to phylogenetic footprinting (3).
It has been proposed that convergent evolution leading to organizational similarities of regulatory elements among different promoters helps facilitate the coordinated regulation required for complex structures and processes (2, 4). The ordered complex of TFBSs that are conserved in distance and orientation has been referred to as a promoter module or framework (5, 6). The hypothesis of “functional context” states that convergent evolution leading to the generation of shared common structures or modules among promoters provides a potential mechanism for the synchronization of the expression of genes whose products must interact within a common biological process (5, 7). Shared promoter modules could help facilitate the orchestrated transcriptional coregulation of functionally interdependent proteins in time and space. In theory, promoter modules/frameworks that have been linked to a specific tissue microenvironment or biologic process could also be used in bioinformatic analyses to detect genes heretofore not associated with the specific biologic process (5).
To test these hypotheses, a unique tissue environment linked to a specific biologic function was selected for a bioinformatic-driven analysis of promoter coregulation as outlined in Fig. 1. Glomerular epithelial cells, also referred to as podocytes, cover the fenestrated capillaries of renal glomeruli in vertebrates and contribute to the kidney filtration barrier (8). Podocytes have a complex phenotype with interdigitating foot processes that are bridged by the final filtration barrier, referred to as the “slit diaphragm.” Currently, it is only possible to study the slit diaphragm in the context of an intact glomerulus. This unique cell–cell contact shares characteristics with adherence junctions (e.g., P-cadherin involvement), tight junctions [expression of tight junction protein 1 (TJP1), also called zonula occludens-1 (ZO-1)], and the “immunological synapse” (participation of CD2AP) (8). A series of molecules [e.g., nephrin and podocin (9, 10)] appear to be uniquely expressed in this functional unit and are key mediators of inherited and acquired human renal disease. We hypothesized that genes with such a restricted expression may share hierarchical features within their promoters, which would help to guide the coregulation of genes involved in this functional complex. The identification of these features would provide a direct link to regulatory pathways and could also, in theory, be used to identify novel interacting partners through bioinformatics-based searches.
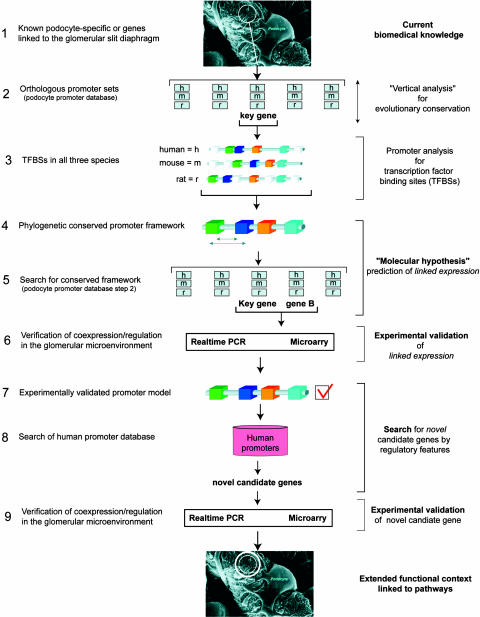
Fig. 1.
Strategy used for comparative promoter analysis. This figure details the strategy that was used to systematically search for promoter frameworks linked to the coregulation of podocyte/slit diaphragm-associated genes. h, human; m, mouse; r, rat. Electron micrograph of podocytes was provided by W. Kriz (University of Heidelberg, Heidelberg).
Results
Identification of an Evolutionarily Shared Promoter Framework in the Nephrin and ZO-1 Genes.
In orthologous promoters, elements important for regulation of a given gene are expected to be conserved over evolution. Starting with a set of previously characterized genes involved in the generation of the slit diaphragm or other important aspects of podocyte biology (Table 1), orthologous promoter regions were identified from three species (Homo sapiens, Mus musculus, and Rattus norvegicus) by using a comparative genomics tool (eldorado; Genomatix). For 18 of the 20 genes associated with podocyte biology, promoter sequences from at least two species could be identified (Table 1).
Table 1.
