Abstract
Arabidopsis thaliana mutants prors1-1 and -2 were identified on the basis of a decrease in effective photosystem II quantum yield. Mutations were localized to the 5′-untranslated region of the nuclear gene PROLYL-tRNA SYNTHETASE1 (PRORS1), which acts in both plastids and mitochondria. In prors1-1 and -2, PRORS1 expression is reduced, along with protein synthesis in both organelles. PRORS1 null alleles (prors1-3 and -4) result in embryo sac and embryo development arrest. In mutants with the leaky prors1-1 and -2 alleles, transcription of nuclear genes for proteins involved in photosynthetic light reactions is downregulated, whereas genes for other chloroplast proteins are upregulated. Downregulation of nuclear photosynthetic genes is not associated with a marked increase in the level of reactive oxygen species in leaves and persists in the dark, suggesting that the transcriptional response is light and photooxidative stress independent. The mrpl11 and prpl11 mutants are impaired in the mitochondrial and plastid ribosomal L11 proteins, respectively. The prpl11 mrpl11 double mutant, but neither of the single mutants, resulted in strong downregulation of nuclear photosynthetic genes, like that seen in leaky mutants for PRORS1, implying that, when organellar translation is perturbed, signals derived from both types of organelles cooperate in the regulation of nuclear photosynthetic gene expression.
INTRODUCTION
Photosynthesis is regulated at multiple levels. In the chloroplast itself, rates of photosynthesis are adjusted by posttranslational modification of proteins (Buchanan et al., 1994; Wollman, 2001; Aro and Ohad, 2003), by structural reorganization of the photosynthetic protein complexes (Niyogi, 1999), and by metabolic regulation within and downstream of the Calvin cycle (Paul and Pellny, 2003). Moreover, the expression of plastome-encoded photosynthetic genes is regulated by transcriptional (Pfannschmidt et al., 1999; Pfannschmidt and Liere, 2005), posttranscriptional (Deng and Gruissem, 1987; Rochaix, 2001), and translational (Danon, 1997; Choquet and Wollman, 2002) mechanisms. In addition, plastid-to-nucleus (retrograde) signaling modulates the expression of nuclear genes coding for photosynthetic proteins (Leister, 2005). The concentrations of carbon metabolites (Rolland et al., 2002), components, and/or products of the light phase of photosynthesis, such as tetrapyrroles (Strand et al., 2003), reactive oxygen species (ROS) (Apel and Hirt, 2004), and the redox state of the thylakoid membranes (Pfannschmidt, 2003), all contribute to retrograde signaling. However, not all signals require light, since inhibition of translation in plastids activates a light-independent retrograde signaling pathway (Gray et al., 2002). For instance, in the albostrians mutant of barley (Hordeum vulgare), which is deficient in plastid ribosomes, the expression of nuclear photosynthetic genes is drastically downregulated (Bradbeer et al., 1979; Hess et al., 1991, 1994). Similar effects are observed when light- or dark-adapted wild-type plants are treated with inhibitors of plastid translation (Oelmüller et al., 1986; Adamska, 1995; Gray et al., 1995; Yoshida et al., 1998; Sullivan and Gray, 1999).
Photosynthesis provides substrates for mitochondrial respiration but also depends on a range of compounds synthesized by mitochondria. Thus, the two organelles are metabolically interdependent (Hoefnagel et al., 1998; Raghavendra and Padmasree, 2003). In the dark, mitochondria are the main source of ATP for cellular processes, including those in the chloroplasts. Moreover, in the dark, mitochondrial ATP maintains the proton gradient across the thylakoid membrane, thus protecting the chloroplast from photoinhibition upon reillumination (Gilmore and Bjorkman, 1995). In the light, mitochondria provide the chloroplast with carbon skeletons (derived from the tricarboxylic acid cycle) for 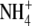 assimilation (Kromer, 1995), while ATP supports various biosynthetic reactions, including the repair and recovery of photosystem II (PSII). Furthermore, mitochondrial respiration protects photosynthesis against photoinhibition by processing redox equivalents exported from chloroplasts (Padmasree et al., 2002). Several pathways contribute to this process, including reactions leading to photorespiration in which the mitochondrial oxidation of Gly ensures the dissipation of excess redox equivalents and generates significant amounts of glycerate for the Calvin cycle (Osmond et al., 1997).
assimilation (Kromer, 1995), while ATP supports various biosynthetic reactions, including the repair and recovery of photosystem II (PSII). Furthermore, mitochondrial respiration protects photosynthesis against photoinhibition by processing redox equivalents exported from chloroplasts (Padmasree et al., 2002). Several pathways contribute to this process, including reactions leading to photorespiration in which the mitochondrial oxidation of Gly ensures the dissipation of excess redox equivalents and generates significant amounts of glycerate for the Calvin cycle (Osmond et al., 1997).
The relationship between mitochondrial function and photosynthesis has been studied using inhibitors of mitochondrial oxidative electron transport (Padmasree and Raghavendra, 1999, 2001). However, published results have been controversial because of the lack of specificity of the inhibitors available. Moreover, analyses of mitochondrion–chloroplast interactions have considered specific mitochondrial mutants, including the non-chromosomal stripe (ncs) mutants of maize (Zea mays), which are defective in mitochondrial genes encoding, for instance, essential subunits of the respiratory electron transport chain (Newton and Coe, 1986; Feiler and Newton, 1987; Newton et al., 1989; Hunt and Newton, 1991). The ncs mutants grow poorly and are characterized by leaves with pale-green or yellow stripes, reflecting defects in chloroplast function. Additional mitochondrial mutants that affect photosynthetic performance include glycine decarboxylase in potato (Solanum tuberosum) (Heineke et al., 2001) and barley (Igamberdiev et al., 2001): here, a reduction in Gly decarboxylase activity impairs photorespiration and leads to excessive reduction and energization of the chloroplast. Furthermore, the tobacco (Nicotiana tabacum) CMSII mutant lacks the major mitochondrial NADH dehydrogenase (Complex I) and exhibits a decrease in the rate of photosynthesis, notably during dark–light transitions or when carbon fixation and photorespiration are simultaneously active (Sabar et al., 2000; Dutilleul et al., 2003a).
Despite the pronounced metabolic interdependence of plastids and mitochondria and the ability of the latter to influence photosynthesis, little is known about the signals that mediate communications between mitochondria and plastids or about the coordination of the activities of the two organelles by signals other than carbon metabolites.
We have identified and characterized mutant alleles of PRORS1, an Arabidopsis thaliana gene coding for a prolyl-tRNA synthetase that is imported into chloroplasts and mitochondria, and used these mutants to elucidate novel aspects of interorganellar crosstalk. Complete absence of PRORS1 disrupts early plant development and causes lethality. Leaky prors1 mutants show defects in photosynthesis due to the simultaneous impairment of translation in plastids and mitochondria. Concomitantly, a specific and marked drop was detected in the levels of transcripts of nuclear genes for proteins involved in the light reactions of photosynthesis. The mutants mrpl11-1 and prpl11-1, which are defective in the ribosomal L11 proteins of mitochondria and plastids, respectively, were employed to separate the contributions of the two organelles to the downregulation of nuclear photosynthetic transcripts. Comparison of the transcript profiles of single prors1, prpl11-1, and mrpl11-1 mutants and the prpl11-1 mrpl11-1 double mutant indicates that plastids and mitochondria are able to generate signals that act synergistically to modulate nuclear photosynthetic gene expression.
