Abstract
AlgE1, AlgE5 and AlgE6 are members of a family of mannuronan C-5 epimerases encoded by the bacterium Azotobacter vinelandii, and are active in the biosynthesis of alginate, where they catalyse the post-polymerization conversion of β-D-mannuronic acid (M) residues into α-L-guluronic acid residues (G). All enzymes show preference for introducing G-residues neighbouring a pre-existing G. They also have the capacity to convert single M residues flanked by G, thus ‘condensing’ G-blocks to form almost homopolymeric guluronan. Analysis of the length and distribution of G-blocks based on specific enzyme degradation combined with size-exclusion chromatography, electrospray ionization MS, HPAEC–PAD (high-performance anion-exchange chromatography and pulsed amperometric detection), MALDI (matrix-assisted laser-desorption ionization)-MS and NMR revealed large differences in block length and distribution generated by AlgE1 and AlgE6, probably reflecting their different degree of processivity. When acting on polyMG as substrates, AlgE1 initially forms only long homopolymeric G-blocks >50, while AlgE6 gives shorter blocks with a broader block size distribution. Analyses of the AlgE1 and AlgE6 subsite specificities by the same methodology showed that a mannuronan octamer and heptamer respectively were the minimum substrate chain lengths needed to accommodate enzyme activities. The fourth M residue from the non-reducing end is epimerized first by both enzymes. When acting on MG-oligomers, AlgE1 needed a decamer while AlgE6 an octamer to accommodate activity. By performing FIA (flow injection analysis)-MS on the lyase digests of epimerized and standard MG-oligomers, the M residue in position 5 from the non-reducing end was preferentially attacked by both enzymes, creating an MGMGGG-sequence (underlined and boldface indicate the epimerized residue).
Keywords: alginate epimerization, G-block formation, mode of action, mannuronan C-5 epimerase, processivity
Abbreviations: DP, degree of polymerization; ESI-MS, electrospray ionization MS; FIA, flow injection analysis; HPAEC–PAD, high-performance anion-exchange chromatography and pulsed amperometric detection; MALDI, matrix-assisted laser-desorption ionization; SEC, size-exclusion chromatography; TOF, time-of-flight
INTRODUCTION
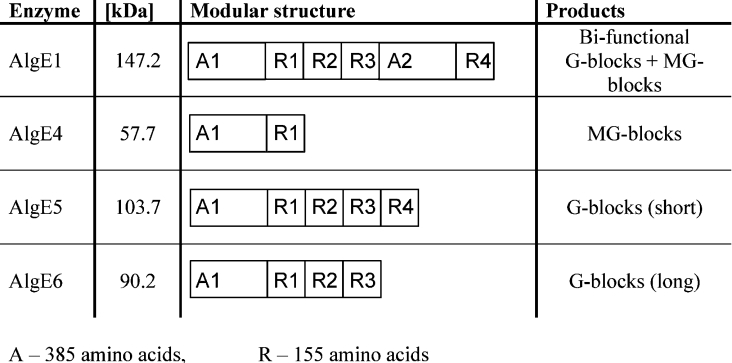
Alginate is a collective term for a family of polysaccharides produced by brown algae [1] and bacteria [2,3]. Chemically, they are linear binary co-polymers of (1→4)-linked β-D-mannuronic acid (M) and α-L-guluronic acid (G) arranged in a block-wise pattern along the chain with homopolymeric regions of M (M-blocks) and G (G-blocks) residues interspersed with regions of alternating structure (MG-blocks). In its biosynthetic pathway, alginate is produced via a post-polymerization reaction involving a C-5 inversion on the M residues of mannuronan. This reaction is catalysed by the mannuronan C-5 epimerases. Recently, it has been found that the genome of the alginate-producing bacterium Azotobacter vinelandii encodes at least seven different mannuronan C-5 epimerase genes. These genes have been sequenced, cloned and expressed in Escherichia coli; the enzymes thus produced have been designated AlgE1–AlgE7 [4–6]. Since all natural alginates are produced from homopolymeric mannuronan by the same basic C-5 inversion from M to G, the remarkable variability in composition and sequence found in the polysaccharide is solely due to the different catalytic properties of the various epimerases. For example, the AlgE4 epimerase forms alginates with long strictly alternating sequences, while AlgE1, AlgE5 and AlgE6 are typical G-block-forming enzymes [6–11]. Structurally these isoenzymes are composed of varying numbers of two modules (Figure 1), the A module (385 amino acids) and R module (155 amino acids). The A modules account for the catalytic activity [12], while the R modules probably modulate the affinity for the substrates [13]. The epimerization reaction is Ca2+-dependent and both the A and R modules bind this cation [12]. AlgE1 comprises two A modules and four R modules (A1R1R2R3A2R4) and has been demonstrated to be a bifunctional enzyme [10]. This epimerase can be divided into two catalytically active parts, where the truncated protein A1R1R2R3 (AlgE1-1) is a G-block-forming enzyme, while A2R4 (AlgE1-2) introduces only single G residues in a similar manner to AlgE4 [10].
Figure 1. Molecular masses (kDa), modular protein structures and product specificities of four of the seven AlgE epimerases of A. vinelandii.
Any enzyme that has more than one substrate-binding subsite, and performs multiple modifications on a substrate, may display processivity. Enzyme processivity refers to the average number of times a reaction is repeated between association and dissociation of an enzyme–substrate complex. Several examples of this type of action have been confirmed for polysaccharides, mainly for exolytic enzymes [14–16]. In alginates, the formation of long blocks (MG or GG) by the mannuronan C-5 epimerases can thus be explained by a high degree of processivity. However, these nonrandom patterns can also result from a preferred attack mechanism where the affinity for the substrates depends on pre-existing G residues. Previously, we have shown that AlgE4 acts processively by sliding along the substrate chain epimerizing every second residue [17]. An average of ten uronic acid residues is epimerized for each enzyme–substrate encounter [17]. By analysing the AlgE4 subsite specificity, a hexameric oligomer was identified as the minimum substrate chain length needed to accommodate enzyme activity, where the third M residue from the non-reducing end is epimerized first. The aim of the present study is to investigate the mode of action of the G-block-forming epimerases AlgE1, AlgE5 and AlgE6 by analysing their substrate requirements and their epimerization patterns. Using specific degrading enzymes, SEC (size-exclusion chromatography), NMR spectroscopy, ESI-MS (electrospray ionization MS), FIA (flow injection analysis)-MS and MALDI (matrix-assisted laser-desorption ionization)-MS, together with HPAEC–PAD (high-performance anion-exchange chromatography and pulsed amperometric detection), the absolute lengths of G-blocks for low conversion of polyMG by AlgE1 and AlgE6 are determined. The subsite specificities of these enzymes are examined when acting on mannuronan and polyMG.
MATERIALS AND METHODS
Alginates
High-molecular-mass mannuronan was isolated from the fermentation broth of an epimerase-negative strain [18] of Pseudomonas fluorescens. Purification and deacetylation were carried out as described previously [19]. No guluronate signals could be detected by 1H-NMR (molar fraction of guluronic acid FG<0.001), indicating a homopolymeric mannuronan. Sodium alginate from Laminaria hyperborea leaves with FG=0.49 and FGG=0.33 was kindly provided by FMC Biopolymer (Drammen, Norway) and was used in epimerization reactions as a G-block-containing substrate. Alginates with strictly alternating structures FG=0.47 and FGG=0.0 were prepared by incubating mannuronan with recombinant AlgE4 for 23 h (S. Holtan, Q. Zhang, W. I. Strand and G. Skjåk-Bræk, unpublished work).
A 13C-1-labelled mannuronan was produced by growing the mannuronan-producing Ps. fluorescens strain on agar plates with D[1-13C]fructose (99%) as carbon source [9]. The mannuronan product was selectively enriched to 59% 13C in the C-1 position [9]. The 13C-1-enriched mannuronan was treated with the epimerase AlgE4 as described above to gain 13C-1-enriched polyalternating MG-alginate with FG=0.47 and FGG=0.0 (S. Holtan, Q. Zhang, W. I. Strand and G. Skjåk-Bræk, unpublished work).
Solid, purified oligomers of mannuronic acid with DP (degree of polymerization) from 2 to 16 were obtained by degrading the high-molecular-mass mannuronan by partial acid hydrolysis at 95 °C (3.5 h at pH 5.6 and 6 h at pH 3.5). After hydrolysis, the sample was neutralized (pH 7), and freeze-dried. The oligomannuronic acid hydrolysis mixture was fractionated according to its chain length on three columns of Superdex 30 preparative grade (HiLoad; 2.6 cm×60 cm, serially connected; Amersham Biosciences) at a flow rate of 0.8 ml/min with 0.1 M ammonium acetate (pH 6.9) at room temperature (20 °C) as described previously [21] (S. Holtan, Q. Zhang, W. I. Strand and G. Skjåk-Bræk, unpublished work). DP and purity of fractions were determined by ESI-MS and 1H-NMR spectroscopy. The same procedure was used to produce MG-oligomers using polyMG as starting material and shorter hydrolysis time (S. Holtan, Q. Zhang, W. I. Strand and G. Skjåk-Bræk, unpublished work). Since the GM glycosidic linkage is more susceptible to acidic hydrolysis than the MG linkage, the even numbered oligomers will predominantly have the same sequential structure with an M residue on the nonreducing end and a G residue on the reducing end, i.e. the hexamer MGMGMG (S. Holtan, Q. Zhang, W. I. Strand and G. Skjåk-Bræk, unpublished work).