Podocyte-expressed genes included in the analysis
| Gene | Relevance | PubMed ID | Mm | Rn | Hs |
|---|---|---|---|---|---|
| ACTN4 | B, C (8) | 10700177 | + | + | |
| CD2AP | A, B, C (8) | 10514378 | + | + | |
| DAG1 | B, C (8) | 10703664 | + | + | |
| DES | C (8) | 2183627 | + | + | + |
| FAT | A, B, C (8) | 11231355 | + | ||
| ILK | C (8) | 11481249 | + | + | + |
| KIRREL | A, B, C (8) | 12865409 | + | ||
| KIRREL2 | A, B, C (39) | 12504092 | + | + | |
| KIRREL3 | A, B, C (40) | 15843475 | + | + | |
| LMX1B | B, C (8) | 11956244, -5 | + | + | |
| MAGI1 | C (8) | 11274227 | + | + | |
| NPHS1 | A, B, C (12) | 9660941 | + | + | + |
| NPHS2 | A, B, C (10) | 10742096 | + | + | |
| PDPN | B, C (8) | 9327748 | + | + | |
| PODXL | B, C (8) | 11457882 | + | + | + |
| PTPRO | B, C (8) | 11086029 | + | + | + |
| SYNPO | B, C (8) | 15841212 | + | + | + |
| TCF21 | B, C (8) | 10572052 | + | + | + |
| TJP1 | A, B, C (8) | 2202736 | + | + | + |
| WT1 | B, C (15) | 11912180 | + | + | + |
Twenty genes were chosen for the initial analysis on the basis of (A) their association with the slit diaphragm, (B) preferential expression in podocytes, or (C) their reported essential role in the biology of this cell type or interaction with known members of the slit diaphragm. Promoter regions were analyzed if they were found phylogenetically conserved in at least two species. Cross-species proximal promoter regions were identified for 18 of these genes. The table displays the gene, relevance leading to inclusion in the study (A–C above), PubMed ID, and the species in which promoter definition was made (+). Please see ref. 8 and references therein for details of podocyte-expressed genes. Mm, Mus musculus; Rn, Rattus norvegicus; Hs, Homo sapiens.
Genes encoding proteins that functionally interact may exhibit conserved organization of promoter elements (5). In an attempt to address this hypothesis for podocyte-associated genes, a systematic approach was applied to identify promoter frameworks shared among podocyte-expressed genes. As described in Materials and Methods, TFBSs common to promoter regions across species were first identified for individual genes (Table 1). Parameter settings of TFBS selection, strand orientation, and order were strictly determined by sequence analysis of the promoter sets. There were only two parameters that were subsequently optimized manually. Minimal distance ranges between matrices were reduced (from default 10 nt to 1 nt) to include conserved TFBSs identified by visual inspection. Matrix similarities were used at default values and fractionally adjusted (default −0.1, −0.2, or −0.5) when the reduction allowed detection of evolutionary conserved TFBS sets that were missed with the default settings (11). The evolutionary conserved frameworks were determined by using frameworker (Genomatix) on the basis of the predefined TFBS subsets. Models, reflecting the frameworks, were then built by using fastm (Genomatix) and optimized for all three species. These models were finally tested for their presence within the promoters of other genes within the podocyte “seed” data set by using modelinspector (Genomatix) (Fig. 1, steps 1–5).
This approach eventually yielded a framework of conserved TFBSs that originated in the nephrin gene (NPHS1) promoter and was also found in the human, mouse, and rat promoters for ZO-1 (TJP1) (Fig. 1, step 5) by searching the podocyte-associated proximal promoter sequences (total of 47 sequences, see Table 1; modelinspector). This nephrin/ZO-1 promoter model comprised four binding sites for the following TFs: E-box binding factors (such as max, c-myc/max, and n-myc), monomeric Meis1 homeodomain protein (MEIS), mouse Krüppel-like factor for the zinc finger protein MOK-2 (MOKF), and homeodomain class recognizing TG motifs (TALE). None of the TFs involved in the model has been formerly linked to the expression of filtration barrier-associated genes. The nephrin/ZO-1 model is graphically presented in Fig. 2A and shows the strand orientation, distance range between elements, and core- and matrix-similarity values determined by direct sequence analysis. This model is found to be conserved across all three species in both slit diaphragm-associated genes, nephrin and ZO-1 (Fig. 2B and Fig. 5, which is published as supporting information on the PNAS web site).
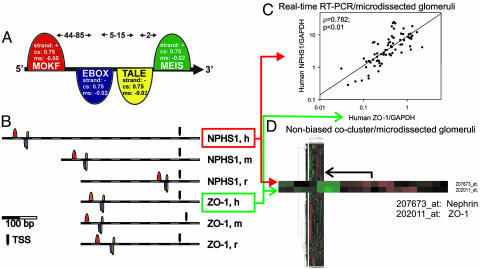
Fig. 2.