RESULTS
Identification and Phenotypic Analysis of Mutants for the PAM15/18 Locus
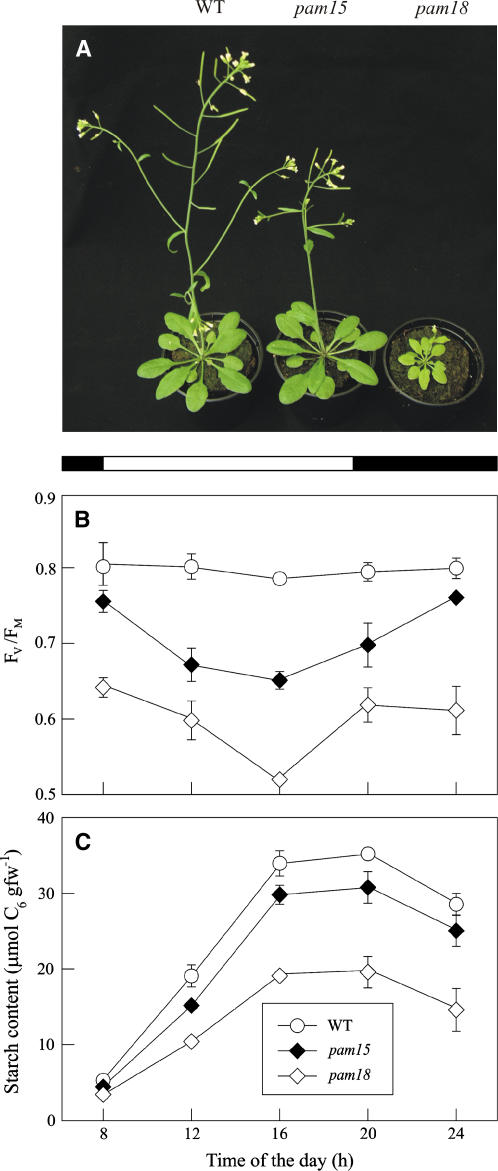
Screening of a collection of T-DNA–mutagenized Arabidopsis lines (ecotype Columbia-0 [Col-0]) for plants with alterations in the effective quantum yield of PSII (ΦII) (Varotto et al., 2000) led to the identification of the mutants photosynthesis altered mutant15 (pam15) and pam18 (Figure 1A). Both photosynthetic phenotypes segregated as monogenic, recessive traits. Crosses between homozygous pam15 and pam18 plants resulted in F1 plants with a pam phenotype, indicating that the same genetic locus was affected in both cases. The phenotype of pam18 was the more severe. Thus, pam18 plants showed a stronger reduction in growth rate than pam15 plants (Figure 1A) and more marked changes in photosynthetic parameters. This is illustrated by the relative decrease in ΦII: wild type, 0.75 ± 0.02; pam15/pam15, 0.58 ± 0.01; pam15/pam18, 0.51 ± 0.02; pam18/pam18, 0.46 ± 0.01. Analysis of chlorophyll fluorescence in light-adapted plants revealed, in addition to the altered ΦII, a significant reduction in the maximum quantum yield of PSII (Fv/Fm) in both mutants (pam15/pam15, 0.65 ± 0.02; pam18/pam18, 0.52 ± 0.01; wild type, 0.79 ± 0.01), implying a defect in energy transfer within PSII. In addition, an increase in the reduction of QA and of the plastoquinone pool, measured as 1-qP, was observed in pam18 leaves (pam15, 0.06 ± 0.02; pam18, 0.09 ± 0.02; wild type, 0.05 ± 0.01). HPLC analysis of light-adapted pam15 and pam18 leaves revealed a decrease in chlorophyll concentration (chlorophyll a+b), indicating a reduction in the abundance of thylakoid antenna proteins (Table 1). Moreover, in leaves of both mutant lines, the reduction in the chlorophyll a/b ratio and the β-carotene content suggested a decrease in levels of PSII core proteins. The relative increase in the level of xanthophyll cycle pigments (violaxanthin + antheraxanthin + zeaxanthin) with respect to neoxanthin and lutein is likely to be the result of increased photooxidative stress in the mutants (see below). This, together with the restoration of photosynthetic efficiency (measured as Fv/Fm) during the night (Figure 1B) and the wild type–like behavior of pam15 plants when grown under low light (data not shown), indicated that illumination exacerbates the molecular defect(s) caused by mutations at the PAM15/18 locus. One direct consequence of the impaired light reactions of photosynthesis in pam15 and pam18 plants on carbohydrate metabolism was a persistent reduction in leaf starch content measured over the course of the day (Figure 1C).
Figure 1.
Phenotypes of Mutant (pam15 and pam18) and Wild-Type Plants.
(A) Six-week-old plants grown in the growth chamber.
(B) and (C) Profiles of maximum quantum yield (Fv/Fm) (B) and leaf starch content (C) during the course of the day. Black bars indicate the time in the dark. Means ± sd of three different experiments are given. fw, fresh weight.
Table 1.
Leaf Pigment Level of Light-Adapted Mutant (pam15 and pam18) and Wild-Type Plants at the Eight-Leaf Rosette Stage
| Pigment/Protein | Wild Type | pam15 | pam18 | Relative Level in pam15 (%)a | Relative Level in pam18 (%)a |
|---|---|---|---|---|---|
| Chlorophyll a/b | 3.03 ± 0.08 | 2.54 ± 0.05 | 2.06 ± 0.08 | 84 | 68 |
| Chlorophyll a+b | 1352 ± 52 | 1122 ± 70 | 676 ± 33 | 83 | 50 |
| Neoxanthin | 49 ± 3 | 41 ± 1 | 31 ± 2 | 84 | 63 |
| Lutein | 190 ± 8 | 149 ± 9 | 107 ± 6 | 79 | 56 |
| β-Carotene | 113 ± 5 | 67 ± 3 | 38 ± 1 | 60 | 34 |
| VAZ | 49 ± 1 | 45 ± 1 | 40 ± 4 | 91 | 82 |
Leaf pigments extracted from pam15, pam18, and wild-type plants were determined by HPLC and reported in nmol g−1 leaf fresh weight. Mean values ± sd are shown. VAZ, violaxanthin + antheraxanthin + zeaxanthin.
Relative values for the mutant genotypes are percentages based on leaf fresh weight.
Expression of Photosynthetic Functions Is Downregulated at Both Protein and RNA Levels in pam15 and pam18
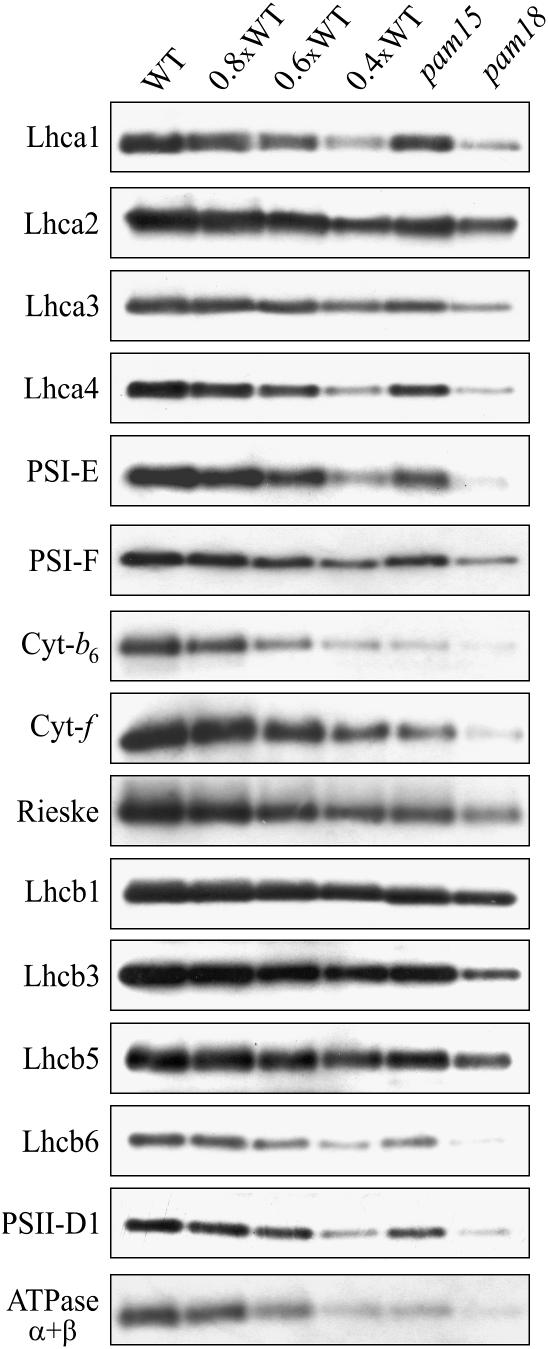
Thylakoid proteins isolated from light-adapted wild-type and mutant plants at the eight-leaf rosette stage were subjected to immunoblot analysis with antibodies specific for proteins of photosystem I (PSI) and PSII, cytochrome b6f, and α- and β-subunits of ATPase (ATPase α+β) (Figure 2). A marked decrease in the amounts of all tested photosynthetic proteins was observed in pam18, while the effects in pam15 were less severe (Table 2). These changes in thylakoid protein composition, which affected both plastid- and nucleus-encoded products (Table 2), can account for the altered kinetics of chlorophyll fluorescence observed in the two mutants.
Figure 2.
Protein Composition of Thylakoid Membranes.
Thylakoid proteins obtained from identical amounts (fresh weight) of light-adapted wild-type and mutant (pam15 and pam18) leaves were fractionated by PAGE. Decreasing levels of wild-type thylakoid proteins were loaded in the lanes marked 0.8xWT, 0.6xWT, and 0.4xWT, and filters were probed with antibodies specific for the proteins indicated on the left. Three independent protein gel blot analyses were performed for each protein.
Table 2.
