Abstract
Multiple-synostosis syndrome is an autosomal dominant disorder characterized by progressive symphalangism, carpal/tarsal fusions, deafness, and mild facial dysmorphism. Heterozygosity for functional null mutations in the NOGGIN gene has been shown to be responsible for the disorder. However, in a cohort of six probands with multiple-synostosis syndrome, only one was found to be heterozygous for a NOGGIN mutation (W205X). Linkage studies involving the four-generation family of one of the mutation-negative patients excluded the NOGGIN locus, providing genetic evidence of locus heterogeneity. In this family, polymorphic markers flanking the GDF5 locus were found to cosegregate with the disease, and sequence analysis demonstrated that affected individuals in the family were heterozygous for a novel missense mutation that predicts an R438L substitution in the GDF5 protein. Unlike mutations that lead to haploinsufficiency for GDF5 and produce brachydactyly C, the protein encoded by the multiple-synostosis–syndrome allele was secreted as a mature GDF5 dimer. These data establish locus heterogeneity in multiple-synostosis syndrome and demonstrate that the disorder can result from mutations in either the NOGGIN or the GDF5 gene.
Multiple-synostosis syndrome (SYNS1 [MIM 186500]) is an autosomal dominant condition characterized by progressive joint fusions of the fingers, wrists, ankles, and cervical spine; characteristic facies; and progressive conductive deafness. Two additional syndromes have very similar phenotypes: proximal symphalangism (SYM1 [MIM 185800]) and tarsal-carpal coalition syndrome (TCC [MIM 186570]). Heterozygosity for NOGGIN (GenBank accession number NM_005450) mutations has been identified in all three disorders (Gong et al. 1999; Dixon et al. 2001). In addition, heterozygosity for mutations in NOGGIN has been identified in stapes ankylosis syndrome without symphalangism (MIM 184460) (Brown et al. 2002). To date, 14 distinct NOGGIN mutations have been reported (Gong et al. 1999; Dixon et al. 2001; Takahashi et al. 2001; Brown et al. 2002; Mangino et al. 2002). The majority (10 of 14) are missense mutations, and the 4 nonsense mutations are predicted to result in premature translation termination codons. Noggin was initially identified in Xenopus as a secreted signal released by the Spemann organizer and is involved in developmental processes, including induction of neural tissue from the ectoderm and dorsalization of the ventral mesoderm (Zimmerman et al. 1996). It also participates in the regulation of chondrogenesis in somites and limb buds, in which it acts as an antagonist to the bone morphogenetic proteins (Zimmerman et al. 1996; Brunet et al. 1998; McMahon et al. 1998).
GDF5 (growth differentiation factor 5, also known as CDMP1 or BMP14), a member of the bone morphogenetic protein and TGF-β families, is a secreted growth factor expressed during several steps in skeletal development, including the formation of the cartilage anlagen (chondrogenesis), chondrocyte differentiation, and joint morphogenesis (Chang et al. 1994; Storm and Kingsley 1999). Several skeletal dysplasias are known to result from mutations in GDF5 (GenBank accession number NM_000557). Brachydactyly C (BDC [MIM 113100]), a disorder characterized by shortened middle phalanges of the second, third, and fifth digits and by hyperphalangia, results from heterozygosity for functional null GDF5 mutations (Polinkovsky et al. 1997). Recently, in the disorder brachydactyly A2 (BDA2 [MIM 112600]), heterozygosity for a GDF5 mutation (L441P) within the active-signaling domain of the molecule was identified (Kjaer et al. 2005). Homozygosity for functional null mutations in GDF5 have been identified in autosomal recessive Hunter-Thompson (MIM 201250), Grebe (MIM 200700), and Du Pan (MIM 228900) chondrodysplasias, as well as in a rare form of BDC in the brachypodism mouse (Storm et al. 1994; Thomas et al. 1996, 1997; Faiyaz-Ul-Haque et al. 2002; Schwabe et al. 2004). Individuals with these disorders have severe abnormalities in the bones and joints of the mesomelic and acromelic limb segments. Interestingly, carriers of the mutations of the recessive forms of GDF5 disorders manifest mild abnormal metacarpohalangeal profiles, which suggests gene-dose sensitivity in the developing phalanges (Schwabe et al. 2004).