Nephrin and ZO-1 share a common promoter model. (A) Nephrin promoter model. The framework of four elements with a maximal total length of 102 bp was found to be highly conserved in the human, mouse, and rat proximal promoter regions of nephrin (NPHS1). Following are the elements, parameters, and their abbreviations. +, Sense strand orientation; −, antisense strand orientation; cs, minimal core similarity; ms, minimal matrix similarity; distance, number of nucleotides to next element (arrows in the upper part of the figure). MOKF (mouse Krüppel-like factor): ribonucleoprotein-associated zinc finger protein MOK-2; strand, +; cs, 0.75; ms, −0.05; distance to next element, 44–85 nt. EBOX (E-box binding factors): (i) upstream stimulating factor, (ii) MYC-MAX binding sites, (iii) MAX, (iv) C-MYC/MAX heterodimer, (v) N-MYC, and (vi) sterol regulatory element binding proteins 1 and 2; strand, −; cs, 0.75; ms, −0.02; distance to next element, 5–15 nt. TALE (homeodomain class recognizing TG motifs): TALE class of homeodomain factors; strand, +; cs, 0.75; ms, −0.02; distance to next element, 2 nt. MEIS (homeodomain factor expressed in myeloid leukemia): monomeric Meis1 homeodomain protein; strand, +; cs, 0.75; ms, −0.02. (B) The nephrin promoter model was found in the mouse, human, and rat promoters for ZO-1 (TJP1). The conserved promoter model for the nephrin gene (NPHS1) was subsequently detected in the proximal promoter for ZO-1 (TJP1) in all three species. h, H. sapiens; m, M. musculus; r, R. norvegicus; TSS, transcription start site. The model’s 3′ end (MEIS binding site) is localized 768, 478, and 48 bp upstream of the TSS of nephrin and 380, 423, or 332 bp upstream of ZO-1 (H. sapiens, M. musculus, and R. norvegicus, respectively; see Fig. 5 for more detail). (C) Nephrin (NPHS1) and ZO-1 (TJP1) steady-state mRNA shows stringent coregulation in human glomerular disease. The steady-state expression of the mRNA templates for nephrin and ZO-1 were analyzed by real-time RT-PCR and showed a significant positive correlation independent of the housekeeper engaged for normalization. Seventy-six biopsies were analyzed from patients with four different proteinuric diseases and two different control groups (see Results). Each dot represents the expression in one patient’s sample of microdissected glomeruli from routine renal biopsies. (D) Genome-wide array analysis of microdissected human glomeruli. The 2D unsupervised clustering of an Affymetrix chip (HG-U133A)-based genome-wide expression analysis of glomerular mRNAs from 22 patients (see Results) demonstrated the close coregulation of nephrin (NPHS1) and ZO-1 (TJP1) (shown enlarged).
Nephrin was the first member of the slit diaphragm identified by positional cloning of one type of hereditary nephrosis found in humans (12). The gene shows highly restricted tissue expression (13, 14). ZO-1 is also a previously identified slit diaphragm-associated gene product (8, 15).
The Nephrin and ZO-1 Genes Are Coregulated Under Diverse Biologic Settings.
The hypothesis of functional context states that genes that share promoter modules may be linked for coregulation of gene expression (Fig. 1, steps 5 and 6). In this example, the biologic context is represented by the podocyte slit membrane associated with glomerular foot processes, a microenvironment that can be studied only in vivo or ex vivo. The potential coregulation of nephrin and ZO-1 was then examined by using real-time RT-PCR analysis of microdissected human glomeruli taken from 76 renal biopsies representing diverse disease states affecting the glomerular filtration barrier [minimal-change disease, n = 13; benign nephrosclerosis, n = 16; membranous glomerulonephropathy, n = 28; focal segmental glomerulosclerosis, n = 9; and control tissue, n = 10 (six tumor nephrectomies and four pretransplant biopsies)] (Fig. 2C). Nephrin and ZO-1 mRNA levels were compared and showed linked regulation over two orders of magnitude of expression, showing a positive correlation across all biopsies studied (ρ, Spearman’s rho correlation) (ρ = 0.78 and P < 0.01 and ρ = 0.49 and P < 0.01 for normalization to GAPDH or 18S rRNA, respectively).
As a second independent approach to test for mRNA coregulation, genome-wide expression profiles using Affymetrix (Santa Clara, CA) oligonucleotide arrays (HG-U133A) and nonbiased clustering of gene expression was generated from human microdissected glomeruli obtained from 22 different patients and controls (Fig. 1, step 6 and Fig. 2D). Control biopsies were taken from pretransplantation kidney biopsies during cold ischemia time (living donors, n = 4) or from tumor nephrectomies (n = 4). Three biopsies with no histological lesions (thin basement membrane disease, n = 3) served as an additional control group. Biopsies with clinical and histopathological diagnosis of diabetic nephropathy (n = 7) (with mild or moderate to severe tubulo-interstitial damage) and biopsies with minimal-change glomerular disease (n = 4) were also included. Among the four groups (control tissue, minimal-change disease, mild diabetic nephropathy, and moderate to severe diabetic nephropathy), 2,162 genes showed significant differential regulation. Consistent with previous studies (16, 17), nephrin mRNA was found to be decreased in severe diabetic nephropathy. Unsupervised cluster analysis was then used to display tightly regulated genes side by side. By using this analysis, ZO-1 was found to be the closest neighbor to nephrin in all 22 samples, demonstrating a tight coregulation of these genes across diverse glomerular diseases (ρ = 0.83, P < 0.01).
The Nephrin/ZO-1 Promoter Model Is Used to Search for Novel Genes Functionally Related to the Slit Diaphragm Complex.