Photosynthetic Proteins and Corresponding mRNAs in Light-Adapted pam15 and pam18 Plants Compared with the Wild Type
|
pam15 versus WT
|
pam18 versus WT
|
||||
|---|---|---|---|---|---|
| Protein | Gene | mRNA | Protein | mRNA | Protein |
| Lhca1 | At3g54890 | 0.50 | 0.87 | 0.26 | 0.33 |
| Lhca2 | At3g61470 | 0.56 | 0.54 | 0.55 | 0.38 |
| Lhca3 | At1g61520 | 0.46 | 0.56 | 0.35 | 0.34 |
| Lhca4 | At3g47470 | 0.58 | 0.75 | 0.30 | 0.33 |
| Lhcb1 | At1g29920 | 0.58 | 0.73 | 0.34 | 0.34 |
| At1g29910 | 0.47 | 0.30 | |||
| At2g29930 | nd | nd | |||
| At2g34430 | 0.58 | 0.33 | |||
| At2g34420 | 0.53 | 0.32 | |||
| Lhcb2 | At2g05100 | 0.48 | nd | 0.09 | nd |
| At2g05070 | 0.49 | 0.08 | |||
| At3g27690 | 1.02ns | 0.56 | |||
| Lhcb3 | At5g54270 | 0.59 | 0.74 | 0.23 | 0.22 |
| Lhcb4 | At5g01530 | 0.92ns | nd | 0.68 | nd |
| At3g08940 | 0.72 | 0.44 | |||
| At2g40100 | 1.49 | 0.98ns | |||
| Lhcb5 | At4g10340 | 0.66 | 0.77 | 0.32 | 0.38 |
| Lhcb6 | At1g15820 | 0.63 | 0.73 | 0.42 | 0.22 |
| PSI-E | At4g28750 | 0.65 | 0.55 | 0.54 | 0.21 |
| At2g20260 | 0.72 | 0.72 | |||
| PSI-F | At1g31330 | 0.42 | 0.67 | 0.39 | 0.34 |
| Rieske protein | At4g03280 | nd | 0.51 | nd | 0.27 |
| PSII-D1a | psbA | 1.85 | 0.53 | 2.10 | 0.21 |
| Cytochrome b6a | petB | nd | 0.39 | nd | 0.22 |
| Cytochrome fa | petA | nd | 0.39 | nd | 0.24 |
| ATPase α+βa | atpA | nd | 0.49 | nd | 0.25 |
| atpB | nd | nd | |||
| RbcLa | rbcL | 0.70 | nd | 1.29 | nd |
Average values were from three independent protein gel blot (see Figure 2), DNA array, and, in the case of psbA, RNA gel blot hybridizations. nd, not determined; ns, not significant; WT, wild type.
Proteins encoded by plastid genes.
To check whether the decrease in the level of nuclear-encoded thylakoid proteins was associated with downregulation of their corresponding genes, DNA array analysis (Kurth et al., 2002; Richly et al., 2003) was performed on a set of 3292 nuclear genes, most of which code for chloroplast-targeted proteins. Comparison of the data obtained from the protein and microarray analyses revealed covariation of photosynthetic proteins and their transcripts in both pam mutants (Table 2) (i.e., the decrease in protein abundance was reflected in reductions in mRNA levels, and the more prominent decrease in photosynthetic protein amounts in pam18 was associated with a more pronounced reduction in transcript levels).
Together, these findings suggest that the reduced amounts of nuclear-encoded photosynthetic proteins seen in the chloroplasts of pam15 and pam18 plants are associated with lower levels of the transcripts of the corresponding nuclear genes.
The Leaky Mutations pam15 and pam18 Are Due to T-DNA Insertions in the 5′-Untranslated Region of PRORS1
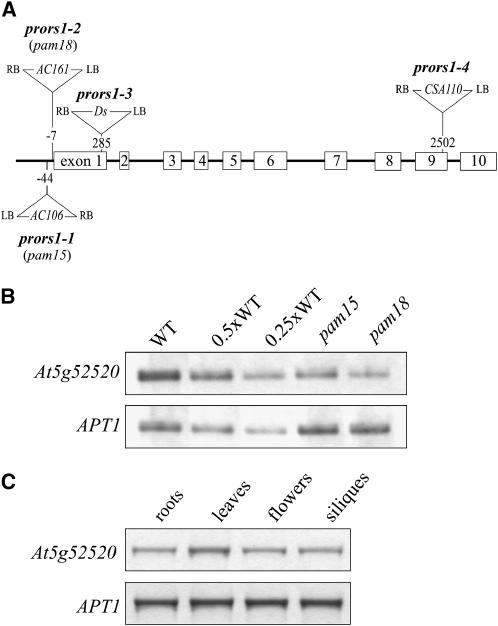
DNA gel blot analyses of populations of 50 plants each, segregating for either pam15 or pam18, were performed using the 5′-ends of the AC106 and the AC161 T-DNAs, respectively, as probes (data not shown). In both cases, only one T-DNA copy that cosegregated with the mutant allele was present. Isolation of genomic sequences flanking the T-DNA termini (Pesaresi et al., 2001) allowed the identification of the insertion sites. Both T-DNAs were inserted in the 5′-untranslated region (UTR) of the gene At5g52520 at positions −44 (pam15) and −7 (pam18) relative to the start codon (Figure 3A). Quantitative RT-PCR (QRT-PCR) analysis showed that the T-DNA insertion in pam15 resulted in a 50% reduction in At5g52520 transcripts and in a 75% reduction in pam18 (Figure 3B).
Figure 3.
T-DNA Tagging and Expression Analysis of the PRORS1 Gene.
(A) The prors1-1, -2, and -4 mutations are due to T-DNA insertions in the PRORS1 locus. In the prors1-3 allele, a copy of the Ds transposon is inserted in the first exon. Insertions are not drawn to scale. LB, left border; RB, right border.
(B) Detection of PRORS1 transcripts in light-adapted mutant (prors1-1 and prors1-2) and wild-type plants. The analysis on 4.5% polyacrylamide gels was performed with 33P-labeled products of QRT-PCR obtained from leaves after PCR for 25 cycles with PRORS1-specific primers and control primers for the APT1 gene. Decreasing levels of wild-type single-stranded cDNA were used in the lanes marked 0.5xWT and 0.25xWT.
(C) Expression of PRORS1 mRNA in different plant organs detected by QRT-PCR performed as in (B).
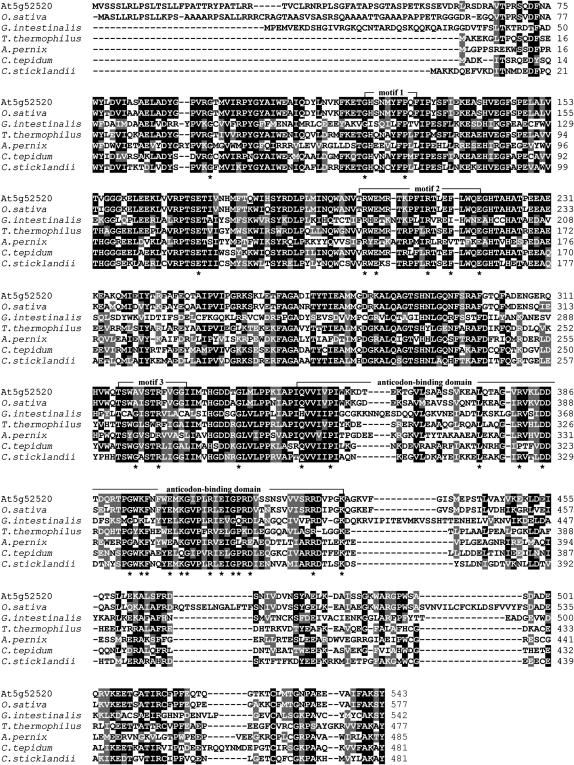
Database analysis of the Arabidopsis genome revealed that At5g52520 is a single-copy gene. Its coding sequence is entirely covered by overlapping ESTs, and transcripts of the gene were detected in green tissues and roots (Figure 3C). The At5g52520 protein contains 543 amino acids and shares significant sequence identity with known prolyl-tRNA synthetases (ProRSs) from eukaryotes and prokaryotes, including Thermus thermophilus (identity/similarity of 55.8/64.2%) (Yaremchuk et al., 2001), Giardia intestinalis (36.7/46.9%) (Bunjun et al., 2000), Aeropyrum pernix (38.5/50.7%) (Yokozawa et al., 2003), and putative ProRS proteins from Oryza sativa (77.8/82.9%), Chlorobium tepidum (53.8/62.3%), and Clostridium sticklandii (52.4/62.1%) (Figure 4). All of these proteins have the catalytic domain (motifs 1 to 3; Eriani et al., 1990) and anticodon binding motif (Cusack et al., 1998) characteristic of class IIa aminoacyl-tRNA synthetases (aaRSs) (reviewed in Ibba and Soll, 2000). Because protein synthesis occurs in three different compartments (cytosol, mitochondria, and chloroplasts) in plants, we searched for additional ProRS sequences in the genome of Arabidopsis. The genes At3g62120 and At5g10880 code for proteins that show moderate homology to At5g52520 (identity/similarity of 43.7/54.8% and 38.2/47.3%, respectively) and are more closely related to eukaryotic bifunctional glutamyl-prolyl-tRNA synthetases.
Figure 4.
Comparison of ProRS Sequences from Eukaryotes and Prokaryotes.
The amino acid sequence of the PRORS1 protein (At5g52520) was compared with ProRS sequences from O. sativa, G. intestinalis (Bunjun et al., 2000), T. thermophilus (Yaremchuk et al., 2001), A. pernix (Yokozawa et al., 2003), and C. tepidum and C. sticklandii. Black boxes indicate strictly conserved amino acids and gray boxes closely related ones. Characteristic conserved sequence stretches in ProRS proteins (motifs 1 to 3, anticodon binding domain) are indicated according to Eriani et al. (1990), Cusack et al. (1998), and Ibba and Soll (2000); the asterisks refer to invariant residues that are typical of ProRSs (Stehlin et al., 1998).