After obtaining institutional approval for human subjects study, we studied a cohort of six genetically independent individuals with SYNS1 diagnosed on the basis of clinical and radiographic evaluations. Five cases occurred sporadically, and one individual was a member of a large four-generation Ashkenazi Jewish family (fig. 1A). In this family, phenotypic findings included a broad hemicyclindrical nose, progressive symphalangism, and carpal, tarsal, and vertebral fusions. Within this family, there was phenotypic variability in the extent of joint fusions and in the presence or absence of equinovarus. To test the hypothesis that heterozygosity for NOGGIN mutations would lead to SYNS1 in this cohort, we determined the sequence of the single exon encoding noggin, using published methods (Gong et al. 1999). One patient (R02-360) was heterozygous for a point mutation (1425G→A) predicted to lead to a premature translation termination codon, W205X (human NOGGIN). This mutation created an Afe1 restriction endonuclease cleavage site, and the mutation was further confirmed by cleavage of a PCR-generated DNA fragment with the enzyme (data not shown). No mutations were identified in the remaining five probands. For two of the patients, normal dosage of the NOGGIN gene was demonstrated by Southern analysis (data not shown).
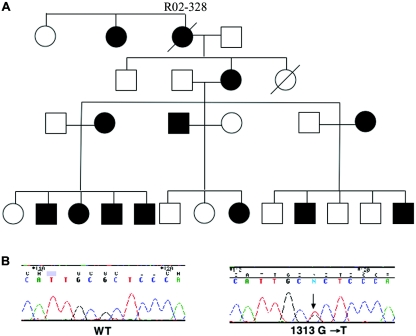
Figure 1.
A, Pedigree of family R02-328 with SYNS1. B, Chromatograms showing the GDF5 wild-type (WT) sequence and the nucleotide substitution, 1313G→T, predicted to lead to the amino acid substitution R438L.
To confirm the exclusion of NOGGIN as the disease gene in the familial case of SYNS1, linkage analysis using polymorphic markers from chromosome 17q21-22 was performed. The data demonstrated exclusion of linkage between the phenotype and the markers D17S787 (maximum LOD score of 0 [θ=0]) and D17S957 (maximum LOD score of 1.0 [θ=0]), which flank the NOGGIN gene, thus excluding NOGGIN as the disease gene. To define the second locus for SYNS1, we considered other genes known to be involved in joint morphogenesis, including the gene encoding GDF5, which is a direct antagonist of noggin. We tested linkage to the marker D20S195, which is located 2.2 Mb centromeric to GDF5. In the family, a single allele cosegregated with disease without exception (maximum LOD score of 2.2 [θ=0]). Since these data supported the candidacy of the GDF5 gene, the sequences of the two exons of GDF5 were determined as described elsewhere (Polinkovsky et al. 1997) for the five NOGGIN mutation–negative probands. Heterozygosity for a nucleotide substitution (1313G→T) (human GDF5) was found in the proband—the index case from the large family—predicting an R438L amino acid change in the protein (fig. 1B). The sequence change was found in all affected members of the family but not in the unaffected individuals, and it is predicted to substitute a hydrophilic arginine with a hydrophobic leucine in a highly conserved residue within the active-signaling domain of the mature protein. However, in four probands, neither a NOGGIN mutation nor a GDF5 mutation was identified, which suggests that other possible loci could lead to SYNS1.