To determine whether this approach could be used for the de novo identification of genes not previously associated with the slit diaphragm, a database of 50,145 human promoter sequences (Genomatix Human Promoter Database) was screened for the occurrence of the nephrin/ZO-1 model (Fig. 1, step 8). The model was found in 79 human promoters (<0.16% of total sequences). Of these 79 sequences, 36 could be associated with genes of known function (Table 2, which is published as supporting information on the PNAS web site). Further characterization revealed that the framework initially found in the human promoters was also evolutionary conserved in at least two species for 6 of the 36 genes. These 6 genes were Rho GDP-dissociation inhibitor β (ARHGDIB), cadherin-5 (CDH5), jun-B (JUNB), rab5C (RAB5C), semaphorin-3A (SEMA3A), and titin (TTN). Screening mRNA isolated from human kidney, microdissected glomeruli and conditionally immortalized podocytes by using real-time RT-PCR demonstrated, with the exception of titin, significant expression of all these genes relative to levels of nephrin mRNA (Fig. 1, step 9 and Table 3, which is published as supporting information on the PNAS web site).
Coregulation and Colocalization of the Candidate Gene Cadherin-5.
On the basis of the following criteria, cadherin-5 was selected for further characterization. First, it was the only candidate gene found to carry the respective promoter model in all three species (Figs. 3 and 5). Second, cadherin-5 is known to be involved in the regulation of cell–cell contacts, an essential aspect of the slit diaphragm, and finally, unlike some other candidate genes (see below), cadherin-5 has not been previously described as being expressed in podocytes.
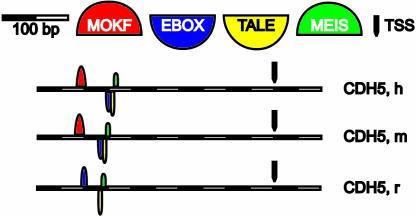
Fig. 3.
The nephrin/ZO-1 model identified the promoter for cadherin-5 (CDH5). The promoter model for the nephrin/ZO-1 genes (Fig. 2 A and B) is found in the promoter region for cadherin-5 (CDH5) and is conserved in all three species (with the exception of MOKF in rat). The 3′ end (MEIS binding site) localizes 344, 335, and 335 bp upstream of the TSS (H. sapiens, M. musculus, and R. norvegicus, respectively; see Fig. 5). h, H. sapiens; m, M. musculus; r, R. norvegicus.
Expression of cadherin-5 in cultured podocytes was confirmed by Western blotting (Fig. 4A). On the basis of the underlying hypothesis, the cadherin-5 protein should be located in the same functional context (podocyte foot processes) as nephrin and ZO-1. Immunogold electron microscopy was used to characterize the subcellular distribution of the cadherin-5 protein in vivo. By using this approach, cadherin-5 protein was localized to the foot processes associated with the slit diaphragm of podocytes (Fig. 4B).
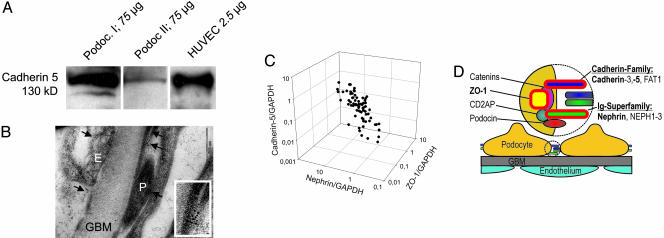
Fig. 4.
Expression of cadherin-5 in podocytes in vitro and in vivo. (A) Western blot for cadherin-5 in cultured podocytes. A monoclonal antibody for human cadherin-5 demonstrated the presence of the protein in two human podocyte cell lines [Podoc. I (36) and Podoc. II (37)]. Human umbilical vascular endothelial cells (HUVEC) served as positive control. The amount of protein loaded is shown for each condition. (B) Immunogold for cadherin-5 on human glomeruli. Immunogold electron microscopy employing a monoclonal antibody for human cadherin-5 demonstrated expression in podocytes (P) and endothelial cells (E). Both cell types are divided by the glomerular basement membrane (GBM). Cadherin-5 localizes in both cell types to the cell–cell contacts, including the slit diaphragm in podocytes. (Inset) Higher magnification of podocyte foot processes showing cadherin-5 at cell–cell contacts. (C) Coregulation of cadherin-5, ZO-1, and nephrin. Testing cadherin-5 mRNA expression in the same cohort of biopsy samples used in Fig. 2C showed that the three mRNA species demonstrated a positive correlation of their expression levels, here normalized to GAPDH: nephrin/cadherin-5, ρ = 0.67 and P < 0.01; ZO-1/cadherin-5, ρ = 0.56 and P < 0.01; nephrin/ZO-1, ρ = 0.78 and P < 0.01. (D) The slit diaphragm is a highly specialized cell–cell contact. Summary showing proteins of the slit diaphragm complex assembled in a zipper-like structure (potentially building the pores of the final filtration barrier) and the recruited cytoplasmatic adapter molecules involved in signal transduction (8, 38). Known members of the slit diaphragm include nephrin, NEPH1–3, podocin, CD2AP, ZO-1, P-cadherin (cadherin-3), and FAT1. The gene products linked through functional context and comparative promoter analysis are highlighted in red (nephrin, ZO-1, and cadherin-5).