The findings described above indicate that the pam15 and pam18 phenotypes are due to the altered transcription of the At5g52520 gene, prompting us to designate At5g52520 as PRORS1, the pam15 mutant as prors1-1, and the pam18 allele as prors1-2.
Complete Loss of PRORS1 Function in the Knockout Mutants prors1-3 and -4 Affects Female Gametogenesis
Because the prors1-1 and -2 mutations allowed residual transcription of the At5g52520 gene, we searched the Cold Spring Harbor Genetrap Database (http://genetrap.cshl.org) for possible null alleles of PRORS1 and identified the GT256 line (ecotype Landsberg erecta) as a candidate. This line, designated prors1-3, carries a Ds element in the first exon of PRORS1 at position +285 relative to the start codon (Figure 3A). In addition, the Garlic_420_E02.b.1a.Lb3Fa line was identified in the Syngenta Arabidopsis Insertion Library (Sessions et al., 2002). In this line (ecotype Col-0), designated prors1-4, the CSA110 T-DNA was inserted in the ninth exon at position +2502 of PRORS1 (Figure 3A).
At least 500 progeny plants obtained by self-fertilizing prors1-3/PRORS1 and prors1-4/PRORS1 plants, either grown on soil or in sterile culture, were genotyped by PCR. No homozygous prors1-3 or -4 plants were identified, implying that disruption of the PRORS1 gene causes lethality. Adult prors1-3/PRORS1 and prors1-4/PRORS1 plants behaved like the wild type with respect to plant size, leaf pigmentation, and photosynthetic performance. Crosses between prors1-3/PRORS1 or prors1-4/PRORS1 plants and prors1-1 or prors1-2 plants resulted in plants that were more severely affected in growth and photosynthesis than homozygous prors1-1 or -2 plants, indicating that a direct correlation between transcript, protein abundance, and phenotype exists in these diploids.
Segregation analyses based on PCR genotyping were performed on the progeny of self-fertilized prors1-3/PRORS1 and prors1-4/PRORS1 plants and revealed that the numbers of heterozygous plants were lower than expected for a 2:1 (heterozygote:wild type) segregation ratio (Table 3). The progenies derived from PRORS1/PRORS1 (♀) × prors1-3/PRORS1 (♂) and PRORS1/PRORS1 (♀) × prors1-4/PRORS1 (♂) crosses comprised homozygous wild-type and heterozygous plants in proportions that were not significantly different from a 1:1 ratio. However, a marked deviation from the expected 1:1 ratio was observed in the reciprocal crosses, in which heterozygous mutants were fertilized with pollen from homozygous wild-type plants, implying that PRORS1 is necessary for normal development of the female gametophyte.
Table 3.
Segregation Analysis of Progenies Derived from Different prors1-3 Crosses Based on PCR Genotyping of Adult Plants
| Genotypes of Adult Plants (Observed/Expected)
|
||||
|---|---|---|---|---|
| Progeny | PRORS1/PRORS1 | prors1-3/PRORS1 | Total | χ2 |
| F1 of selfed prors1-3/PRORS1 | 295/190 | 275/380 | 570 | 87.00* |
| F1 of prors1-3/PRORS1 (♀) × PRORS1/PRORS1 (♂) | 174/111 | 48/111 | 222 | 71.40** |
| F1 of PRORS1/PRORS1 (♀) × prors1-3/PRORS1 (♂) | 111/105 | 99/105 | 210 | 0.68** |
Because homozygous mutants are supposed to be lethal at the embryo stage, χ2 values for a heterozygote:homozygous wild type segregation of 2:1 (*) and 1:1 (**) were calculated based on two genotypic classes (1 degree of freedom). The 95% confidence limit for rejecting the expected 2:1 or 1:1 segregation is ≥3.84 and the 99% limit ≥6.64. The prors1-4 mutation had similar segregation ratios.
Development of the Embryo Sac and the Embryo Is Arrested in Null Mutants for PRORS1
We used scanning electron microscopy to analyze prors1-3/PRORS1 siliques at different developmental stages. Siliques containing seeds at the globular stage of normal embryo development were found to include small abnormal seeds and aborted ovule-like structures (Figures 5A and 5B). Visual inspection of ∼300 siliques at this stage showed that, in all cases, two to four abnormal seeds (corresponding to 4 to 8% of the total structures present) were observed per silique, whereas the frequency of the aborted ovule-like structures ranged from 20 to 30%. In younger siliques, abnormal seeds were difficult to distinguish from the wild type, whereas in older siliques, a fraction of seeds had disintegrated or were in the process of disintegrating. To further examine the phenotypic consequences of absence of the PRORS1 protein during seed development, 94 abnormal seeds were investigated as cleared whole mounts. Microscopy comparison revealed that all of the abnormal embryos scored were arrested at preglobular stages (Figures 5C and 5D; data not shown). In addition, the endosperm of abnormal seeds was drastically reduced (Figures 5C and 5D). Identical results were obtained for siliques of self-fertilized heterozygous prors1-4 plants. These observations indicate that the PRORS1 protein is necessary for normal seed development. In combination with the results of the PCR genotyping of the progenies of self-fertilized heterozygous prors1 knockout plants (see previous section), the findings allowed us to conclude that absence of the PRORS1 protein results in lethality prior to the seedling stage.
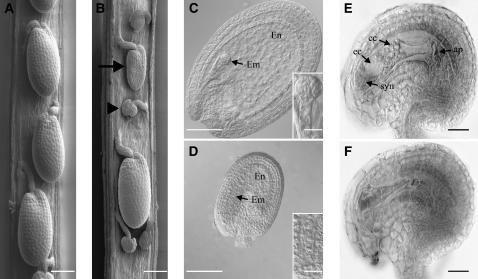
Figure 5.
Effects of Loss of PRORS1 Function on Early Plant Development.
(A) and (B) Wild-type (A) and prors1-3/PRORS1 (B) siliques at the globular stage of normal embryo development analyzed by scanning electron microscopy. In prors1-3/PRORS1 siliques, abnormal seeds (arrow) and aborted ovule-like structures (arrowhead) are visible. Bars = 100 μm.
(C) Cleared whole mount of wild-type seed containing an embryo (Em) at the globular stage (see inset) and the large endosperm (En).
(D) Cleared whole mount of an abnormal seed containing an embryo at the single cell stage (see inset) and a reduced endosperm. Bars in (C) and (D) = 100 μm; inset bars = 20 μm.
(E) and (F) Optical sections of cleared whole mounts of wild-type (E) and abnormal (F) ovules stained with Mayer's hemalaun. The wild-type ovule exhibits the two synergids (syn), the egg cell (ec), the central cell (cc) with its large vacuole and its fused polar nuclei, and the antipodal cells (ap), which have already degenerated. Bars = 20 μm.
The images in (C) to (F) were taken with a Zeiss Axiophot microscope equipped with differential interface contrast optics.
To further characterize the aborted ovule-like structures, optical sections of cleared, whole-mount ovules at stages 3-I and 3-VI (Schneitz et al., 1995) were stained with Mayer's hemalaun. At stage 3-I, only wild type–like ovules carrying the degenerating tetrad and the mononuclear embryo sac were observed in siliques of self-fertilized heterozygous prors1-3 plants, indicating that early megaspore development and meiosis proceed normally (data not shown). By contrast, of the 879 ovules scored at stage 3-VI, only ∼70% had a normal-looking seven-nuclear embryo sac (Figure 5E); the others had a patch of rather nondescript tissue in its place (Figure 5F). Because formation of a mononuclear embryo sac was always observed, this patch is likely to represent an embryo sac that failed to develop any further and degenerated. While these data indicate that the PRORS1 protein is essential for efficient embryo sac development, a fraction of the mutant embryo sacs look like the wild type, indicating incomplete penetrance of the mutation. Since 50% of the ovules in siliques of self-fertilized heterozygous prors1-3 and -4 plants should carry the wild-type allele, but ∼70% of the ovules scored at stage 3-VI had a normal-looking seven-nuclear embryo sac, we conclude that ∼40% of mutant ovules behave like the wild type. Indeed, these mutant ovules can be still fertilized by wild-type pollen and develop into embryos that are heterozygous for the mutation, which in turn give rise to the unexpected ratio of homozygous wild-type to heterozygous plants when fertilized with pollen from homozygous wild-type plants (as reported in the previous section). Conversely, complete absence of the PRORS1 protein (prors1-3 or -4 ovules fertilized by mutant pollen grains) leads to development arrest at preglobular stages.