Enzymes
The mannuronan C-5 epimerases AlgE1, AlgE5 and AlgE6 with molecular masses of 147.2, 103.7 and 90.2 kDa respectively were produced by fermentation of recombinant E. coli strains: AlgE1 and AlgE5 in JM 109 [10,11,22], and AlgE6 in SURE [6]. The epimerases were partially purified by ion-exchange chromatography on Q-Sepharose FF (Amersham Biosciences) and desalted by diafiltration in a Pellicon Cassette System (Millipore). The activities of the enzymes were assayed by measuring the release of 3H to water, when 3H-5-labelled mannuronan was incubated with the respective enzymes [19]. The two truncated derivatives of AlgE1; the N-terminal (AlgE1-1, 89 kDa) and the C-terminal (AlgE1-2, 59 kDa) parts were expressed separately in E. coli [10] and kindly provided by Dr Helga Ertesvåg (Department of Biotechnology, NTNU, Trondheim, Norway). Alginate α-L-guluronic acid endolyase (G-lyase) was isolated from Klebsiella pneumoniae as described in [23]. Haliotis tuberculata alginate lyase, a specific mannuronate β-eliminase [24], was isolated from abalones as described in [25]; it was kindly provided by Håvard Sletta (SINTEF Applied Chemistry, Trondheim, Norway). The activity and specificity of the two alginate lyases were measured by monitoring the increase in absorbance (A) at 230 nm and 25 °C [26] for three different substrates: mannuronan (FG=0.0), polyMG (FG=0.47) and polyG (FG=0.95) when treated with the lyases, and by 1H-NMR spectroscopy and ESI-MS of the degraded alginate samples. One unit of lyase activity was defined as described by Ertesvåg et al. [27].
C-5 epimerization
Kinetic experiments
Alginates were dissolved in MQ-water [deionized water purified with the MilliQ system from Millipore (Bedford, MA, U.S.A.)] overnight before a concentrated stock solution of Mops buffer (pH 6.9) with CaCl2·2H2O and NaCl was added and the mixtures were preheated at 37 °C. The respective enzymes were dissolved in MQ-water and immediately added to the alginate solutions. Final concentrations of the reaction mixtures were 0.25% (w/v) alginate, 50 mM Mops, 0.8 mM (AlgE1), 2 mM (AlgE6) or 3 mM (AlgE5) CaCl2, and 20 mM NaCl. Molar ratios of enzyme: uronic acid residues were 1:152000 for AlgE1, 1:27000 for AlgE5 and 1:9500 for AlgE6. The mixtures were kept at 37 °C for 0.5, 1, 2, 4, 8/10 and 23–25 h before the reactions were terminated on ice by addition of HCl until the pH was below 3. After epimerization, the samples were purified by dialysis against 0.05 M HCl and finally against MQ-water until the conductance was below 4 μS at 4 °C. The pH was then adjusted to 6.8, and the alginates were freeze-dried.
PolyMG was epimerized with AlgE1 up to 7 days in an identical manner with that described above, except for a final concentration of 100 mM NaCl and with the addition of extra enzyme (equal amount to the starting amount) every day until day 5 after reaction start. Chloroform (0.12%, v/v) was added to prevent bacterial growth during the reaction.
Large batches of polyMG were modestly epimerized with AlgE1 or AlgE6 to FG=0.55 according to the procedure described above, except for a final concentration of 75 mM NaCl in these reactions.
Oligomers
Solutions (0.3%, w/v) of M- (DP6–DP11) and MG- (DP6, DP8, DP10, DP12 and DP14) oligomers were incubated with AlgE1 or AlgE6 in 50 mM ammonium acetate buffer (pH 6.9) with 1 mM CaCl2 (AlgE1) or 2 mM CaCl2 (AlgE6). Molar ratios of enzyme/uronic acid residues were 1:9100 for AlgE1 and 1:1900 for AlgE6. The solutions were left at 37 °C for 24 h. The reactions were stopped by heating the samples at 95 °C for 5 min. The denatured enzymes were removed by centrifugation and the supernatants were freeze-dried.
Lyase degradation
Alginate samples were treated with G-lyase from K. pneumoniae (7.5×10−3 unit/mg of alginate for standard and epimerized Moligomers with low degree of conversion) or H. tuberculata M-lyase (0.33 unit/mg of alginate for native and epimerized polymers, 0.2 unit/mg of alginate for standard and epimerized MG-oligomers and 8.0×10−3 unit/mg of alginate for epimerized M-oligomers with FG>0.12) in 100 mM ammonium acetate buffer (pH 7.1) for 1.5 h at 30 °C. The enzymes were then inactivated by heating the samples in a water bath at 95 °C for 5 min. The degraded samples were freeze-dried three times to remove acetate.
SEC
The lyase digests of native and epimerized polymers were fractionated according to their chain lengths on three columns of Superdex 30 preparative grade (HiLoad; 2.6 cm×60 cm, serially connected; Amersham Biosciences) at a flow rate of 0.8 ml/min with 0.1 M ammonium acetate (pH 6.9) at room temperature as described in our unpublished work (S. Holtan, Q. Zhang, W. I. Strand and G. Skjåk-Bræk, unpublished work). DP and composition (sequence) of fractions were determined by ESI-MS, MALDI-MS or HPAEC–PAD and 1H-NMR spectroscopy.
1H-NMR spectroscopy
1H-NMR spectra were recorded on a Bruker Avance DPX 300, 400 or DMX 500 spectrometer in 2H2O at 90 °C. To reduce the viscosity of the high-molecular-mass polymers for NMR analysis, the samples were degraded by mild acid hydrolysis [19] to a final average DPn of approx. 35. The acid-degraded alginate samples were neutralized and freeze-dried; 5 mm sample tubes were used and 3-(trimethylsilyl)-propionic-2,2,3,3-d4 acid sodium salt (Aldrich, Milwaukee, WI, U.S.A.) was used as internal standard for the chemical shift. For sample concentrations of 10 mg/ml in 2H2O, 20 μl of 0.3 M triethylenetetra-amine hexa-acetate (Sigma) was added to chelate any remaining Ca2+ from the epimerization. The peaks were assigned according to Grasdalen [28], Heyraud et al. [29] and our unpublished work (S. Holtan, Q. Zhang, W. I. Strand and G. Skjåk-Bræk, unpublished work).
13C-NMR monitoring of epimerization reaction
Individual solutions in 2H2O of 13C-1-enriched polyMG (average DPn of approx. 30) CaCl2, NaCl, Mops (Sigma; p2H 7 at 40 °C) and epimerase were prepared separately and calculated volumes transferred into a 5 mm (Norell) NMR sample tube. Before addition of epimerase, a spectrum of the starting material was recorded. The final concentrations in the NMR sample (500 μl total volume) were: 20 mg/ml 13C-1-polyMG, 5 mg/ml AlgE6 (as total protein), 20 mM Mops, 5 mM CaCl2 and 20 mM NaCl. To monitor the progress of the epimerase experiment, series of 65 successive 13C-NMR spectra were recorded on a Bruker DPX 400 spectrometer (100 MHz) at 40 °C. The procedure was automated by using the multi-zg command. Spectra were obtained using a 30° pulse, a spectral width of 20080 Hz and a data-block size of 32k; 400 scans were accumulated after four dummy scans. The resulting time interval between two successive spectra was 15.8 min and total recording time was 17.1 h.
HPAEC–PAD
Samples and a hydrolysate of G-block seaweed alginate (FG=0.96) [30] of 1 mg/ml in MQ-water were analysed on a Dionex BioLC system (Dionex, Sunnyvale, CA, U.S.A.) coupled with a Dionex AS50 autosampler. The HPLC system was equipped with an IonPac AS4A (4 mm×250 mm) anion-exchange column connected to an IonPac AG4A (4 mm×50 mm) guard-column. Chromatography was performed at room temperature and at a flow rate of 1 ml/min. Buffer A was 0.10 M NaOH, prepared from a carbonate-free 50% (w/w) NaOH solution, and buffer B was 1 M ammonium acetate in 0.10 M NaOH (buffer A). Linear gradients of acetate were produced to elute the samples by increasing the concentration of buffer B from 0 to 80% over 80 min. Column effluent was monitored with a pulsed amperometric detector on an Au working electrode and Ag/AgCl reference electrode, as described previously [21]. Data acquisition and integration were performed using Dionex PeakNet software. By plotting the retention time as a function of chain length for the G-block hydrolysate [30], the G-block lengths of the samples could be determined.
ESI-MS
Samples were dissolved in MQ-water and diluted in aq. 50% methanol containing formic acid (0.5%) in positive mode and aq. 50% methanol containing ammonia (1%) in negative mode. Sample concentrations of 0.01–0.02% (w/v) were analysed by direct infusion (0.6 ml/h) into an Agilent MSD Trap SL mass spectrometer equipped with an electrospray ion source. The drying gas flow rate was 4 litres/min, the drying gas temperature was 325 °C, and the nebulizer pressure was 15 lb/in2. The capillary voltage was 3.5 kV in negative-ion mode with an endplate offset of −0.5 kV. In positive-ion mode, capillary voltages of −3.5 or −4 kV were used.
FIA-MS
FIA-MS was performed on an Agilent 1100 high-pressure liquid chromatograph connected to an Agilent TOF (time-of-flight) mass spectrometer with ESI in negative mode. Automatic calibration of the mass axis was performed with continuous infusion of an Agilent reference mixture through a second nebulizer needle in the ion source. The mobile phase consisted of 10 mM ammonium acetate (pH 9.2) (nine parts) and methanol (one part). A flow rate of 0.25 ml/min and an injection of 20 μl were used. Samples were prepared by diluting freeze-dried alginate oligomer samples in HPLC-grade water and further dilution to appropriate concentration (0.2 mg/ml) in a methanol/water mixture (1:1) with 1% ammonia. Analyst™ QS for Agilent TOF was used for data analysis. Quantitative analysis was performed by integrating a ±10 p.p.m. window for each m/z and adding −1 to −4 ions of each oligomer. Owing to overlapping m/z for unsaturated (Δ) oligomers, the isotope peaks had to be used for the quantitative analysis.