The predicted coregulation of cadherin-5 with nephrin and ZO-1 was then investigated (Fig. 1, step 9). The low signal strength of cadherin-5 on the gene array precluded cluster analysis. Real-time RT-PCR was again engaged for quantification of the three genes (nephrin, ZO-1, and cadherin-5) using glomerular mRNA from the cohort of 76 patients (minimal-change disease, benign nephrosclerosis, membranous glomerulonephropathy, focal segmental glomerulosclerosis, and control tissue) described in Fig. 2. The results show a stringent coregulation of cadherin-5 mRNA with nephrin and ZO-1 in human glomerular disease (Fig. 4C) (nephrin/cadherin-5: ρ = 0.67, P < 0.01 and ρ = 0.45, P < 0.01; ZO-1/cadherin-5: ρ = 0.56, P < 0.01 and ρ = 0.23, P < 0.05, for normalization to GAPDH or 18S rRNA, respectively).
Discussion
Understanding the orchestration of gene networks is a fundamental issue in the characterization of complex biological processes. Analysis of promoters for organizational features provides a crucial link between the static nucleotide sequence of the genome and the dynamic aspects of gene regulation and expression.
Genes expressed in the same tissue under similar conditions can share a common organization of regulatory binding elements that represent a “footprint” of the transcriptional regulatory mechanisms at work in a specific biologic context. Pilpel et al. (18) used microarray data and bioinformatics approaches to detect functional combinations of promoter elements in Saccharomyces cerevisiae that serve to control gene expression during the cell cycle, sporulation, and the response to stress. Beer and Tavazoie (19) used a combinatorial approach for characterizing regulatory DNA elements in yeast that could be used to predict gene expression patterns. Studies in higher organisms have also used computational searches to identify TFBSs combinations in phylogenetically conserved sequences controlling cell cycle-dependent transcription of G2/M genes (20). A slightly different approach was used by Dohr et al. (6) to define potential regulatory networks by in silico promoter analysis by first finding potentially coregulated subgroups without a priori knowledge. Pairs of TFBSs conserved in orthologous genes as well as in promoter sequences of coregulated genes were used to represent potential coregulation. The approach was applied to a maturity-onset diabetes of the young (MODY)-associated gene list, which yielded models that functionally connected interacting genes within MODY-related insulin/glucose signaling pathways.
The approach detailed here expands upon these observations to specifically address the hypothesis of shared transcriptional regulation of genes with common biological function (2, 5). The approach uses a priori knowledge of specific tissue microenvironments, here molecules functionally interacting in the slit diaphragm or involved in podocyte biology. Evolutionarily conserved promoter features linked the nephrin promoter to the promoter of ZO-1, both slit membrane-associated proteins. The demonstration of coregulation of these genes was used to verify the promoter model. Coregulation is a specific event tied to a common mechanism, whereas coexpression can occur by unrelated events. The experimental model was then used to search for novel interacting partners.
A group of five additional genes was identified by using this approach. Phylogenetic conservation of TFBS frameworks was the only criterion used for the selection of these promoters from the set of 79 promoters initially found in the human promoter database. The five respective candidate genes can each be functionally linked to podocytes: semaphorin 3A is expressed in podocytes (21) and is part of the VEGF system, which is important for nephrin signaling (22); Rab5C is a member of the RAS superfamily whose synaptic member Rab3A shares with nephrin functional expression between neurons and podocytes (23); ARHGDIB, the Rho GDP dissociation inhibitor β, controls the cytoskeletal organization essential for podocyte biology and slit diaphragm function (8, 24); and jun-B is a member of the AP1-family of TFs shown to be essential elements in nephrin signaling (25). Although the podocyte association described above was an independent observation for the genes grouped into this short list, a “hit” in the database could be considered “verified” only if the candidate gene showed coregulation with the “seed” gene and could be placed into the correct functional context by independent evidence. On the basis of a series of criteria, cadherin-5 was selected for more detailed study.
Cadherin-5, also known as vascular endothelial cadherin (VE-cadherin), is a unique member of the cadherin protein family and shows predominant (but not exclusive) endothelial expression (26–29). Importantly, cadherin-5 has not been previously described in podocytes. The biology of cadherin-5 shares functional aspects with nephrin and ZO-1: Nephrin and cadherin-5 are associated with zipper-like cell–cell contacts, and both genes are involved in regulation of the permeability of intercellular barriers (8, 30). ZO-1, nephrin, and several different cadherins (P-cadherin and FAT1) are known to participate in a multiprotein complex at the slit diaphragm in podocytes (31). The bioinformatics methodology detailed here correctly identified cadherin-5 as a slit diaphragm-associated molecule and effectively links the coexpression and coregulation of nephrin, ZO-1, and cadherin-5 (summarized in Fig. 4D).