PRORS1 Is Targeted to Both Mitochondria and Chloroplasts
The Arabidopsis PRORS1 and O. sativa ProRS proteins share a long N-terminal sequence that is not found in prokaryotic ProRSs (see Figure 4) and may function as a targeting peptide. Computational prediction of subcellular localization by the ChloroP (Emanuelsson et al., 1999), TargetP (Emanuelsson et al., 2000), and PCLR (Schein et al., 2001) programs suggested a chloroplast localization for ProRS1, while Predotar (version 1.03 at www.inra.fr/predotar/), MITOPROT (Claros and Vincens, 1996), PSORT (Nakai and Kanehisa, 1992), and iPSORT (Bannai et al., 2002) predicted a mitochondrial localization. Because Christensen et al. (2005) highlighted the importance of 5′-UTRs for correct protein targeting, we considered the possibility that the PRORS1 5′-UTR could be translated. The presence of a stop codon just before the predicted ATG start codon, however, indicated that no alternative translation initiation site is present in PRORS1. Taken together, it seems to be clear that PRORS1 has an N-terminal sequence that might be recognized as an import signal by both chloroplasts and mitochondria. Indeed, proteins involved in organellar gene expression, including aaRSs, have previously been reported to be targeted to both organelles (for review, see Peeters and Small, 2001; Silva-Filho, 2003).
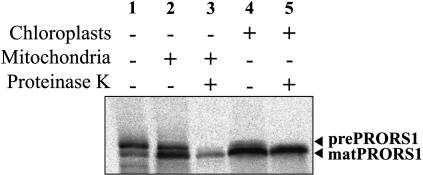
To experimentally investigate the subcellular localization of PRORS1, the precursor protein was synthesized and radiolabeled in vitro and mixed with import-competent mitochondria and chloroplasts according to the procedure described by Rudhe et al. (2002). After incubation to allow for import and digestion of the nonimported precursor by proteinase K, organelles were recovered by centrifugation on a Percoll gradient, and the imported products were analyzed (Figure 6). The results clearly show that the PRORS1 protein can be imported into both chloroplasts and mitochondria (Figure 6). Recently, a systematic analysis of the targeting of organellar aaRSs in Arabidopsis has confirmed the mitochondrial and plastid localization of PRORS1 (Duchene et al., 2005).
Figure 6.
In Vitro Import of PRORS1 into Mitochondria and Chloroplasts.
The precursor protein was incubated together with mitochondria and chloroplasts from spinach (Spinacia oleracea) leaf. Proteinase K (15 μg/mL) was added to degrade extraorganellar proteins where indicated. Mitochondria and chloroplasts were reisolated on a 4% Percoll gradient after the incubation as described by Rudhe et al. (2002). Lane 1, precursor protein alone; lane 2, mitochondrial fraction; lane 3, as lane 2 except that the import reaction was treated with proteinase K; lane 4, chloroplast fraction; lane 5, as lane 4 except that the import reaction was treated with proteinase K. The prefixes “pre” and “mat” designate the precursor and mature forms of the corresponding protein, respectively. For controls of the specificity of the import assay, see Supplemental Figure 3 online.
A Decrease in PRORS1 Level Is Associated with Partial Loss of Proteins of the Mitochondrial Respiratory Chain and Perturbs Protein Synthesis in Plastids
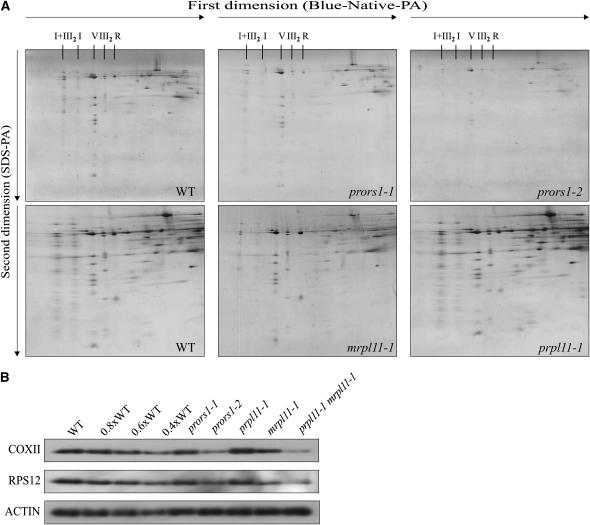
The role of PRORS1 in mitochondrial function was investigated by analyzing the polypeptide composition of the respiratory chain in prors1-1 and -2 mitochondria. Digitonin-solubilized mitochondria were fractionated by Blue-native PAGE in the first dimension and by SDS-PAGE in the second dimension (Figure 7A). A decrease in the abundance of inner membrane complexes was noted in both prors1-1 (15% reduction) and prors1-2 (50% reduction) mitochondria. The levels of all of the visualized complexes were decreased to the same extent, implying a general impairment of oxidative phosphorylation. Similar results were obtained by probing total protein extracts with antibodies raised against the mitochondrial encoded cytochrome oxidase II subunit (COXII) and the ribosomal protein S12 (RPS12) (Figure 7B).
Figure 7.
Two-Dimensional Resolution of Mitochondrial Protein Complexes.
(A) Digitonin-solubilized mitochondrial protein complexes and supercomplexes from light-adapted wild-type, prors1-1, prors1-2, mrpl11-1, and prpl11-1 leaves were fractionated by Blue-native PAGE (Blue-Native-PA) in the first dimension and SDS-PAGE (SDS-PA) in the second. The protein complexes are designated as follows: I, NADH dehydrogenase; III2, cytochrome c reductase (functional dimer); V, ATP synthase; I+III2, supercomplex comprising complex I and dimeric complex III. R, ribulose-1,5-bis-phosphate carboxylase/oxygenase contaminant. Note that mitochondrial analyses in the three top panels (WT, prors1-1, and prors1-2) were performed separately from analyses in the bottom panels (WT, mrpl11-1, and prpl11-1), making it necessary to use two wild-type controls.
(B) Protein gel blot analysis of total protein extracts from wild-type and mutant plants. Antibodies raised against mitochondrial COXII and mitochondrial RPS12 were used. An antibody raised against ACTIN was employed to control for equal loading.
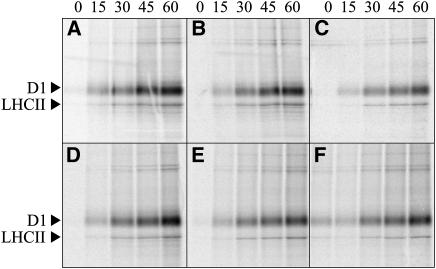
To measure the rates of protein synthesis in mutant and wild-type plastids, in vivo translation assays were performed (Figures 8A to 8C). Leaves were collected from plants at the eight-leaf rosette stage, incubated with [35S]Met and illuminated for different times (0, 15, 30, 45, and 60 min). Subsequently, thylakoid membranes were isolated and polypeptides fractionated by SDS-PAGE. Accumulation of labeled D1 protein was reduced by ∼20% in prors1-1 and by ∼70% in prors1-2 plants.
Figure 8.
Analysis of Protein Synthesis in Plastids from Wild-Type and Mutant Plants.
Autoradiogram of thylakoid membrane proteins isolated from wild-type (A), prors1-1 (B), prors1-2 (C), mrpl11-1 (D), prpl11-1 (E), and prpl11-1 mrpl11-1 (F) leaves, after being pulse-labeled with [35S]Met for 0, 15, 30, 45, and 60 min and subsequently fractionated by SDS-PAGE.
mRNA Expression Profiles in the Leaky prors1 Mutant Reveal Specific Downregulation of Photosynthetic Genes
To study the effects of the altered functional state of mitochondria and chloroplasts on nuclear gene expression, the data obtained with the 3292-gene DNA array, used for the analysis of transcript abundance of nuclear genes encoding photosynthetic proteins (see above), were evaluated in more detail. Because in both leaky mutants the severity of the photosynthetic lesions seen in the light and dark differed (see Figure 1B), expression profiles were analyzed in light- and dark-adapted plants (see Methods). In light-adapted prors1-1 plants, 2170 genes displayed significant differential expression relative to wild-type plants. A similar number of genes (2124) showed differential expression in light-adapted prors1-2 plants. Direct comparison of the prors1-1 and -2 expression profiles showed that 1805 genes were differentially expressed in both genotypes, and 1763 of these showed the same trend in expression (1631 upregulated and 132 downregulated), as expected for mutations that reduce the expression of the same gene. In dark-adapted mutant plants, slightly fewer genes displayed significant differential expression (prors1-1, 2090; prors1-2, 1862). Both the number of genes differentially expressed in both genotypes (1286) and the fraction of genes exhibiting the same trend in expression (1219) were smaller than in light-grown plants (the entire set of data is available online at the Gene Expression Omnibus repository, http://www.ncbi.nlm.nih.gov/geo/; see Methods for accession numbers).
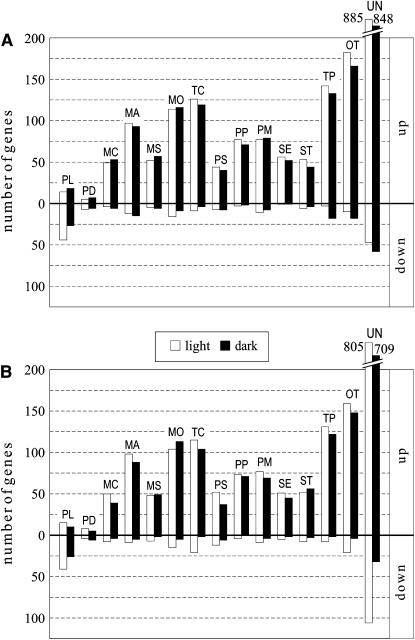
When the differentially expressed genes were grouped into 15 functional categories (Figure 9), 80 to 90% of the genes in each category were upregulated in both mutants, irrespective of whether the plants were light- or dark-adapted. By contrast, in light-adapted mutant plants ∼70% of genes coding for proteins involved in the light reactions of photosynthesis, including antenna and photosystem core proteins, were predominantly downregulated. This downregulation of photosynthetic genes goes against the general trend and was noted also in dark-adapted plants (Figure 9).