MALDI-MS
The MALDI–TOF-MS analyses were performed employing a Bruker Daltonics MALDI–TOF Reflex IV (Bremen, Germany). Ionization occurred in dry nitrogen at 337 nm at a pulse of 20 Hz. TOF was determined in linear mode with a 20 kV acceleration voltage and 17.9 kV secondary grid voltage. Both positive and negative ion spectra were determined. The number of laser shots for each spectrum was varied from 50 to 200, depending on the sample signal intensities. The MALDI probe was coated with a film by applying 1 μl of Nafion 5 wt% solution (Sigma–Aldrich, St. Louis, MO, U.S.A.) [31] and removing the solvent by evaporation. A saturated solution of 2,5-dihydroxybenzoic acid (Sigma–Aldrich) in water (32 mg/ml) was used as matrix. A 1 μl portion of the sample (10 mg/ml in MQ-water) and 1–2 μl of the matrix solution were mixed and applied to the probe, after which solvent was removed by evaporation. In some experiments, the sample was recrystallized on the probe surface to improve the signal intensity.
RESULTS AND DISCUSSION
Influence of pre-existing G residues in the substrate on the epimerization rate
We have previously suggested that the non-random distribution of G residues in alginates arises from epimerases acting according to a multiple attack mechanism [32]. Whether the enzymes slide along the polymer chain during the epimerization process (processive mode) or leave the substrate after each single productive event and then preferentially re-attack an M residue neighbouring a G (preferential attack model) is however difficult to distinguish from the NMR and polymer statistical calculations alone, since both models could account for the observed dyad and triad frequencies [8]. If the enzymes act according to a preferred attack model, one should expect an increased conversion rate when the enzymes act on an alginate with pre-existing G residues, either as single G residues in a polyMG sequence or as G-blocks.
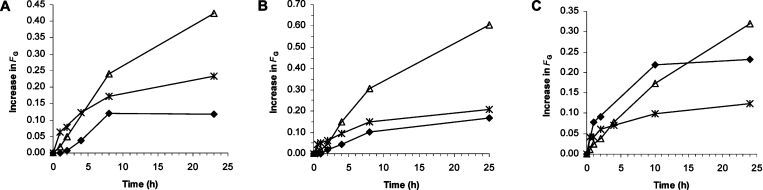
Kinetic data from epimerization of mannuronan (FM=1.0), a polyMG-alginate (FG=0.47 and FGG=0.0) and an alginate from L. hyperborea (FG=0.49 and FGG=0.33) with the epimerases AlgE1, AlgE5 and AlgE6 are shown in Figure 2. Results were obtained by integration of 1H-NMR spectra at selected time points for the epimerization experiments. All three enzymes expressed activity on all three substrates. The initial reaction rate for both AlgE1 and AlgE6 (Figures 2A and 2B) was highest on the substrate with pre-existing G-blocks, where the reaction rates follow typical hyperbolic curves. On the other two substrates mannuronan and polyMG, the initial reaction rates were lower and the reactions follow sigmoid curves, suggesting that the affinity for the substrates increases upon the increment of G-blocks. Taken together, this suggests that G-block formation is partly due to a preferred attack on a -G-G-M-M- or -G-G-M-G- sequence. Whether the enzymes propagate along the chain after the initial GGM sequence is formed, and thus are partly preferred and partly processive can however not be established from the present experiments. In Figure 2(C), kinetic data from the action of AlgE5 on the same substrates are shown. This epimerase differs from the other two by its preference for polyMG. This enzyme is highly homologous with AlgE2 [5] (amino acid sequence identity 98%), which exhibits both epimerase and a weak lyase activity [22]. The lyase activity of AlgE5 was confirmed by incubating a high-molecular-mass mannuronan with the enzyme and monitoring the decline in molecular mass by viscometry. After 48 h incubation, signals from the 4-deoxy-L-erythro-hex-4-enepyranosyl uronate residues on the non-reducing ends appeared in the 1H-NMR spectra (results not shown). This might complicate the analysis of the epimerization pattern and in the following investigation only AlgE1 and AlgE6 are examined further.
Figure 2. Increase in FG as a function of epimerization time by AlgE1 (A), AlgE6 (B) and AlgE5 (C) on various substrates.
Mannuronan (FM=1.0) (△), L. hyperborea leaf alginate (FG=0.49 and FGG=0.33) (*) and polyMG (FG=0.47 and FGG=0.0) (◆) as substrates.
Activity and time-resolved NMR
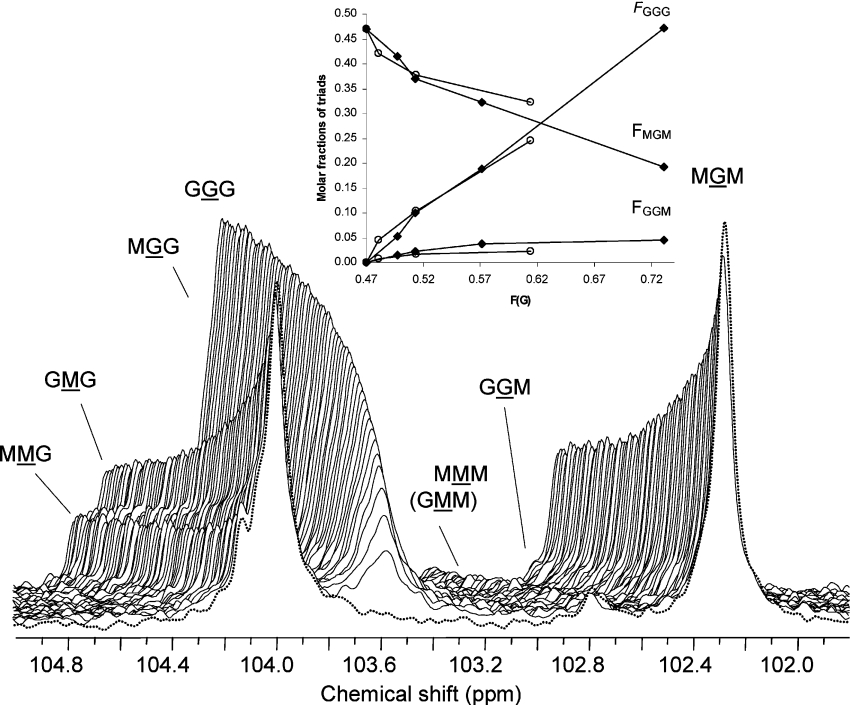
As shown in Figures 2(A) and 2(B), AlgE1 and AlgE6 act effectively on polyMG. Since this substrate contains only a minor fraction of MM (FMM=0.06), the dominating reaction is therefore the conversion of single M residues flanked by G residues. To obtain a direct and better assessment of G-block formation, we monitored the evolution of the G centred triads by 13C-NMR spectroscopy (100 MHz) using a polyMG enriched in 13C-1 as substrate for the enzyme AlgE6. Owing to the low sensitivity of 13C spectroscopy based on the naturally occurring isotope, we prepared a mannuronan enriched in 13C-1 by growing mannuronan-producing bacteria Ps. fluorescence on 13C-l-labelled D-fructose [9]. The labelled mannuronan was then converted into polyMG by in vitro epimerization with AlgE4 (FG=0.47 and FGG=0.0). In the time-resolved experiment with AlgE6 shown in Figure 3, we could monitor the change in the anomeric resonance signals both from the G centred triads GGG, GGM, (MGG) and MGM, and the M centred triads MMM, MMG, GMM and GMG. The generation of G-blocks can thus be directly visualized from the increment in the resonance signals from GGG and GGM, and the concomitant decrease in the GMG and MGM signals. In this experiment, FG increased from 0.47 to 0.77 over 17 h, after which the measurement had to be aborted. By plotting the experimental triad frequencies FGGG, FMGM and FGGM obtained from 1H- and 13C-NMR as a function of epimerization (FG) by AlgE1 and AlgE6 as shown in Figure 3 (inset), it is evident that after an initial increase, FGGM remains constant during the epimerization process for both enzymes. Since the GGM resonance arises from the end of a G-block, a constant FGGM signifies that the number of G-blocks remains constant during the epimerization, at least for low to moderate degrees of conversion. This indicates that epimerization takes place as elongation of existing G-blocks or that introduction of new blocks are compensated for by block condensation. Moreover, since the molar fraction FGGM is slightly lower in the AlgE1- than in the AlgE6-epimerized sample, the former enzyme probably generates longer G-blocks.
Figure 3. 13C-NMR monitored AlgE6 epimerization of polyMG.
A stacked plot of the anomeric region of 13C-NMR spectra (100 MHz) recorded during AlgE6 epimerization of 13C-1-enriched polyMG (FG=0.47 and FG=0.0) at 40 °C and p2H 7 of buffer solution (Mops) in 2H2O. The enzymatic reaction was carried out inside the NMR spectrometer. The final concentrations in 500 μl total volume in the NMR tube were 20 mg/ml polyMG-alginate, 5 mg/ml AlgE6 (as total protein) and 5 mM Ca2+. Molar ratio of enzyme/mannuronic acid residues susceptible to epimerization (FM=0.53) was 1:999. Spectra recorded after 19.9 min (FG=0.51) to 17.1 h reaction time (FG=0.77) with a time interval of 15.8 min between successive recordings are shown. Front spectrum (dotted line) is recorded before addition of epimerase. Inset: molar fractions of chain trisaccharide combinations versus degree of conversion (FG) for polyMG epimerized with the mannuronan C-5 epimerases AlgE1 (○) and AlgE6 (◆). Molar fractions were determined from the NMR spectra after 0, 4, 8 and 23 h reaction time for AlgE1, and after 0, 2, 4, 8 and 25 h reaction time for AlgE6. The epimerization reactions were then terminated. FGGM represents the molar fraction of G-block termination, i.e. the increase in number of G-blocks in the polymer chain during the epimerization reaction.