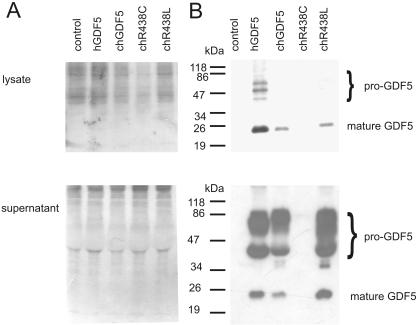
To determine the consequences of the substitution on dimerization and secretion of GDF5, we used an RCAS viral construct (Lehmann et al. 2003) to transiently transfect and express a chicken GDF5 cDNA carrying the R438L sequence in DF-1 cells. The data were compared with control constructs carrying either the wild-type sequence or a cDNA with an R438C mutation, which causes BDC (Everman et al. 2002). The BDC mutation is known to lead to inefficient dimerization and secretion of the mature protein, which leads to functional haploinsufficiency for GDF5 (Everman et al. 2002). Proteins secreted into the medium and within the infected cells were each separated by PAGE under nonreducing conditions, were transferred to membranes, and were incubated with a monoclonal antibody to the aminoterminal portion of GDF5 (kind gift from Biopharm GmbH). The antibody recognizes the disulfide-linked full dimer (pro-GDF5) and the mature, active-domain dimer (GDF5) (Wang et al. 2004). The R438L protein was assembled into mature GDF5 dimers and was secreted in a similar way as wild-type GDF5 (fig. 2B). In contrast, but consistent with prior studies (Everman et al. 2002), the R438C construct did not lead to efficient formation or secretion of either pro-GDF5 or mature GDF5 dimers. Thus, although the same amino acid residue is altered in both BDC and SYNS1, the different substitutions lead to distinct fates for the mutant protein. It is likely that altered GDF5 activity due to the R438L substitution, rather than due to haploinsufficiency, produces SYNS1, revealing an alternative pathway to disease resulting from a GDF5 mutation (Seeman et al. 2005)
Figure 2.
Synthesis and secretion of normal and mutant GDF5 proteins. DF-1 cells transfected were control vector, human GDF5, chicken (ch) GDF5, ch R438C-GDF5, and ch R438L-GDF5. A, Ponceau red-stained western blot demonstrating protein expression and uniform loading in samples derived from the cell lysate and supernatant. B, Western blot analysis of lysate from ch GDF5 transfected cells showing pro-GDF5 in the human control lane and mature GDF5 in wild-type ch GDF5 and in ch R438L-GDF5 but not in ch R438C-GDF5 (top); western blot analysis of supernatant from ch GDF5–transfected cells showing pro-GDF5 in the human control lane and mature GDF5 in wild-type ch GDF5 and in ch R438L-GDF5 but not in ch R438C-GDF5 (bottom).
To date, all reported mutations causing SYNS1 have been heterozygous NOGGIN mutations (Marcelino et al. 2001). This report identifies GDF5 as a second locus for the SYNS1 phenotype. A role for GDF5 in joint formation is further supported by the report of a large family with features similar to SYNS1, described in abstract form only (Akarsu et al. 1999). In the family, an S475N substitution, also in the highly conserved active-signaling domain of GDF5, was found to segregate with the phenotype. The effects of the substitution on the protein are unknown.
One of the questions raised by the identification of a second locus for SYNS1 is whether there are phenotypic differences between the individuals in whom NOGGIN versus GDF5 mutations have been identified. Detailed review of the phenotypes of the individuals in the family described here show characteristic clinical and radiographic findings for the disorder, including a broad hemicyclindrical nose, progressive symphalangism, and carpal, tarsal, and vertebral fusions. Analysis of hand radiographs of affected individuals showed no findings characteristic of BDC, indicating that the substitution does not result in an overlapping SYNS1/BDC phenotype. Detailed evaluation of this family did not reveal any distinguishing clinical features between SYNS1 patients with NOGGIN mutations and patients with GDF5 defects.
NOGGIN and GDF5 are both required for proper joint morphogenesis. In the absence of noggin, BMP growth factors are unregulated, resulting in chondrocyte hyperplasia instead of apoptosis in the developing joint and, thus, leading to lack of normal joint formation. Noggin−/− mice had a lethal skeletal phenotype with a very abnormal skeleton, characterized by joint fusions with a complete absence of limb joints, costovertebral defects, and cartilage spurs, and with up-regulation of GDF5 in the areas of the presumptive joints (Brunet et al. 1998). Human NOGGIN mutations that lead to SYNS1 are due to functional haploinsufficiency and a presumed lack of appropriate antagonism of GDF5. Since the GDF5 R438L protein is able to form mature secreted dimers, we suggest the hypothesis that the mutation leads to increased GDF5 activity and to SYNS1 (Seeman et al. 2005). This is mechanistically similar to the consequences of haploinsufficiency for NOGGIN, in that the resultant effect is the up-regulation of GDF5 in the mesenchyme surrounding the developing joint. This study identifies GDF5 as a locus for SYNS1. In addition, mutations were not identified in either NOGGIN or GDF5 in the remaining four patients in the cohort, suggesting the possibility of further locus heterogeneity.