Figure 9.
Effects of the Leaky prors1-1 and -2 Mutations on the Expression of Nuclear Genes.
An array bearing sequence tags derived from 3292 nuclear genes, most of them coding for chloroplast proteins, was used to determine the levels of the corresponding transcripts in the prors1-1 (A) and -2 (B) mutants relative to the wild type. Expression ratios were determined in light- (open bars) and dark-adapted (closed bars) plants. Total RNA isolated from mutant and wild-type leaves was used to probe the DNA arrayed on nylon filters. Only genes that showed significant differential expression in prors1-1 and -2 relative to the wild type were considered (see Methods). Upregulated and downregulated genes were grouped into 15 functional categories: PL, photosynthesis/light reactions; PD, photosynthesis/dark reactions; MC, carbohydrate metabolism; MA, amino acid metabolism; MS, secondary metabolism; MO, other metabolic functions; TC, transcription; PS, protein synthesis; PP, protein phosphorylation; PM, protein modification and fate; SE, sensing, signaling, and cellular communication; ST, stress response; TP, transport; OT, other functions; UN, unknown function.
Taken together, the mRNA profiling data indicate that the mitochondrial and chloroplast defects caused by partial loss of PRORS1 result in downregulation of genes for the light reactions of photosynthesis. This type of regulation is seen in both light- and dark-adapted prors1 plants, indicating that the transcriptional response is independent of light and of photooxidative stress.
Mutational Dissection of the Specific Contributions of Mitochondria and Chloroplasts to the Modulation of Nuclear Photosynthetic Gene Expression
To investigate the specific roles of protein synthesis in mitochondria and chloroplasts in the regulation of nuclear photosynthetic gene expression, we first searched for Arabidopsis mutants that are specifically affected in the function of either mitochondrial or plastid ribosomes. The mutational analysis of plastid and mitochondrial ribosomal proteins has the advantage that these polypeptides are known to be specifically targeted to the respective organelle.
Screening of the insertion flanking database SIGnAL (http://signal.salk.edu/cgi-bin/tdnaexpress) led to the isolation of the Salk_090016 line (ecotype Col-0), which carries a T-DNA element inserted at position +484 relative to the start codon of the single-copy gene At4g35490 (see Supplemental Figure 1A online). This line, designated as mrpl11-1, shows a 90% reduction in MRPL11 transcripts (see Supplemental Figure 1B online) coding for the protein MRPL11, a component of the 50S subunit of the mitochondrial ribosome. The MRPL11 protein contains 155 amino acid residues, shares significant sequence homology with other plant and prokaryotic L11 proteins (see Supplemental Figure 2 online; Porse et al., 1999; Yamaguchi and Subramanian, 2000), and is specifically targeted to mitochondria (see Supplemental Figure 3A online).
As a consequence of the marked reduction of MRPL11 gene expression, mutant plants are reduced in size and show a darker leaf coloration than the wild type (Figure 10D). Agrobacterium tumefaciens–mediated transformation of mrpl11-1 plants with MRPL11 cDNA fused to the 35S promoter of Cauliflower mosaic virus fully restored wild-type leaf coloration and growth. Analysis of the mitochondrial electron transport chain revealed a marked and general decrease in the abundance of the protein complexes in mutant plants with respect to the wild type (Figure 7A), implying that mitochondrial activity is altered. The reduction in mitochondrial protein abundance was confirmed by protein gel blot analysis of total protein extracts (Figure 7B). However, mrpl11-1 plants showed essentially normal photosynthetic performance, as indicated by the parameters Fv/Fm (mrpl11-1, 0.80 ± 0.02; wild type, 0.79 ± 0.01), ΦII (mrpl11-1, 0.75 ± 0.02; wild type, 0.75 ± 0.02), and 1-qP (mrpl11-1, 0.05 ± 0.01; wild type, 0.05 ± 0.01). Moreover, no alteration in plastid translation rate (Figure 8D) or thylakoid protein composition (Figure 11B) was detectable in the mutant. Interestingly, the absence of any photosynthetic phenotype in mrpl11 leaves is in contrast with the evident chloroplast alterations observed in the maize ncs and tobacco cytoplasmic male sterility (cms) plants. However, while the latter mutants were impaired in specific subunits of the mitochondrial electron transport chain leading to a block in the electron transport flow, the mrpl11 leaves show a reduction in the abundance of all mitochondrial electron transport complexes that limits but does not arrest at any point the electron flow. Evidently, this limited electron transport in mrpl11 leaves is still able to fully support chloroplast activities.
Figure 10.
Phenotypes of Mutant and Wild-Type Plants.
Four-week-old wild-type and mutant plants propagated in the growth chamber.
(A) Wild-type Col-0.
(B) prors1-1.
(C) prors1-2.
(D) mrpl11-1.
(E) prpl11-1.
(F) prpl11-1 mrpl11-1.
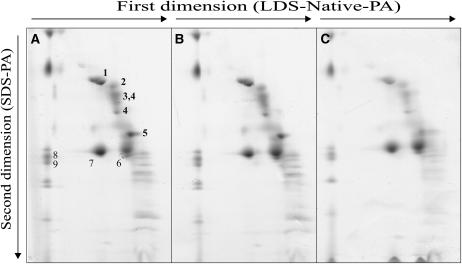
Figure 11.
Protein Composition of Thylakoid Membranes.
Thylakoid proteins from identical amounts (fresh weight) of light-adapted wild-type (A), mrpl11-1 (B), and prpl11-1 mrpl11-1 (C) leaves were fractionated first by electrophoresis on a nondenaturing lithium dodecyl sulfate polyacrylamide (LDS-Native-PA) gel and then on a denaturing SDS-polyacrylamide (SDS-PA) gel. Positions of wild-type thylakoid proteins identified previously by protein gel blot analyses with appropriate antibodies are indicated (Pesaresi et al., 2001): 1, α- and β-subunits of the ATPase complex; 2, D1-D2 dimer; 3, CP47; 4, CP43; 5, oxygen-evolving complex; 6, LHCII monomer; 7, LHCII trimer; 8, PSI-D; 9, PSI-F.
The prpl11-1 mutation affects the plastid counterpart of MRPL11 and has been described previously (Pesaresi et al., 2001). PRPL11 is a component of the 50S subunit of the chloroplast ribosome. Its amino acid sequence contains a typical chloroplast transit peptide (see Supplemental Figure 2 online), and the protein is specifically imported into chloroplasts (see Supplemental Figure 3B online; Aronsson and Jarvis, 2002). The prpl11-1 plants have pale-green leaves and are drastically reduced in size in comparison with the wild type (Figure 10E). Analysis of leaf chlorophyll fluorescence revealed a marked reduction of Fv/Fm in mutant plants (prpl11-1, 0.69 ± 0.01; wild type, 0.79 ± 0.01), implying a defect in energy transfer within PSII. In addition, the ΦII value is markedly decreased in prpl11-1 (0.55 ± 0.01 versus 0.75 ± 0.02 in the wild type), whereas the reduction state of the plastoquinone pool (1-qP) is only slightly increased (prpl11-1, 0.06 ± 0.01; wild type, 0.05 ± 0.01). Two-dimensional PAGE analysis revealed a drastic reduction (between 40 and 50%) in the level of plastome-encoded thylakoid proteins (Pesaresi et al., 2001) as a consequence of the impairment of translation in the plastids (Figure 8E). Analysis of the polypeptide composition of the mitochondrial electron transport chain, however, did not show any major difference between prpl11-1 and wild-type plants (Figures 7A and 7B), indicating that the mitochondrial activity remains unaffected.
The prpl11-1 mrpl11-1 double mutant, generated by crosses between prpl11-1 and mrpl11-1 plants, exhibited pale-green leaves and a drastic reduction in size, like prpl11-1 (Figure 10F). Furthermore, the photosynthetic performance of the double mutant is identical to that of prpl11-1 plants (Fv/Fm, 0.69 ± 0.03 versus 0.79 ± 0.01 in the wild type; ΦII, 0.53 ± 0.03 versus 0.75 ± 0.02 in the wild type; 1-qP, 0.06 ± 0.01 versus 0.05 ± 0.01 in the wild type), implying that the mitochondrial defect does not have an additive effect on plastid function. As a matter of fact, the reduction in the plastid translation rate (Figure 8F) and the concomitant drop in the abundance of the major thylakoid photosynthetic complexes to between 40 and 60% of wild-type levels in the double mutant were similar to the decrease reported in prpl11-1 leaves (Figure 11C).