Analysis of the length and distribution of the G-blocks in AlgE1- and AlgE6-epimerized polyMG
The average length of G-blocks given as the average number of consecutive G residues can be analysed by NMR spectroscopy. From the molar fractions of the G-centred triad frequencies FGGG, FGGM and FMGM, the average length of G-blocks can be estimated from: NG>1=(FG−FMGM)/FGGM [33]. However, to get information about the real G-block lengths and their distribution in the polymer, we degraded the epimerized polymers with a specific alginate lyase. The digests were then fractionated by SEC and the molecular mass and composition (sequence) of each fraction were determined by means of ESI-MS and 1H-NMR spectroscopy. The DP of fractions with high molecular mass (>2200 Da) was analysed with HPAEC–PAD or MALDI-MS. The alginate lyase isolated from H. tuberculata is a mannuronate β-eliminase, specific for cleavage of M–M and G–M glycosidic linkages [24]. We examined the rate of the β-elimination reaction of this lyase as the relative increase in A230 and 25 °C on mannuronan and polyMG, and also on polyG as a negative control. As previously reported [24,26,29,34], we found that the lyase has a preference for performing a β-elimination reaction of an M residue following an M, as in mannuronan, and with approximately four times higher initial activity compared with the degradation rate of polyMG with FG=0.47. For polyG (FG=0.95), no degradation was observed after a small initial activity due to the 5% M in the polymer. When the lyase attacks a sequence of alternating M and G interspaced with homopolymeric G-blocks of various lengths, the blocks will appear as Δ-(G)n-Gred where the Δ signifies a 4-deoxy-L-erythro-hex-4-enepyranosyl uronate residue at the non-reducing end, arising in this case from an M, while the polyalternating sequences will be degraded into dimers (ΔG) and tetramers (ΔGMG) [17].
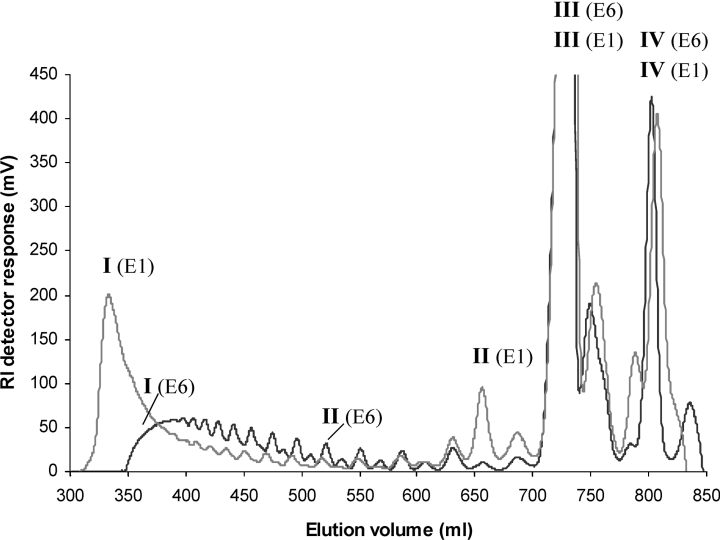
PolyMG (FG=0.47) was epimerized with AlgE1 and AlgE6 to FG=0.55, FGG=0.19, NG>1=7 and FG=0.55, FGG=0.17, NG>1=5 respectively. The polymers were degraded with the lyase and the digests were separated by SEC on Superdex 30. The chromatograms are given in Figure 4. The fractions were analysed by 1H-NMR, ESI-MS and HPAEC–PAD or MALDI-MS, and the values are given in Table 1. Both samples were heavily degraded, and the NMR analysis of the total digests and fractions showed that G residues at the reducing ends and guluronate-linked unsaturated non-reducing ends (Δ-G) were the dominating resonance signals. The most conspicuous difference between the two chromatograms of the separated digests in Figure 4 is the chain length distribution of the oligomers: the AlgE1 sample displays a very clear bimodal distribution with a void fraction [I (E1)] and two major low molecular mass peaks [III (E1) and IV (E1)]. Analysis with HPAEC–PAD and ESI-MS shows that the void comprises chain lengths of 52–58 residues, while the low-molecular-mass fractions are tetramers and dimers. From the 1H-NMR spectrum (see Figure 5A), it is evident that the high-molecular-mass (void) fraction consists of pure G-blocks (FG≥0.97), while the two major low-molecular-mass fractions are mainly composed of the tetramer Δ-G-M-G (see Figure 5B) and the dimer Δ-G. A hexamer fraction [II (E1)] and traces of larger oligomers with an odd number of uronic acid residues (5, 7, 9, 11 etc.) could also be identified. In contrast with AlgE1, the digest of the AlgE6-epimerized sample displays a broader distribution of oligomers rather than a clear-cut biomodality (see Figure 4). The major peaks in this case are also from the tetramer Δ-G-M-G [III (E6)] and the dimer Δ-G [IV (E6)] as expected degradation products from a regular polyMG sequence, but the larger oligomers are G-blocks ranging from 5 to 35 residues [I (E6)].
Figure 4. SEC of AlgE1- and AlgE6-epimerized polyMG (FG=0.55 for both enzymes) specific degraded by M-lyase from H. tuberculata.
The oligo-uronic acids were chromatographed on three columns of Superdex 30 at a flow rate of 0.8 ml/min with 0.1 M ammonium acetate at room temperature. The eluent was monitored on-line with a refractive index detector. The DP and composition (sequence) of marked peaks [I−IV (E1) and (E6)] are given in Table 1. RI, refractive index.
Table 1. DP and composition of selected fractions obtained by SEC of lyase digests of epimerized polyMG.
PolyMG (FG=0.47 and FGG=0.0) was epimerized with AlgE1 and AlgE6 to FG=0.55 for both enzymes. For the AlgE1-epimerized sample, FGG=0.19 and NG>1=7, while for the AlgE6-epimerized sample, FGG=0.17 and NG>1=5. These MG/GG- alginates were degraded by a specific M-lyase from H. tuberculata and separated by SEC on Superdex 30. Fractions with low molecular mass were characterized by ESI-MS, while those with high molecular mass by HPAEC-PAD or MALDI-MS. The composition of fractions were analysed by 1H-NMR (300 MHz) spectroscopy.
| DP | |||
|---|---|---|---|
| SEC fraction | ESI-MS | MALDI-MS/HPAEC-PAD | Composition/sequence 1H-NMR |
| I (AlgE1) | DP52Δ−DP58Δ | FG=0.97 | |
| II (AlgE1) | DP6Δ | ΔGMGMG | |
| III (AlgE1) | DP4Δ | ΔGMG | |
| IV (AlgE1) | DP2Δ | ΔG | |
| I (AlgE6) | DP27Δ−DP35Δ | FG=0.95 | |
| II (AlgE6) | DP13Δ | DP13Δ | FG=0.83 |
| III (AlgE6) | DP4Δ | ΔGMG | |
| IV (AlgE6) | DP2Δ | ΔG | |
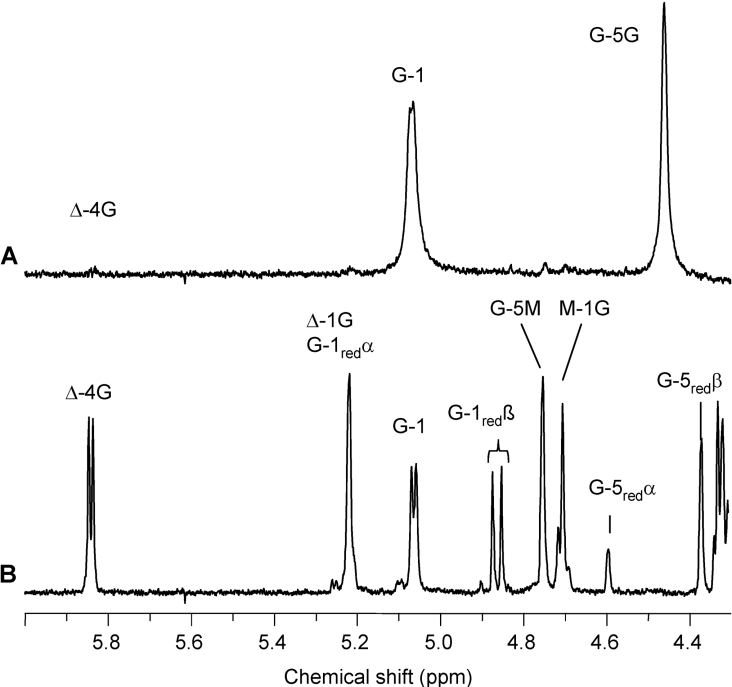
Figure 5. 1H-NMR (400 MHz) spectra of the void (A) and major low-molecular-mass fraction (B) from SEC of the lyase digest of AlgE1-epimerized polyMG (FG=0.55).
The void and the low-molecular-mass fraction are denoted I (E1) and III (E1) respectively in the SEC chromatogram in Figure 4. Δ-4G and Δ-1G signals are produced upon lyase degradation. Δ denotes a 4-deoxy-L-erythro-hex-4-enepyranosyl uronate residue. Resonance signals from G residues (G-1red, G-5redα and G-5redβ) are dominating at the reducing end (B). The void fraction in (A) has FG≥0.97. The low-molecular-mass fraction has FG=0.5 and an alternating structure, determined as Δ-G-M-G with additional ESI-MS analysis confirming the molecular mass of DP4Δ (Table 1).