The finding that nephrin, ZO-1, and cadherin-5 all belong to the slit diaphragm, a protein complex composed of approximately eight known gene products (see Table 1), shows that the model was able to identify ≈30% of known slit diaphragm-associated genes. The probability of finding by chance two additional slit diaphragm-associated molecules (ZO-1 and cadherin-5) in addition to the seed gene (nephrin) was tested by a standardized difference score (Z score; genes under a hypergeometric distribution). When the result [two slit diaphragm-associated genes of seven known slit diaphragm-associated genes (class A genes from Table 1, but excluding nephrin)] was compared with the 79 original matches of the model in 50,145 human promoters (or 0.15%), a Z score of 19.0 was calculated, demonstrating that the finding is significant (a Z score of >2.0 indicates significance).
Although the identification of the promoter structure described here strongly suggests an active role for this framework in podocyte-directed expression, a recent study has elegantly verified the specific biological function of the nephrin promoter sequence where the framework was identified. Guo et al. (32) demonstrated that a partially conserved “enhancer” region identified in the murine nephrin gene could direct podocyte-specific expression of transgenes. The respective 186-bp enhancer region identified overlaps significantly with the nephrin/ZO-1/cadherin-5 promoter model identified in the present study. Although the study by Guo et al. specifically addresses the functionality of a WT1 binding site (visually identified by this group and not part of the framework identified here), the nephrin/ZO-1/cadherin-5 promoter model appears to describe an additional aspect of gene coregulation relevant for a different “context” of podocyte/slit diaphragm biology. The region appears to be of general importance for podocyte gene expression.
The apparent transcriptional coregulation of three slit diaphragm-associated genes with a common promoter model is consistent with the central role of these genes in the maintenance of a unique biological activity. The stringent coregulation of these genes is impressive because transcriptional control is only one mechanism linked to gene expression. The promoter model identified here most likely represents the endpoint of one set of regulatory pathways at work in normal and diseased podocyte biology, which can now be further elucidated starting from the TFs identified in this study. These pathways represent unique targets for the development and analysis of therapeutic agents for an intractable set of human diseases.
Materials and Methods
Computational Promoter Analysis.
Promoter regions were identified by using the software tool eldorado. The proximal promoter regions used were generally defined as 500 nt upstream of and 100 nt downstream from the transcription start site (TSS). TSSs were automatically assigned to genes on the basis of 5′ cap site databases integrated into promoter identification programs (eldorado). If no phylogenetically conserved elements could be defined, the area was enlarged by an additional 500 nt upstream of the TSS. If again no conserved elements were identified in the sequence, it was excluded from further analysis. The sequences were retrieved from three species if available (H. sapiens, M. musculus, and R. norvegicus) and compared by alignment (dialign; Genomatix) and by definition of a similar pattern (framework) of TFBSs (frameworker) (11). Position weight matrices were used to represent the TFBSs using default parameters (33). A total of 356 matrices from 138 families (MATRIX FAMILY LIBRARY, Version 4.1; Genomatix) were used for the analysis. To define specificity and selectivity of the respective promoter sequences, the orthologous promoters were searched for two or more common TFBSs conserved in space and orientation. If found, the respective patterns of common TFBSs were tested on promoter libraries compiled from all three species (total of 154,389 sequences, i.e., 50,145 promoter regions for human, 70,868 for mouse, and 33,376 for rat; Genomatix Promoter Database) using the program modelinspector [gems launcher, Genomatix (34)]. Selectivity was taken as sufficient if the framework was detected in <15 promoter regions of the total of 154,389 sequences (<0.01%). If the input sequences allowed no framework of this selectivity, the corresponding genomic regions were analyzed for the presence of 5′ UTRs or 5′ exons missing from annotation to predict the correct TSS, and these sequences were tested as above.
A framework is defined as a set of two or more TFBSs with a specific order, strand orientation, and distance range between the individual TFBSs. To detect common frameworks in more than one seed gene of podocyte-related molecules, the following approach was used: the common tf function was selected to reduce the number of TFBSs to be considered for automatic frameworker analysis. This step was performed because initial analyses using the whole matrix library produced too many potential frameworks for subsequent study. fastm was applied to optimize the models produced by frameworker from the matrices preselected by common tf. modelinspector was then used to identify models that matched additional promoters in the podocyte seeds set (frameworker, common tf, fastm, and modelinspector are all software tools in the gems launcher).
Gene Expression Analysis on Human Renal Biopsies.