Thus, while mrpl11-1 specifically impairs mitochondrial gene expression without affecting photosynthesis, the converse is true of the prpl11-1 mutation. The prpl11-1 mrpl11-1 double mutants (like prors1 plants) are defective in both mitochondrial (Figure 7B) and plastid gene expression.
Chloroplasts and Mitochondria Act Synergistically in Regulating the Expression of Photosynthetic Genes
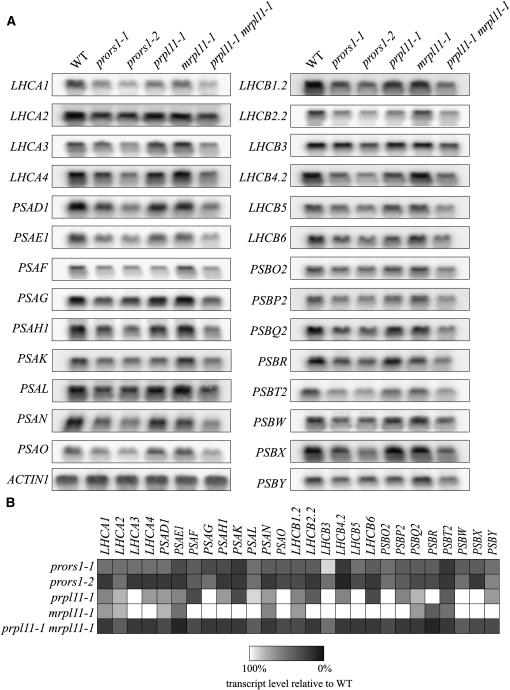
The prpl11-1 and mrpl11-1 mutants, specifically affected in either chloroplast or mitochondrial activities, and the prpl11-1 mrpl11-1 and the prors1 mutants, in which protein synthesis in both organelles is impaired, represent valuable genetic tools that allow us to assess the relative contributions of chloroplasts and mitochondria to the regulation of nuclear photosynthetic genes. Probes for 13 nuclear genes encoding subunits of PSI and 14 nuclear genes encoding subunits of PSII (Figure 12A) were hybridized to RNA gel blots loaded with RNA isolated from the genotypes listed above. A marked downregulation of the entire set of these genes was observed in prors1-1 and prors1-2 plants, confirming the trend in expression revealed by DNA array hybridization (Table 2, Figure 12).
Figure 12.
Expression Analysis of Nuclear Photosynthetic Genes.
(A) Forty-microgram samples of total RNA from light-adapted wild type, prors1-1, prors1-2, prpl11-1, mrpl11-1, and prpl11-1 mrpl11-1 mutants were size-fractionated by agarose gel electrophoresis, transferred to nitrocellulose filters, and probed with cDNA fragments specific for transcripts encoding PSI (LHCA1, -2, -3, and -4 and PSAD1, -E1, -F, -G, -H1, -K, -L, -N, and -O) and PSII (LHCB1.2, -2.2, -3, -4.2, -5, and -6 and PSBO2, -P2, -Q2, -R, -T2, -W, -X, and -Y) subunits. To control for RNA loading, the blots were reprobed with a cDNA fragment derived from the ACTIN1 gene. Three independent RNA gel blot analyses were performed for each photosystem gene.
(B) Quantification of transcript levels determined in (A). The color/shading code indicates the levels of transcripts relative to the wild type.
In the prpl11-1 mutant, transcripts of only 18 of the 27 genes analyzed were downregulated, most of them accumulating to 60 to 80% of wild-type levels; the exceptions were PSAF, PSAK, LHCB2.2, LHCB6, and PSBT2, which were more strongly affected. Levels of transcripts for the antenna proteins Lhca3, Lhcb3, and Lhcb5, the G and O subunits of PSI (PsaG and PsaO), the O subunit of the oxygen-evolving complex (PsbO2), and the low molecular weight PSII subunits PsbR, PsbW, and PsbX were not affected by the prpl11-1 mutation. In mrpl11-1 leaves, fewer genes were downregulated: five encoding subunits of PSI (Lhca1, Lhca2, PsaD1, PsaE1, and PsaN) and four coding for subunits of PSII (Lhcb1.2, PsbQ2, PsbR, and PsbT2). Moreover, the abundance of transcripts was reduced by only 20 to 40%, implying that alterations in mitochondrial activity alone do not have a major influence on the transcription of nuclear photosynthetic genes. However, when mitochondrial and chloroplast functions are simultaneously impaired, as in the case of the prpl11-1 mrpl11-1 double mutant, all the nuclear photosynthetic genes analyzed were markedly downregulated to a similar extent to that seen in prors1 plants. Thus, in prpl11-1 mrpl11-1 leaves, transcript levels of most of the genes tested were reduced by up to 80%.
Taken together, the data clearly demonstrate that only the concomitant impairment of mitochondrial and plastid protein synthesis, in prors1 mutants and the prpl11 mrpl11 double mutant, results in a drastic downregulation of most of the nuclear photosynthetic genes. This suggests the existence of a synergistic contribution of plastid and mitochondrial protein synthesis to the transcriptional regulation of nuclear photosynthetic genes.
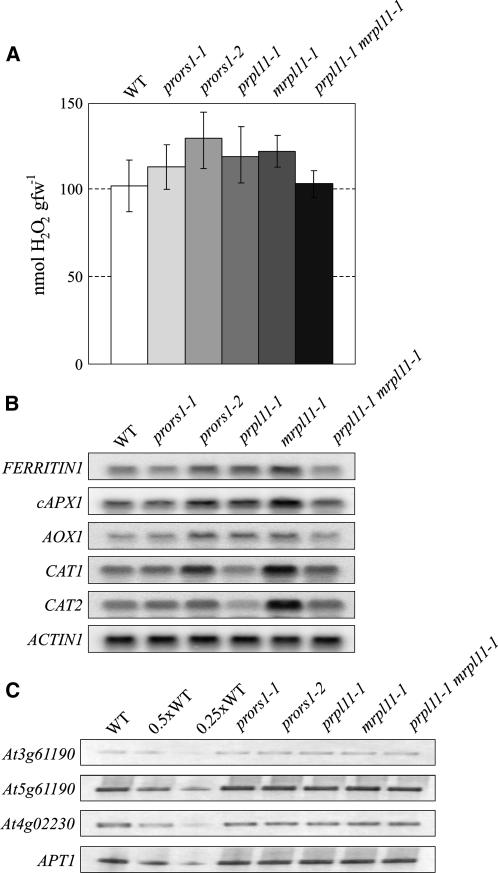
Downregulation of Nuclear Photosynthetic Genes Is Not Correlated with Increased Levels of ROS
In prors1 plants, the progressive reduction of PSII activity under illumination (Figure 1B) suggests that prors1 thylakoid membranes are sustaining photooxidative damage (Constant et al., 1997; Kettunen et al., 1997; reviewed in Yamamoto, 2001). Superoxide anion radicals (O2·−), hydroxyl radicals (OH·), and hydrogen peroxide (H2O2) are the major ROS molecules leading to oxidative damage (reviewed in Mittler, 2002). We therefore determined the H2O2 content in leaves according to Veljovic-Jovanovic et al. (2002). The assay showed a slight increase in H2O2 levels in light-adapted prors1-2 and, to a lesser extent, also in prors1-1 plants (Figure 13A). Comparable levels of hydrogen peroxide were detected in prpl11-1 and mrpl11-1 leaves, while the prpl11-1 mrpl11-1 double mutant did not show any increase in H2O2 content. Dark-adapted mutant and wild-type plants showed comparable levels of H2O2 (data not shown). In addition, expression of genes involved in preventing the formation or in scavenging of ROS was investigated (Dutilleul et al., 2003b). The levels of transcripts of the FERRITIN1 and cytosolic ASCORBATE PEROXIDASE1 (cAPX1) genes, expression of which has been reported to be stimulated by increases in H2O2 production (Karpinski et al., 1997; Briat and Lobreaux, 1998), were markedly increased in light-adapted mrpl11-1, prors1-2, and prpl11-1 mutants, while no major changes in expression were detected in prors1-1 or prpl11-1 mrpl11-1 leaves (Figure 13B).
Figure 13.
Quantification of H2O2 Content in Leaves and Induction of Antioxidant Defenses in Mutant and Wild-Type Leaves.
(A) Levels of H2O2 in light-adapted leaves from wild-type, prors1-1, prors1-2, prpl11-1, mrpl11-1, and prpl11-1 mrpl11-1 plants. Values are means ± sd from five independent experiments. fw, fresh weight.
(B) Quantification of antioxidant transcripts. Forty micrograms of total RNA from light-adapted wild-type and mutant plants grown on soil were subjected to RNA gel blot analysis. Autoradiograms were obtained using probes specific for FERRITIN1, cAPX1, AOX1, and CAT1 and CAT2. To control for RNA loading, the filter was reprobed with a cDNA fragment specific for ACTIN1. Results of one out of three independent RNA gel blot analyses are presented.
(C) Expression analysis of singlet oxygen-induced genes. The analysis was performed as in Figure 3B. Specific primers for At3g61190 (Bonsai-associated protein), At5g64870 (nodulin-like protein), and At4g02230 (protein similar to pectinesterase) were used. The expression of the APT1 gene was monitored as loading control. Decreasing levels of wild-type single-stranded cDNA were used in the lanes marked 0.5xWT and 0.25xWT.