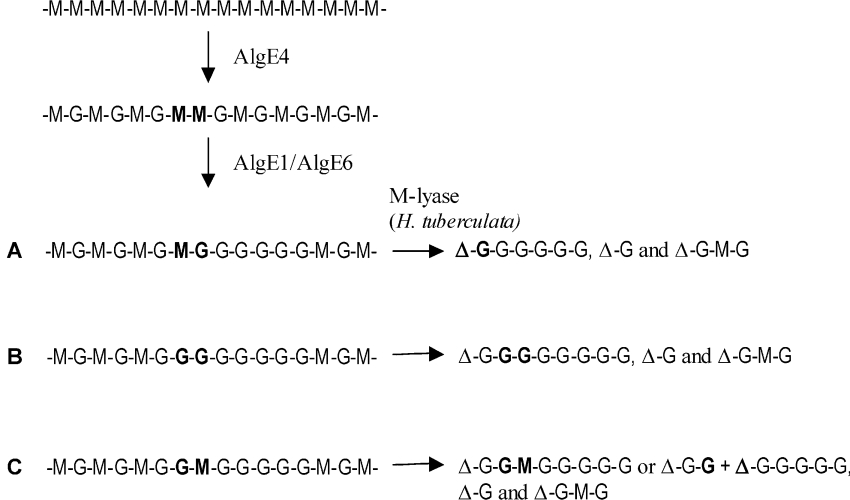
If the epimerases were attacking a strictly polyalternating sequence and propagating along the chain epimerizing M in GMG sequence, only G-blocks with an odd number of G residues would be generated. The isolation of these blocks using the G–M, M–M specific lyase would thus produce oligomers with an even number of residues due to the unsaturated residue (Δ) originating from an M on the non-reducing end of the G-blocks. In contrast, most of the oligomers with a DP>4Δ in the lyase digests of the AlgE1- and AlgE6-epimerized polyMG are composed of an odd number of uronic acid residues (5, 7, 9, 11 etc.). This is confirmed by both ESI- and MALDI-MS analyses of the prominent SEC-fractions. 1H-NMR spectroscopy shows that the odd number oligomers have ΔG and Gred at the non-reducing and reducing end respectively. To explain these results, we must take into account that the polyMG substrate is not completely homogeneous but contains some MM sequences. From the 13C-NMR spectra of 13C-1-enriched polyMG, the average M-block length of the extra 6% MM content in the polymer can be calculated to be approx. 2, resulting in average blocks of [MG] larger than 20 [17] interspersed with MM sequences (-M-G-M-M-G-M-). A statistical simulation of the length and distribution of these extra M residues in the polyMG [30] suggests an average MG-block length of approx. 38 monomer units and a poly-disperse distribution of M-blocks with [M]n=3.4, where the dimer (MM) fraction is the most abundant, constituting 41% of total M-blocks. A preferred initial attack of AlgE1 and AlgE6 on the -MM- neighbouring an alternating sequence, leading to a subsequent epimerization towards the reducing end of the polymer chains, as illustrated in Figure 6, would explain the generation of odd numbered oligomers. This mode of action (Figure 6) is also supported by the kinetic experiments shown in Figures 2(A) and 2(B), where both AlgE1 and AlgE6 prefer M-blocks to polyMG. Moreover, as the initial activity of the lyase was four times greater on mannuronan (FM=1.0) than polyMG (FG=0.47) as substrate, thus confirming a preferential cleavage of M–M to G–M glycosidic linkages, the absence of 1H-NMR resonance signals from M-1red, ΔM and M-1M in the lyase digests (spectra not shown) indicates that the MM sequences are epimerized initially.
Figure 6. Illustration of the preferential enzyme attack by AlgE1 or AlgE6 of interspersed MM-sequences in polyMG (FG=0.47) and the following degradation products produced after treatment with M-lyase from H. tuberculata.
(A, B) Most probable epimerization patterns achieving homogeneous G-blocks with an odd number of uronic acid residues (including the 4-deoxy-L-erythro-hex-4-enepyranosyl uronate residue), in addition to ΔG-M-G and Δ-G as main degradation products after lyase treatment of low epimerized polyMG-alginates. (C) Heterogeneous blocks with an odd number of residues, or shorter homogeneous G-blocks but with an even number of residues produced upon lyase degradation of epimerized material.
Size of the substrates and subsite analysis
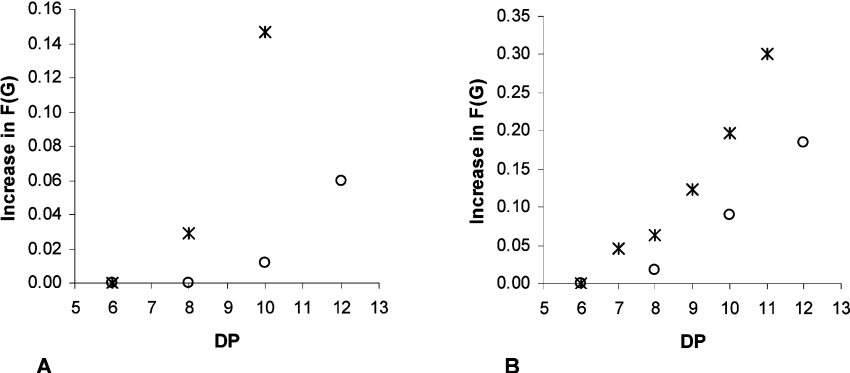
A polymer-modifying enzyme that acts according to a multiple attack mechanism, independent of whether it acts processively or in a preferred mode, would have to interact with more than one residue in the polymer substrate. We have previously demonstrated that AlgE4 needs a hexameric mannuronan oligomer as a minimum size of the substrate to support epimerase activity [17]. Oligomers with uniform chain lengths ranging from 6 to 14 uronic acid residues were made from mannuronan and polyMG by acid hydrolysis and separation by SEC (S. Holtan, Q. Zhang, W. I. Strand and G. Skjåk-Bræk, unpublished work) [21]. The oligomers were epimerized with AlgE1 and AlgE6 for 24 h and their degree of epimerization was analysed by 1H-NMR. As shown in Figure 7, the minimum size of a mannuronan (M) oligomer is 7–8 for AlgE6 and 8–10 for AlgE1 epimerization. The NMR spectra reveal that AlgE1 introduces mainly single G residues, while AlgE6 also generates GG-dyads. In both cases, the non-reducing and reducing ends of the oligomers remain unaltered. From the 1H-NMR spectra of an AlgE6-epimerized 11-mer of mannuronan (Figure 8) with FG=0.30 and FGG=0.15, the non-reducing end is M–M, while on the reducing end, M–Mred is dominating, although a weak signal from G–Mred indicates that the penultimate residue can be epimerized.
Figure 7. Minimum chain lengths (DP) of M- (*) and MG- (○) oligomers to accommodate activity of AlgE1 (A) and AlgE6 (B) epimerases.
Oligomer samples were treated with the individual epimerases for 24 h, freeze-dried and analysed by 1H-NMR (300 and 500 MHz) spectroscopy in 2H2O at 90 °C. Initial values of FG were 0.0 and 0.47 for M- and MG-oligomers respectively.
Figure 8. 1H-NMR (500 MHz) spectra of control (non-epimerized) (A) and AlgE6-epimerized (B) mannuronate undecamer.
The oligomannuronic acid sample in (A) was treated with the epimerase for 24 h and an increase in FG from 0.0 to 0.30 (FGG=0.15) (B). The α-anomeric resonance signal of the reducing end at 5.22 p.p.m. (A) shifts down-field to 5.24 p.p.m. when the penultimate residue is epimerized (B).
To analyse the epimerization patterns of the two enzymes more carefully and ultimately identify which uronic acid residues are being converted, the epimerized M-oligomers (DP6–DP9) were degraded with the G-lyase from K. pneumonia, specific for G–G and G–M glycosidic linkages [35]. New signals from G reducing ends could be observed from 1H-NMR spectra after the treatment with lyase, indicating a specific degradation of the epimerized oligomers. The digests were then analysed by ESI-MS and HPAEC–PAD. In Figure 9, the mass spectrum of the AlgE6-epimerized mannuronan octamer treated with lyase is shown. The dominating saturated oligomer is the tetramer at m/z 721.1, with minor peaks from saturated trimers and pentamers, as well as heptamers and octamers, the latter two from non-degraded epimerized and/or non-epimerized materials. Other peaks attributable to the oligo-uronates containing double bonds could be observed with lower signal intensities such as DP4 with an unsaturation at m/z 703.0, corresponding to the dominating saturated tetramer in the octamer sample. The peak at m/z 527.0, relative to DP3 with an unsaturation, could originate from degradation of epimerized heptamer contained in the DP8 sample. Peak assignments are reported in the Figure. This suggests that the M-residue at position 4 from the non-reducing end is preferentially epimerized. This was confirmed by HPAEC–PAD analysis of the same sample, where the MMMG-oligomer is the dominating peak (results not shown). Similar degradation patterns, with a saturated tetramer as dominating fragment, was also observed for the AlgE6-epimerised heptamer and nonamer, see Table 2. Owing to a low degree of epimerization or possibly non-degraded epimerized sample, original peaks for DP7 and DP9 were also detected for these two samples. A non-epimerized heptamer of mannuronan used as a negative control was not degraded by the G-lyase.