For array analysis of microdissected glomeruli, total RNA was isolated from the microdissected glomeruli (average of eight glomeruli). RNA quality and quantity were controlled by microfluid electrophoresis using the RNA 6000 Nano LabChip kit on a 2100 bioanalyzer (Agilent Technologies, Waldbronn, Germany), yielding ≈15–30 ng of total RNA. Total RNA was reverse-transcribed and quantitative RT-PCR was performed by using the European Renal cDNA Bank protocol described in ref. 35. The real-time PCR probes used and the fragmentation, hybridization, staining, and imaging analysis performed for the Affymetrix DNA chip expression analysis are described in Supporting Text, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank S. Irrgang, K. Frach, I. Edenhofer, and S. Kaden for excellent technical assistance; B. Luckow for his support; and D. Schlondorff for his suggestion, support, and critical reading of the manuscript. We thank all members of the European Renal cDNA Bank (ERCB) and their patients for their support and cooperation (for details, see Supporting Text). This work was supported by the State of Bavaria “Bayerischer Habilitationsförderpreis” and Friedrich-Baur-Stiftung (C.D.C.), Deutsche Forschungsgemeinschaft Grants SFB 571 (to P.J.N.) and Research Group 406 (to H.-J.G.), European Union Fifth Framework Programme Grant QLG1-CT-2002-01215 (“Chronic renal disease”) (to M.K., H.-J.G., and K.-P.K.), Nationales Genomforschungsnetz-2, and the Else-Kröner-Fresenius Foundation (M.K.). This work was also supported, in part, by the Bioinformatics for the Functional Analysis of Mammalian Genomes Ring funding project of the Federal Ministry for Education and Research of Germany (Grant 031U112B/031U212B, “Analysis of regulatory regions”).
Abbreviations
- TF
transcription factor
- TFBS
TF binding site
- ZO-1
zonula occludens-1
- TSS
transcription start site.
Footnotes
Conflict of interest statement: A.K. and T.W. are employed by Genomatix GmbH, a bioinformatics company that has developed tools for the comparative genomic analysis described here. As such, they may gain or lose financially through publication of this paper.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Qiu P. Biochem. Biophys. Res. Commun. 2003;309:495–501. doi: 10.1016/j.bbrc.2003.08.052. [DOI] [PubMed] [Google Scholar]
- 2.Werner T., Fessele S., Maier H., Nelson P. J. FASEB J. 2003;17:1228–1237. doi: 10.1096/fj.02-0955rev. [DOI] [PubMed] [Google Scholar]
- 3.Qiu P., Qin L., Sorrentino R. P., Greene J. R., Wang L., Partridge N. C. J. Mol. Biol. 2003;326:1327–1336. doi: 10.1016/s0022-2836(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 4.Halfon M. S., Grad Y., Church G. M., Michelson A. M. Genome Res. 2002;12:1019–1028. doi: 10.1101/gr.228902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fessele S., Maier H., Zischek C., Nelson P. J., Werner T. Trends Genet. 2002;18:60–63. doi: 10.1016/s0168-9525(02)02591-x. [DOI] [PubMed] [Google Scholar]
- 6.Dohr S., Klingenhoff A., Maier H., Hrabe de Angelis M., Werner T., Schneider R. Nucleic Acids Res. 2005;33:864–872. doi: 10.1093/nar/gki230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu R., McEachin R. C., States D. J. Genome Res. 2003;13:654–661. doi: 10.1101/gr.911803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavenstadt H., Kriz W., Kretzler M. Physiol. Rev. 2003;83:253–307. doi: 10.1152/physrev.00020.2002. [DOI] [PubMed] [Google Scholar]
- 9.Beltcheva O., Kontusaari S., Fetissov S., Putaala H., Kilpeläinen P., Hökfelt T., Tryggvason K. J. Am. Soc. Nephrol. 2003;14:352–358. doi: 10.1097/01.asn.0000043081.65110.c4. [DOI] [PubMed] [Google Scholar]
- 10.Boute N., Gribouval O., Roselli S., Benessy F., Lee H., Fuchshuber A., Dahan K., Gubler M. C., Niaudet P., Antignac C. Nat. Genet. 2000;24:349–354. doi: 10.1038/74166. [DOI] [PubMed] [Google Scholar]
- 11.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 12.Kestila M., Lenkkeri U., Mannikko M., Lamerdin J., McCready P., Putaala H., Ruotsalainen V., Morita T., Nissinen M., Herva R., et al. Mol. Cell. 1998;1:575–582. doi: 10.1016/s1097-2765(00)80057-x. [DOI] [PubMed] [Google Scholar]
- 13.Moeller M. J., Kovari I. A., Holzman L. B. J. Am. Soc. Nephrol. 2000;11:2306–2314. doi: 10.1681/ASN.V11122306. [DOI] [PubMed] [Google Scholar]
- 14.Wong M. A., Cui S., Quaggin S. E. Am. J. Physiol. 2000;279:F1027–F1032. doi: 10.1152/ajprenal.2000.279.6.F1027. [DOI] [PubMed] [Google Scholar]