Acknowledgments
We thank the families and their physicians for participating in the studies described here. The work was supported in part by grants from the National Institutes of Health (NIH) (HD22657) and from the Joseph Drown Foundation (to D.K.) and by the NIH-sponsored General Clinical Research Center grant M01-RR00425 (to Cedars-Sinai Medical Center).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for NOGGIN [accession number NM_005450] and GDF5 [accession number NM_000557])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SYNS1, SYM1, TCC, stapes ankylosis syndrome without symphalangism, BDC, BDA2, and Hunter-Thompson, Grebe, and Du Pan chondrodysplasias)
References
- Akarsu AN, Rezaie T, Demirtas M, Farhud DD III, Sarfarazi M (1999) Multiple synostosis type 2 (SYNS2) maps to 20q11.2 and caused by a missense mutation in the growth/differentiation factor 5 (GDF5). Am J Hum Genet Suppl 65:A281 [Google Scholar]
- Brown DJ, Kim TB, Petty EM, Downs CA, Martin DM, Strouse PJ, Moroi SE, Milunsky JM, Lesperance MM (2002) Autosomal dominant stapes ankylosis with broad thumbs and toes, hyperopia, and skeletal anomalies is caused by heterozygous nonsense and frameshift mutations in NOG, the gene encoding noggin. Am J Hum Genet 71:618–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet LJ, McMahon JA, McMahon AP, Harland RM (1998) Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science 280:1455–1457 10.1126/science.280.5368.1455 [DOI] [PubMed] [Google Scholar]
- Chang SC, Hoang B, Thomas JT, Vukicevic S, Luyten FP, Ryba NJ, Kozak CA, Reddi AH, Moos M Jr (1994) Cartilage-derived morphogenetic proteins: new members of the transforming growth factor-beta superfamily predominantly expressed in long bones during human embryonic development. J Biol Chem 269:28227–28234 [PubMed] [Google Scholar]
- Dixon ME, Armstrong P, Stevens DB, Bamshad M (2001) Identical mutations in NOG can cause either tarsal/carpal coalition syndrome or proximal symphalangism. Genet Med 3:349–353 [DOI] [PubMed] [Google Scholar]
- Everman DB, Bartels CF, Yang Y, Yanamandra N, Goodman FR, Mendoza-Londono JR, Savarirayan R, White SM, Graham JM Jr, Gale RP, Svarch E, Newman WG, Kleckers AR, Francomano CA, Govindaiah V, Singh L, Morrison S, Thomas JT, Warman ML (2002) The mutational spectrum of brachydactyly type C. Am J Med Genet 112:291–296 10.1002/ajmg.10777 [DOI] [PubMed] [Google Scholar]
- Faiyaz-Ul-Haque M, Ahmad W, Wahab A, Haque S, Azim AC, Zaidi SH, Teebi AS, Ahmad M, Cohn DH, Siddique T, Tsui LC (2002) Frameshift mutation in the cartilage-derived morphogenetic protein 1 (CDMP1) gene and severe acromesomelic chondrodysplasia resembling Grebe-type chondrodysplasia. Am J Med Genet 111:31–37 10.1002/ajmg.10501 [DOI] [PubMed] [Google Scholar]
- Gong Y, Krakow D, Marcelino J, Wilkin D, Chitayat D, Babul-Hirji R, Hudgins L, Cremers CW, Cremers FPM, Brunner HG, Reinker K, Rimoin DL, Cohn DH, Goodman FR, Reardon W, Patton M, Francomano CA, Warman ML (1999) Heterozygous mutations in the gene encoding noggin affect human joint morphogenesis. Nat Genet 21:302–304 10.1038/6821 [DOI] [PubMed] [Google Scholar]
- Kjaer KW, Eiberg H, Hansen L, van der Hagen CB, Rosendahl K, Tommerup N, Mundlos S (2005) A mutation in the receptor binding site of GDF5 causes Mohr-Wriedt brachydactyly type A2. J Med Genet (http://jmg.bmjjournals.com/cgi/content/full/43/2/111) (electronically published Jul 13, 2005; accessed February 23, 2006) [DOI] [PMC free article] [PubMed]