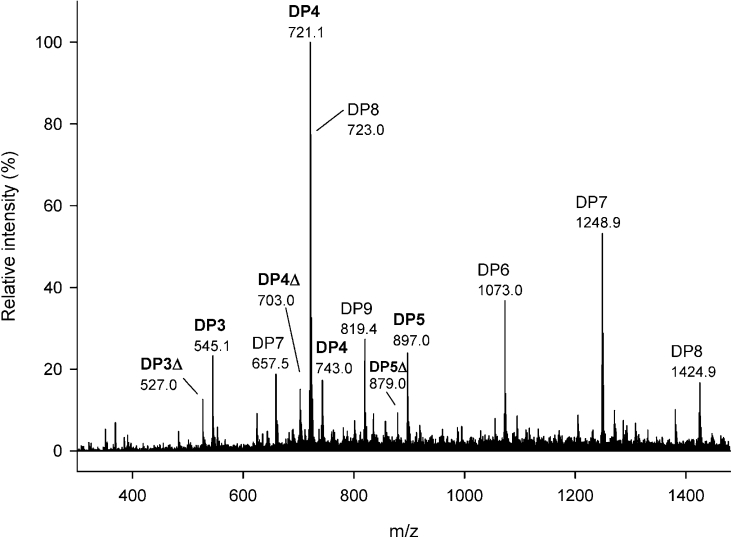
Figure 9. Negative ion ESI mass spectrum of mannuronate octamer after treatment with AlgE6 epimerase and G-lyase.
Peak attributions: saturated tetramer, 721.1 [M−H+]− and 743.0 [M+Na+−2H+]−; saturated (unchanged) octamer, 723.0 [M+Na+−3H+/2]2− and 1424.9 [M−H+]−; saturated pentamer 897.0 [M−H+]−; saturated trimer, 545.1 [M−H+]−; unsaturated tetramer, 703.0 [M−H+]−; unsaturated trimer, 527.0 [M−H+]−; unsaturated pentamer, 879.0 [M−H+]−; saturated (unchanged) heptamer, 657.5 [M+3Na+–5H+/2]2− and 1248.9 [M−H+]−; saturated (unchanged) hexamer, 1073.0 [M−H+]−; and saturated (unchanged) nonamer, 819.4 [M+Na++NH4+−4H+/2]2−. Capillary voltage: 3.5 kV; solvent: aq. 50% methanol containing 1% ammonia.
Table 2. Composition and G-lyase degradation patterns of AlgE1- and AlgE6-epimerized mannuronate oligomers.
Molar fractions FG and FGG of the samples after 24 h reaction time were obtained by integration of 1H-NMR spectra (400 MHz). The molecular masses of saturated and unsaturated oligomers in the G-lyase digests of the samples were obtained by ESI-MS. A non-epimerized heptamer of mannuronan was used as a negative control of the specificity of the G-lyase from K. pneumoniae.
| Molar fraction | ||||
|---|---|---|---|---|
| Mannuronate oligomer (DP) | FG | FGG | Number of residues in saturated oligomers after G-lyase digestion (ESI-MS) | Number of residues in unsaturated oligomers after G-lyase digestion (ESI-MS) |
| After AlgE6 epimerization | ||||
| 7 | 0.05 | 0.01 | 4 (3) | Δ3 and Δ4 |
| 8 | 0.06 | 0.01 | 4 (3 and 5) | Δ3 and Δ4 (Δ5) |
| 9 | 0.12 | 0.03 | 4 (3, 5 and 6) | Δ3, Δ4 and Δ5 (Δ6) |
| After AlgE1 epimerization | ||||
| 8 | 0.03 | 0.01 | 4 and 3 | Δ4 (Δ3 and Δ5) |
| Control | ||||
| 7 | 0.00 | 0.00 | 7 | 0 |
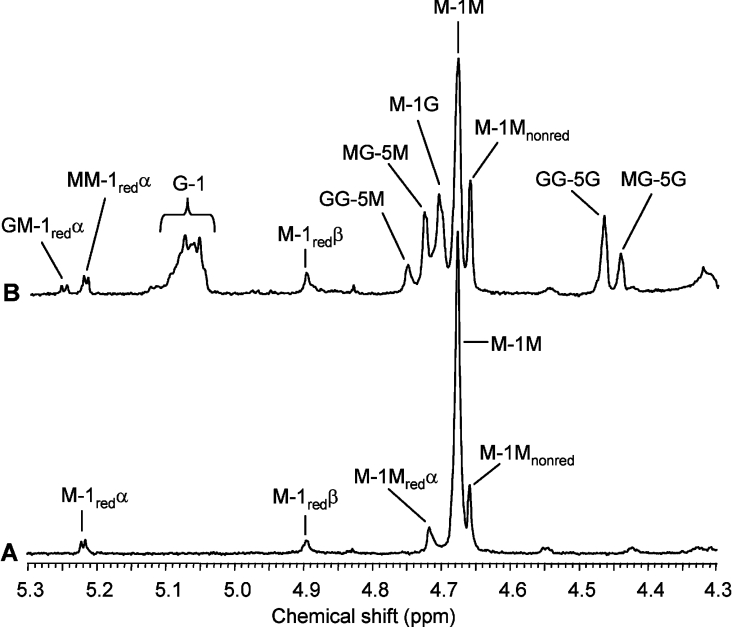
No change in FG could be detected by 1H-NMR for a DP6 mannuronan oligomer upon AlgE6 epimerization, see Figure 7. However, small peaks for saturated DP4 (at m/z 721.1) and DP3 (at m/z 545.1) could be detected in the G-lyase digest by ESI-MS analysis, suggesting a low level of epimerase activity on a hexamer (mass spectrum not shown). The AlgE6-epimerized 11-mer shown in Figure 8 was degraded by M-lyase from H. tuberculata with specificity for M–M and G–M glycosidic linkages. The digest was analysed by ESI-MS and signals attributable to all saturated oligomers (DP2–DP11) but with distinctly lower signal intensities for DP5, DP6 and DP7 were observed (mass spectrum not shown). For the unsaturated oligomers, trimers and tetramers were dominant in the digest in addition to DP7 with an unsaturation. This confirms that the ends of the oligomer are mainly unaltered (see Figure 8), and that the formed G-block (from NMR; NG>1=4) is positioned in the middle of the chain. All the ESI-MS data supported by NMR and HPAEC–PAD indicate that AlgE6 acts on the fourth sugar in an oligomannuronic acid chain with DP equal or higher than 7, regardless of the number of monomeric units, although AlgE6 activity increases with the chain length. This could fit with a subsite model with seven binding sites, where site number 4 expresses catalytic activity, leading to the observed G-lyase degradation patterns for the epimerized heptamer, octamer and nonamer shown in Table 2.
For AlgE1-epimerized and lyase-degraded samples, only the mannuronan octamer was successfully analysed, because of residual protein influencing the ESI mass analysis. As seen in Table 2, the main saturated oligomers after lyase digestion are the tetramer and trimer. In addition, unchanged heptamer and octamer were also present in the sample. The main unsaturated oligomer is DP4, corresponding to an epimerization of M residue number 4 from the non-reducing end in the octamer and specific cleavage at this site. AlgE1 thus displays the same epimerization pattern as AlgE6 when considering site of epimerization, although the relatively higher content of the saturated trimer might be attributed to AlgE1-2, which is very similar to AlgE4, previously shown to epimerize the third residue in an M-octamer [17].
In the case of MG-oligomers, taking into account that only 50% of the residues can be converted, AlgE6 shows similar activity dependence on chain lengths as demonstrated for mannuronan. For AlgE1, larger oligomers are required to obtain epimerization (see Figure 7). Whether this reflects the differences in size between the two enzymes is not clear. Since AlgE1 possesses two catalytic domains AlgE1-1 and AlgE1-2, it is conceivable that binding of the substrates to both sites is necessary for enzyme action. To investigate this, the two truncated enzymes were expressed separately and their activity assayed on both mannuronan and polyMG. In the former case, both enzymes expressed activity, AlgE1-1 by introducing G-block structures, and AlgE1-2 by exclusive introduction of single G residues, as would be expected from previous work using high M alginates as substrates [10]. In contrast, polyMG could not serve as substrate for either of the two truncated enzymes, indicating that an intact AlgE1 is necessary to epimerase polyMG, see Table 3. A probable role for AlgE1-2 in this matter may be the recognition and binding of the alternating sequences. The epimerization of every second M residue itself suggests a larger subsite, and Ertesvåg et al. [10] imply that AlgE1 and AlgE1-1 are able to epimerize smaller M-blocks in the chain than AlgE1-2. This could explain the requirement for larger MG- than M-oligomers for AlgE1 epimerization.
Table 3. Epimerization characteristics of AlgE1-1 and AlgE1-2.