- 15.Schnabel E., Anderson J. M., Farquhar M. G. J. Cell Biol. 1990;111:1255–1263. doi: 10.1083/jcb.111.3.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baelde H. J., Eikmans M., Doran P. P., Lappin D. W., de Heer E., Bruijn J. A. Am. J. Kidney Dis. 2004;43:636–650. doi: 10.1053/j.ajkd.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 17.Toyoda M., Suzuki D., Umezono T., Uehara G., Maruyama M., Honma M., Sakai T., Sakai H. Nephrol. Dial. Transplant. 2004;19:380–385. doi: 10.1093/ndt/gfg545. [DOI] [PubMed] [Google Scholar]
- 18.Pilpel Y., Sudarsanam P., Church G. M. Nat. Genet. 2001;29:153–159. doi: 10.1038/ng724. [DOI] [PubMed] [Google Scholar]
- 19.Beer M. A., Tavazoie S. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- 20.Garten Y., Kaplan S., Pilpel Y. Nucleic Acids Res. 2005;33:605–615. doi: 10.1093/nar/gki166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villegas G., Tufro A. Mech. Dev. 2002;119(Suppl. 1):S149–S153. doi: 10.1016/s0925-4773(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 22.Foster R. R., Saleem M. A., Mathieson P. W., Bates D. O., Harper S. J. Am. J. Physiol. 2005;288:F48–F57. doi: 10.1152/ajprenal.00146.2004. [DOI] [PubMed] [Google Scholar]
- 23.Rastaldi M. P., Armelloni S., Berra S., Li M., Pesaresi M., Poczewski H., Langer B., Kerjaschki D., Henger A., Blattner S. M., et al. Am. J. Pathol. 2003;163:889–899. doi: 10.1016/S0002-9440(10)63449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X. L., Kilpelainen P., Hellman U., Sun Y., Wartiovaara J., Morgunova E., Pikkarainen T., Yan K., Jonsson A. P., Tryggvason K. FEBS J. 2005;272:228–243. doi: 10.1111/j.1432-1033.2004.04408.x. [DOI] [PubMed] [Google Scholar]
- 25.Huber T. B., Kottgen M., Schilling B., Walz G., Benzing T. J. Biol. Chem. 2001;276:41543–41546. doi: 10.1074/jbc.C100452200. [DOI] [PubMed] [Google Scholar]
- 26.Vincent P. A., Xiao K., Buckley K. M., Kowalczyk A. P. Am. J. Physiol. 2004;286:C987–C997. doi: 10.1152/ajpcell.00522.2003. [DOI] [PubMed] [Google Scholar]
- 27.Hendrix M. J., Seftor E. A., Meltzer P. S., Gardner L. M., Hess A. R., Kirschmann D. A., Schatteman G. C., Seftor R. E. Proc. Natl. Acad. Sci. USA. 2001;98:8018–8023. doi: 10.1073/pnas.131209798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y., Damsky C. H., Fisher S. J. J. Clin. Invest. 1997;99:2152–2164. doi: 10.1172/JCI119388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim I., Yilmaz O. H., Morrison S. J. Blood. 2005;106:903–905. doi: 10.1182/blood-2004-12-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corada M., Liao F., Lindgren M., Lampugnani M. G., Breviario F., Frank R., Muller W. A., Hicklin D. J., Bohlen P., Dejana E. Blood. 2001;97:1679–1684. doi: 10.1182/blood.v97.6.1679. [DOI] [PubMed] [Google Scholar]
- 31.Lehtonen S., Lehtonen E., Kudlicka K., Holthofer H., Farquhar M. G. Am. J. Pathol. 2004;165:923–936. doi: 10.1016/S0002-9440(10)63354-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo G., Morrison D. J., Licht J. D., Quaggin S. E. J. Am. Soc. Nephrol. 2004;15:2851–2856. doi: 10.1097/01.ASN.0000143474.91362.C4. [DOI] [PubMed] [Google Scholar]
- 33.Quandt K., Frech K., Karas H., Wingender E., Werner T. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frech K., Danescu-Mayer J., Werner T. J. Mol. Biol. 1997;270:674–687. doi: 10.1006/jmbi.1997.1140. [DOI] [PubMed] [Google Scholar]
- 35.Cohen C. D., Frach K., Schlondorff D., Kretzler M. Kidney Int. 2002;61:133–140. doi: 10.1046/j.1523-1755.2002.00113.x. [DOI] [PubMed] [Google Scholar]
- 36.Delarue F., Virone A., Hagege J., Lacave R., Peraldi M. N., Adida C., Rondeau E., Feunteun J., Sraer J. D. Kidney Int. 1991;40:906–912. doi: 10.1038/ki.1991.292. [DOI] [PubMed] [Google Scholar]
- 37.Saleem M. A., O’Hare M. J., Reiser J., Coward R. J., Inward C. D., Farren T., Xing C. Y., Ni L., Mathieson P. W., Mundel P. J. Am. Soc. Nephrol. 2002;13:630–638. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- 38.Benzing T. J. Am. Soc. Nephrol. 2004;15:1382–1391. doi: 10.1097/01.asn.0000130167.30769.55. [DOI] [PubMed] [Google Scholar]
- 39.Ihalmo P., Palmen T., Ahola H., Valtonen E., Holthofer H. Biochem. Biophys. Res. Commun. 2003;300:364–370. doi: 10.1016/s0006-291x(02)02854-1. [DOI] [PubMed] [Google Scholar]
- 40.Gerke P., Sellin L., Kretz O., Petraschka D., Zentgraf H., Benzing T., Walz G. J. Am. Soc. Nephrol. 2005;16:1693–1702. doi: 10.1681/ASN.2004060439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.