- Lehmann K, Seemann P, Stricker S, Sammar M, Meyer B, Suring K, Majewski F, Tinschert S, Grzeschik KH, Muller D, Knaus P, Nurnberg P, Mundlos S (2003) Mutations in morphogenetic protein receptor 1B cause brachydactyly type A2. Proc Natl Acad Sci USA 100:12277–12282 10.1073/pnas.2133476100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangino M, Flex E, Digilio MC, Giannotti A, Dallapiccola B (2002) Identification of a novel NOG gene mutation (P35S) in an Italian family with symphalangism. Hum Mutat 19:308 10.1002/humu.9016 [DOI] [PubMed] [Google Scholar]
- Marcelino J, Sciortino CM, Romero MF, Ulatowski LM, Ballock RT, Economides AN, Eimon PM, Harland RM, Warman ML (2001) Human disease-causing NOG missense mutations: effects on noggin secretion, dimer formation, and bone morphogenetic protein binding. Proc Nat Acad Sci USA 98:11353–11358 10.1073/pnas.201367598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon JA, Takada S, Zimmerman LB, Fan CM, Harland RM, McMahon AP (1998) Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev 12:1438–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polinkovsky A, Robin NH, Thomas JT, Irons M, Lynn A, Goodman FR, Reardon W, Kant SG, Brunner HG, van der Burgt I, Chitayat D, McGaughran J, Donnai D, Luyten FP, Warman ML (1997) Mutations in CDMP1 cause autosomal dominant brachydactyly type C. Nat Genet 17:18–19 10.1038/ng0997-18 [DOI] [PubMed] [Google Scholar]
- Schwabe GC, Turkmen S, Leschik G, Palanduz S, Stover B, Goecke TO, Mundlos S (2004) Brachydactyly type C caused by a homozygous missense mutation in the prodomain of CDMP1. Am J Med Genet A 124:356–363 10.1002/ajmg.a.20349 [DOI] [PubMed] [Google Scholar]
- Seemann P, Schwappacher R, Kjaer KW, Krakow D, Lehmann K, Dawson K, Stricker S, Pohl J, Ploger F, Staub E, Nickel J, Sebald W, Knaus P, Mundlos S (2005) Activating and deactivating mutations in the receptor interaction site of GDF5 cause symphalangism or brachydactyly type A2. J Clin Invest 115:2373–2381 10.1172/JCI25118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Huynh TV, Copeland NG, Jenkins NA, Kingsley DM, Lee SJ (1994) Limb alterations in brachypodism mice due to mutations in a new member of the TGF beta-superfamily. Nature 368:639–643 10.1038/368639a0 [DOI] [PubMed] [Google Scholar]
- Storm EE, Kingsley DM (1999) GDF5 coordinates bone and joint formation during digit development. Dev Biol 209:11–27 10.1006/dbio.1999.9241 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Takahashi I, Komatsu M, Sawaishi Y, Higashi K, Nishimura G, Saito H, Takada G (2001) Mutations of the NOG gene in individuals with proximal symphalangism and multiple synostosis syndrome. Clin Genet 60:447–451 10.1034/j.1399-0004.2001.600607.x [DOI] [PubMed] [Google Scholar]
- Thomas JT, Kilpatrick MW, Lin K, Erlacher L, Lembessis P, Costa T, Tsipouras P, Luyten FP (1997) Disruption of human limb morphogenesis by a dominant negative mutation in CDMP1. Nat Genet 17:58–64 10.1038/ng0997-58 [DOI] [PubMed] [Google Scholar]
- Thomas JT, Lin K, Nandedkar M, Camargo M, Cervenka J, Luyten FP (1996) A human chondrodysplasia due to a mutation in a TGF-beta superfamily member. Nat Genet 12:315–317 10.1038/ng0396-315 [DOI] [PubMed] [Google Scholar]
- Wang W, Gu W, Wang Q, Piao Z, Piao YJ (2004) Cloning of integral mature peptide gene of human GDF-5. J Huazhong Univ Sci Technolog Med Sci 24:212–213 [DOI] [PubMed] [Google Scholar]
- Zimmerman LB, De Jesus-Escobar JM, Harland RM (1996) The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell 86:599–606 10.1016/S0092-8674(00)80133-6 [DOI] [PubMed] [Google Scholar]