Enzyme activity on different substrates of the two separate parts of AlgE1, AlgE1-1 (A1R1R2R3) and AlgE1-2 (A2R4) expressed separately in E. coli [10]. Samples were analysed by 1H-NMR (300 MHz) spectroscopy, in 2H2O at 90 °C.
| AlgE1-1 (A1R1R2R3) | AlgE1-2 (A2R4) | ||||||
|---|---|---|---|---|---|---|---|
| Substrate | Reaction time (h) | FG | FGG | FGM | FG | FGG | FGM |
| Mannuronan | 0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 3 | 0.67 | 0.54 | 0.13 | 0.37 | 0.00 | 0.37 | |
| 23 | 0.72 | 0.61 | 0.11 | 0.42 | 0.00 | 0.42 | |
| PolyMG | 0 | 0.46 | 0.00 | 0.46 | 0.46 | 0.00 | 0.46 |
| 3 | 0.46 | 0.00 | 0.46 | 0.46 | 0.00 | 0.46 | |
| 23 | 0.46 | 0.00 | 0.46 | 0.46 | 0.00 | 0.46 | |
| L. hyperborea leaf | 0 | 0.49 | 0.33 | 0.16 | 0.49 | 0.33 | 0.16 |
| 3 | 0.65 | 0.47 | 0.18 | 0.59 | 0.36 | 0.23 | |
| 23 | 0.69 | 0.51 | 0.18 | 0.59 | 0.35 | 0.24 | |
Subsite analysis on MG-oligomers, using G-lyase degradation and ESI-MS, is more complex than with pure mannuronan oligomers, since G-specific lyases will degrade the starting material as well as the epimerized oligomers. To solve this problem, we compared the degradation pattern of the starting material with the epimerized samples using the lyase from H. tuberculata, specific for cleavage of G–M and M–M glycosidic linkages. FIA-MS was performed on the lyase digests to achieve a quantitative analysis of the degradation products before and after epimerization. In Figure 10, the relative intensity of the saturated oligomers in the digests of AlgE1- and AlgE6-epimerized DP12 MG-oligomer compared with control non-epimerized sample is given. The saturated oligomers arise from lyase cleavage of susceptible linkages close to the non-reducing end of the sugar chains. The non-epimerized DP12 MG-oligomer is degraded into oligomers with an even number of residues, mainly di (MG) and tetra (MGMG), the latter with approximately double intensity (light-grey bars in Figure 10). In the epimerized samples, the amount of tetramer is reduced with respect to the dimer (dark-grey and black bars in Figure 10). This effect is more pronounced for the AlgE6-epimerized sample (FG=0.65), with DP4 constituting less than half the intensity of DP2, than for the AlgE1-epimerized sample (FG=0.52). This is probably due to the higher degree of epimerization in the former, which is also indicated by the presence of saturated oligomers with longer chain lengths in this sample. The ionization efficiency for MG-oligomers of various chain lengths was tested to validate the analysis results. The dimer (MG) and tetramer (MGMG) have approximately equal efficiencies, within the same magnitude. However, for larger oligomers there is a linear decrease in ionization efficiency up to DP16 tested (results not shown). The observed degradation patterns indicate that both AlgE1 and AlgE6 preferentially attack an M in position 5 from the non-reducing end, creating an MGMGGG-sequence (underlined and bold face indicate the epimerized residue). Since position 4 from the non-reducing end is occupied by a G residue in the case of MG-oligomers, the proposed subsite model and position of the active site for AlgE6 are valid by shifting the subsite one residue towards the reducing end. Thus, for AlgE6, the minimum size of an MG-oligomer for binding to all the indicated seven sites is an octamer (see Figure 7).
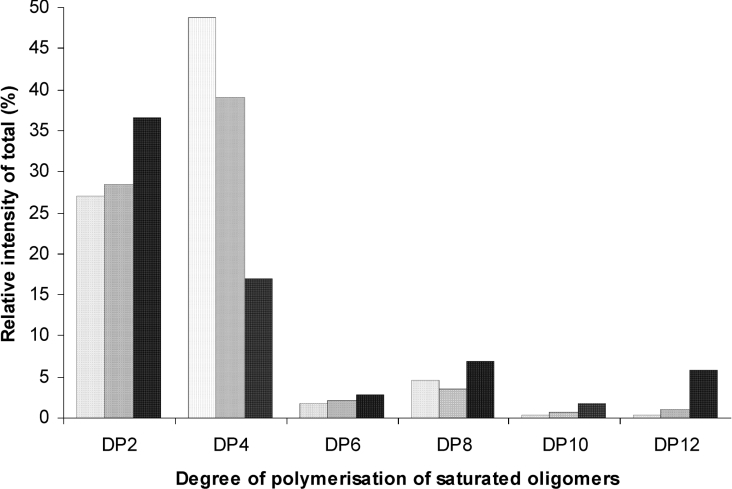
Figure 10. Relative intensities of total of saturated oligomers in the M-lyase (H. tuberculata) digests of DP12 MG-oligomers.
Control (non-epimerized), FG=0.47 (light-grey bars to the left); AlgE1-epimerized, FG=0.52 (dark-grey bars in the middle); AlgE6-epimerized, FG=0.65 (black bars to the right). The digests were analysed by FIA-MS in negative ion mode. Quantitative analysis was performed by integrating a ±10 p.p.m. window for each m/z and the contributions from the generated ions, [M−H+]−, [M−2H+]2−, [M−3H+]3−, [M−4H+]4−, were summed for each chain length. Only the chain lengths with an even number of residues are shown, due to the degradation pattern of the lyase and low intensities of odd numbered oligomers.
In Figure 10, only the saturated oligomers are given. Of the unsaturated oligomers, the tetramer (DP4Δ) is dominant in both standard and epimerized oligomers. Similar characteristics were also observed for epimerized and standard DP14 MG-oligomer (results not shown).
Has the epimerization reaction any end point?
Since naturally occurring alginates in brown algae and in bacteria, as well as in vitro epimerized alginates, still contain >20% mannuronic acid, it has been suggested that there exists some kind of end point for the epimerase reaction because a minimum length of M-block is necessary to support enzyme activity [10,36]. If this were the case, condensation of G-blocks would not be permitted. Since we show in the present paper that both AlgE1 and AlgE6 act effectively upon a polyalternating substrate and thus epimerize an M residue flanked by G residues, there is apparently no lower limit for the M-block length in chain to support epimerase activity. An alternative explanation for the apparent end point of epimerization is that calcium ions, required by the enzyme, are gradually depleted due to their co-operative binding to the long G-blocks. Moreover, binding of calcium ions could lead to gel formation rendering the molecules less accessible as substrate for the enzyme. To investigate this, high-molecular-mass mannuronan was incubated with AlgE1 and AlgE6. When epimerization took place with a concentration of calcium ions that gives optimum reaction rate (2.5 mM), the reaction stopped at approx. 50% conversion due to precipitation of Ca-alginate (results not shown). By reducing the calcium level to 0.8 mM and adding 100 mM NaCl, which serves as an anti-gelling salt, a conversion of 90% was obtained with both AlgE1 and AlgE6. By replacing mannuronan with polyMG as substrate and adding enzyme several times during the incubation process, the degree of epimerization was extended to >97% G after 4 days of epimerization (NMR spectra not shown). Since the reducing and non-reducing ends cannot be attacked by the enzyme, this probably represents the end point of the epimerization, as no further increase in G content could be observed from 4 to 7 days of epimerization. The molecular mass of the starting material consequently determines the deviation from 100% polyG. The functional properties of these extreme alginates are currently being investigated in our laboratory.
Concluding remarks
The results reported in the present paper demonstrate that the three mannuronan C-5 epimerases AlgE1, AlgE5 and AlgE6 introduce G-blocks in various types of alginates, either by elongating existing blocks or by condensing them, as demonstrated by their action on polyMG. Still, and in spite of their genetic homology [5,6], they act differently on various substrates. While AlgE5, which also exhibits lyase activity, prefers substrates containing single G residues, the other two prefer a substrate containing G-blocks. Moreover, AlgE1 and AlgE6 differ in the length and distribution of the G-blocks they introduce into polyMG. This probably reflects their different degrees of processivity. The alternating [4)-β-D-ManpA-(1→4)-α-L-GulpA(1→]n linkages in polyMG imply that each residue is rotated nearly 180° with respect to its neighbours. Since only every second residue can be epimerized, the enzyme can slide along the substrate chain without rotation, analogous to the action of AlgE4 on mannuronan [17]. The knowledge of the epimerization patterns of the G-block-forming mannuronan C-5 epimerases AlgE1 and AlgE6 here examined on polyMG as substrate opens up a more specific tailoring of alginate than with the composition of alternating (AlgE4) and homogeneous (AlgE1 and AlgE6) block structures in the polysaccharide only. Controlling the lengths and distribution of blocks in the alginate and thus the physical properties, the performances in a large number of applications can be improved.
Acknowledgments
This work was funded by a grant from the Norwegian Research Council. We are very grateful to Wenche Iren Strand (NTNU) for recording the 1H-NMR spectra, Lars Hagen (NTNU) for assistance with MALDI-MS analysis, Håvard Sletta for providing M-lyase from abalone, Dr Helga Ertesvåg for providing AlgE1-1 and AlgE1-2, Cristiana Campa (Bracco Industries, Trieste, Italy) for useful discussions regarding MS analysis, Olav Andreas Aarstad (NTNU) for performing HPAEC–PAD analysis and Catherine Taylor (NTNU) for a critical reading of this paper. We also thank Mona Senneset, Kåre Andre Kristiansen and Hilde Sofie Larsen (all at NTNU) for technical assistance.
References
- 1.Painter T. J. Algal polysaccharides. In: Aspinall G. O., editor. The Polysaccharides, vol. 2. New York: Academic Press; 1983. pp. 195–285. [Google Scholar]
- 2.Gorin P. A. J., Spencer J. F. T. Exocellular alginic acid from Azotobacter vinelandii. Can. J. Chem. 1966;44:993–998. [Google Scholar]
- 3.Govan J. R. W., Fyfe J. A. M., Jarman T. R. Isolation of alginate-producing mutants of Pseudomonas fluorescens, Pseudomonas putida and Pseudomonas mendocina. J. Gen. Microbiol. 1981;125:217–220. doi: 10.1099/00221287-125-1-217. [DOI] [PubMed] [Google Scholar]
- 4.Ertesvåg H., Doseth B., Larsen B., Skjåk-Bræk G., Valla S. Cloning and expression of an Azotobacter vinelandii mannuronan C-5 epimerase gene. J. Bacteriol. 1994;176:2846–2853. doi: 10.1128/jb.176.10.2846-2853.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ertesvåg H., Høidal H. K., Hals I. K., Rian A., Doseth B., Valla S. A family of modular type mannuronan C-5 epimerase genes controls alginate structure in Azotobacter vinelandii. Mol. Microbiol. 1995;16:719–731. doi: 10.1111/j.1365-2958.1995.tb02433.x. [DOI] [PubMed] [Google Scholar]
- 6.Svanem B. I. G., Skjåk-Bræk G., Ertesvåg H., Valla S. Cloning and expression of three new Azotobacter vinelandii genes closely related to a previously described gene family encoding mannuronan C-5 epimerases. J. Bacteriol. 1999;181:68–77. doi: 10.1128/jb.181.1.68-77.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Høidal H. K., Ertesvåg H., Skjåk-Bræk G., Stokke B. T., Valla S. The recombinant Azotobacter vinelandii mannuronan C-5 epimerase AlgE4 epimerizes alginate by a nonrandom attack mechanism. J. Biol. Chem. 1999;274:12316–12322. doi: 10.1074/jbc.274.18.12316. [DOI] [PubMed] [Google Scholar]
- 8.Hartmann M., Holm O. B., Johansen G.-A. B., Skjåk-Bræk G., Stokke B. T. Mode of action of recombinant Azotobacter vinelandii mannuronan C-5 epimerases AlgE2 and AlgE4. Biopolymers. 2002;63:77–88. doi: 10.1002/bip.10017. [DOI] [PubMed] [Google Scholar]
- 9.Hartmann M., Duun A. S., Markussen S., Grasdalen H., Valla S., Skjåk-Bræk G. Time-resolved 1H and 13C NMR spectroscopy for detailed analyses of the Azotobacter vinelandii mannuronan C-5 epimerase reaction. Biochim. Biophys. Acta. 2002;1570:104–112. doi: 10.1016/s0304-4165(02)00195-2. [DOI] [PubMed] [Google Scholar]
- 10.Ertesvåg H., Høidal H. K., Skjåk-Bræk G., Valla S. The Azotobacter vinelandii mannuronan C-5-epimerase AlgE1 consists of two separate catalytic domains. J. Biol. Chem. 1998;273:30927–30932. doi: 10.1074/jbc.273.47.30927. [DOI] [PubMed] [Google Scholar]
- 11.Ertesvåg H., Høidal H. K., Schjerven H., Svanem B. I. G., Valla S. Mannuronan C-5-epimerases and their application for in vitro and in vivo design of new alginates useful in biotechnology. Metab. Eng. 1999;1:262–269. doi: 10.1006/mben.1999.0130. [DOI] [PubMed] [Google Scholar]
- 12.Ertesvåg H., Valla S. The A modules of the Azotobacter vinelandii mannuronan C-5-epimerase AlgE1 are sufficient for both epimerization and binding of Ca2+ J. Bacteriol. 1999;181:3033–3038. doi: 10.1128/jb.181.10.3033-3038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sletmoen M., Skjåk-Bræk G., Stokke B. T. Mapping enzymatic functionalities of mannuronan C-5 epimerases and their modular units by dynamic force spectroscopy. Carbohydr. Res. 2005;340:2782–2795. doi: 10.1016/j.carres.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 14.Greenwood C. T., Milne E. A. Studies on starch-degrading enzymes. A comparison of α-amylases from different sources – their properties and action patterns. Starke. 1968;20:139–144. [Google Scholar]
- 15.Ernst S., Rhomberg A. J., Biemann K., Sasisekharan R. Direct evidence for a predominantly exolytic processive mechanism for depolymerization of heparin-like glycosaminoglycans by heparinase I. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4182–4187. doi: 10.1073/pnas.95.8.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breyer W. A., Matthews B. W. A structural basis for processivity. Protein Sci. 2001;10:1699–1711. doi: 10.1110/ps.10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campa C., Holtan S., Nilsen N., Bjerkan T. M., Stokke B. T., Skjåk-Bræk G. Biochemical analysis of the processive mechanism for epimerization of alginate by mannuronan C-5 epimerase AlgE4. Biochem. J. 2004;381:155–164. doi: 10.1042/BJ20031265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gimmestad M., Sletta H., Ertesvåg H., Bakkevig K., Jain S., Suh S., Skjåk-Bræk G., Ellingsen T. E., Ohman D. E., Valla S. The Pseudomonas fluorescens AlgG protein, but not its mannuronan C-5-epimerase activity, is needed for alginate polymer formation. J. Bacteriol. 2003;185:3515–3523. doi: 10.1128/JB.185.12.3515-3523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ertesvåg H., Skjåk-Bræk G. Modification of alginates using mannuronan C-5 epimerases. In: Bucke C., editor. Methods in Biotechnology, Vol. 10. Totowa, NJ: Humana Press; 1999. pp. 71–78. [Google Scholar]
- 20. Reference deleted.
- 21.Campa C., Oust A., Skjåk-Bræk G., Paulsen B. S., Paoletti S., Christensen B. E., Ballance S. Determination of average degree of polymerisation and distribution of oligosaccharides in a partially acid-hydrolysed homopolysaccharide: a comparison of four experimental methods applied to mannuronan. J. Chromatogr. A. 2004;1026:271–281. doi: 10.1016/j.chroma.2003.11.045. [DOI] [PubMed] [Google Scholar]
- 22.Ramstad M. V., Ellingsen T. E., Josefsen K. D., Høidal H. K., Valla S., Skjåk-Bræk G., Levine D. W. Properties and action pattern of the recombinant mannuronan C-5-epimerase AlgE2. Enzyme Microb. Technol. 1999;24:636–646. [Google Scholar]
- 23.Østgaard K., Knutsen S. H., Dyrset N., Aasen I. M. Production and characterization of guluronate lyase from Klebsiella pneumoniae for applications in seaweed biotechnology. Enzyme Microb. Technol. 1993;15:756–763. doi: 10.1016/0141-0229(93)90006-n. [DOI] [PubMed] [Google Scholar]
- 24.Heyraud A., Colin-Morel P., Girond S., Richard C., Kloareg B. HPLC analysis of saturated or unsaturated oligoguluronates and oligomannuronates. Application to the determination of the action pattern of Haliotis tuberculata alginate lyase. Carbohydr. Res. 1996;291:115–126. doi: 10.1016/s0008-6215(96)00138-3. [DOI] [PubMed] [Google Scholar]
- 25.Boyen C., Kloareg B., Polnefuller M., Gibor A. Preparation of alginate lyases from marine mollusks for protoplast isolation in brown-algae. Phycologia. 1990;29:173–181. [Google Scholar]
- 26.Østgaard K. Enzymatic microassay for the determination and characterization of alginates. Carbohydr. Polymers. 1992;19:51–59. [Google Scholar]
- 27.Ertesvåg H., Erlien F., Skjåk-Bræk G., Rehm B. H. A., Valla S. Biochemical properties and substrate specificities of a recombinantly produced Azotobacter vinelandii alginate lyase. J. Bacteriol. 1998;180:3779–3784. doi: 10.1128/jb.180.15.3779-3784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grasdalen H. High-field, H-1-NMR spectroscopy of alginate sequential structure and linkage conformations. Carbohydr. Res. 1983;118:255–260. [Google Scholar]
- 29.Heyraud A., Gey C., Leonard C., Rochas C., Girond S., Kloareg B. NMR spectroscopy analysis of oligoguluronates and oligomannuronates prepared by acid or enzymatic hydrolysis of homopolymeric blocks of alginic acid. Application to the determination of the substrate specificity of Haliotis tuberculata alginate lyase. Carbohydr. Res. 1996;289:11–23. doi: 10.1016/0008-6215(96)00060-2. [DOI] [PubMed] [Google Scholar]
- 30.Ballance S., Holtan S., Aarstad O. A., Sikorski P., Skjåk-Bræk G., Christensen B. E. Application of high-performance anion-exchange chromatography with pulsed amperometric detection and statistical analysis to study oligosaccharide distributions – a complementary method to investigate the structure and some properties of alginates. J. Chromatogr. 2005;1093:59–68. doi: 10.1016/j.chroma.2005.07.051. [DOI] [PubMed] [Google Scholar]
- 31.Jacobs A., Dahlman O. Enhancement of the quality of MALDI mass spectra of highly acidic oligosaccharides by using a Nafion-coated probe. Anal. Chem. 2001;73:405–410. doi: 10.1021/ac001222i. [DOI] [PubMed] [Google Scholar]
- 32.Larsen B., Skjåk-Bræk G., Painter T. Action pattern of mannuronan C-5-epimerase: generation of block-copolymeric structures in alginates by a multiple-attack mechanism. Carbohydr. Res. 1986;146:342–345. [Google Scholar]
- 33.Grasdalen H., Larsen B., Smidsrød O. C-13-NMR studies of monomeric composition and sequence in alginate. Carbohydr. Res. 1981;89:179–191. [Google Scholar]
- 34.Haugen F., Kortner F., Larsen B. Kinetics and specificity of alginate lyases: part I, a case study. Carbohydr. Res. 1990;198:101–109. doi: 10.1016/0008-6215(90)84280-8. [DOI] [PubMed] [Google Scholar]
- 35.Boyd J., Turvey J. Structural studies of alginic acid, using a bacterial poly-alpha-L-guluronate lyase. Carbohydr. Res. 1978;66:187–194. [Google Scholar]
- 36.Skjåk-Bræk G., Smidsrød O., Larsen B. Tailoring of alginates by enzymatic modification in vitro. Int. J. Biol. Macromol. 1986;8:330–336. [Google Scholar]