Because in plants significant amounts of ROS are also generated in peroxisomes and mitochondria, expression of genes encoding the mitochondrial alternative oxidase (AOX1) and catalases (CAT1 and CAT2) was investigated. AOX1 transcripts were increased by approximately threefold in prors1-2, prpl11-1, and mrpl11-1 leaves, but no increase was detected in prors1-1 or prpl11-1 mrpl11-1 leaves. The abundance of CAT1 transcripts did not change markedly in prors1-1, prpl11-1, or prpl11-1 mrpl11-1 leaves, while a threefold to fourfold increase was observed in prors1-2 and mrpl11-1. Furthermore, CAT2 was strongly upregulated in mrpl11-1 leaves; in prpl11-1, a reduction in transcript accumulation was detected. The markedly enhanced expression of antioxidant genes in mrpl11-1, together with the essentially wild-type content of H2O2 in leaves, is consistent with a previous report showing that an increase in antioxidant gene expression is associated with a reduction in leaf H2O2 (Dutilleul et al., 2003b). Furthermore, it is interesting to note that prors1-1 and prpl11-1 mrpl11-1 plants did not show any increase in antioxidant gene expression, suggesting that the concomitant impairment of both organelles might result in less oxidative stress than a defect in either organelle. The increased accumulation of antioxidant transcripts in prors1-2 might be attributable to the more severe phenotype.
Singlet oxygen (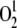 ) has been also reported to influence the expression of photosynthesis-related nuclear genes (op den Camp et al., 2003). In order to monitor
) has been also reported to influence the expression of photosynthesis-related nuclear genes (op den Camp et al., 2003). In order to monitor 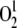 production, the transcript levels of three genes (At4g02330, At3g61190, and At5g64870) that have been reported to be upregulated by singlet oxygen were investigated. QRT-PCR analysis clearly showed that the expression of these genes was not altered in any of the mutants analyzed (Figure 13C), implying that defects in chloroplast and/or mitochondrial translation do not increase
production, the transcript levels of three genes (At4g02330, At3g61190, and At5g64870) that have been reported to be upregulated by singlet oxygen were investigated. QRT-PCR analysis clearly showed that the expression of these genes was not altered in any of the mutants analyzed (Figure 13C), implying that defects in chloroplast and/or mitochondrial translation do not increase 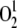 amount in these genotypes.
amount in these genotypes.
Taken together, the data do not reveal any association between ROS production in leaves and downregulation of nuclear photosynthetic genes. Although prors1-2 and prpl11-1 mrpl11-1 mutant leaves display similar patterns of downregulation of nuclear photosynthetic genes, they contain different amounts of hydrogen peroxide and antioxidant transcripts. Furthermore, the highest expression of ROS-induced genes was observed in mrpl11-1 leaves, in which the expression of photosynthetic genes was virtually unaffected.
DISCUSSION
Photosynthesis is strictly dependent on the coordinated activity of the chloroplast, mitochondrial, and nuclear compartments (Raghavendra and Padmasree, 2003). This, in turn, implies the existence of signaling pathways that serve to integrate nuclear and organellar gene expression (Rodermel and Park, 2003). The preeminence of the nucleus in this interorganellar exchange is indisputable: nuclear proteins imported into plastids and mitochondria regulate the transcription and translation of organellar genes. Nevertheless, it is now well established that perturbation of a number of plastid processes, including tetrapyrrole biosynthesis, protein synthesis, and photosynthesis, influence the expression of nuclear genes encoding photosynthetic proteins (Leister, 2005). In addition, investigations of mitochondrial mutants, such as cms (Noctor et al., 2004) and ncs (Newton and Coe, 1986), have clearly indicated that mitochondrial performance influences the activities of plastids and nucleus. However, modeling of mitochondrion–plastid crosstalk has been hampered by the inability of available experimental strategies to unequivocally define individual signaling pathways (Gray et al., 2002). Norflurazon, which was used to initially define the plastid-to-nucleus signal, for instance, results in a broad range of secondary effects, including ROS production (Mullineaux and Karpinski, 2002), and impairment of transcription (Tonkyn et al., 1992) and translation (Frosch et al., 1979; Reiss et al., 1983) in plastids. Studies employing inhibitors of prokaryotic-like 70S ribosomes, such as lincomycin, chloramphenicol, erythromycin, and streptomycin, could not distinguish unambiguously between effects on plastid and mitochondrial protein synthesis. Furthermore, the genetic dissection of organelle signaling has often been hindered by the pleiotropic nature of many mutant phenotypes.
In this study, the role of chloroplast and mitochondrial protein synthesis in controlling nuclear photosynthetic gene expression has been investigated genetically by employing mutants primarily affected in either the mitochondrial (mrpl11) or chloroplast (prpl11) compartment or in both (prors1). The PRORS1 locus encodes a ProRS with an ambiguous targeting presequence that is recognized as an import signal by both mitochondria and chloroplasts (Figure 6). Many proteins have now been shown to be localized in both mitochondria and chloroplasts, and most of them belong to the aaRS family: MetRS (Menand et al., 1998), HisRS (Akashi et al., 1998), CysRS and AspRS (Peeters et al., 2000), GlyRS (Duchene et al., 2001), and LysRS and TrpRS (Peeters and Small, 2001; Duchene et al., 2005). The action of Arabidopsis PRORS1 in mitochondria and chloroplasts prompts the question of which ProRS(s) operates in the other protein-synthesizing compartment, the cytosol. The best candidates are At3g62120 and At5g10880, which are homologous to eukaryotic bifunctional glutamyl-prolyl-tRNA synthetases and lack a canonical transit peptide.
Since all proteins encoded by the chloroplast and mitochondrial genomes of Arabidopsis contain at least one Pro residue, a complete lack of ProRS activity in these compartments should result in lethality. Indeed, absence of PRORS1 impairs ovule development and arrests embryo development at preglobular stages, implying that neither of the other two putative ProRS enzymes, At3g62120 and At5g10880, can efficiently replace PRORS1. In addition to the prors1-3 and -4 mutations described in this article, knockout mutants for other mitochondrial aaRSs have been described; however, these were only impaired in embryo development. In the acd mutant, which shows reduced expression of the ALARS gene coding for an alanyl-tRNA synthetase targeted to both cytosol and mitochondria (Mireau et al., 1996), abortion of embryos at the globular stage is observed (Ge et al., 1998). In the edd1 mutant, inactivation of the glycyl-tRNA synthetase GLYRS caused the arrest of embryo development between the globular and heart stages. A plastid location was initially proposed for GLYRS by Uwer et al. (1998), but it was later shown to be targeted to both plastids and mitochondria (Duchene et al., 2001). The variation in timing of the developmental block between the three mitochondrial aaRS mutants, prors1 (block in preglobular stage), edd1 (between globular and heart stages), and acd (globular stage), could be due to the incomplete penetrance of edd1 (Uwer et al., 1998) and acd (Ge et al., 1998) and/or the existence of a second aaRS that partially compensates for function in the same compartment. Indeed, a second mitochondrial glycyl-tRNA synthetase has been reported (Duchene et al., 2001), and the At5g22800 gene encodes an alanyl-tRNA synthetase that is predicted to be located in the mitochondrion.
The two leaky prors1 alleles, prors1-1 and -2, were identified because of their photosynthetic phenotype. The drop in photosynthetic performance in prors1-1 and, to a greater extent, in prors1-2 mutants could be attributed to the direct impairment of chloroplast and mitochondrial translation. As an additional consequence of the mutation, however, a marked and specific downregulation of nuclear photosynthetic gene expression was detected in prors1-2 (and to a lesser extent also in prors1-1). The contribution of each organelle to the repression of nuclear photosynthetic gene expression was assessed by analyzing mutants that are defective specifically in translation in mitochondria or plastids. A defect in protein synthesis in either mitochondria (in the mrpl11 mutant) or chloroplasts (in the prpl11 mutant) failed to elicit a transcriptional response like that observed in prors1 leaves (Figure 12). Only when both organelles were affected (in the prpl11-1 mrpl11-1 double mutant) was a prors1-like transcriptional response observed, implying that the two organelles cooperate in regulating the expression of nuclear photosynthetic genes. Clearly, the prpl11-1 mrpl11-1 transcriptional response was not due to an additive effect of impaired mitochondria on plastid activity, since the prpl11-1 and prpl11-1 mrpl11-1 leaves were identical in terms of thylakoid protein composition and photosynthetic performance. Moreover, prors1-1 leaves showed a more general and marked downregulation of nuclear photosynthetic gene expression than prpl11-1, although the prpl11-1 chloroplast activities were more drastically impaired. The fact that a comparable decrease in the level of photosynthetic transcripts is observed in pea seedlings treated with lincomycin and erythromycin, which inhibit translation in both chloroplasts and mitochondria (Sullivan and Gray, 1999), supports this idea.
In addition to organellar protein synthesis per se, the prors1 and prpl11 mrpl11 mutations also affect photosynthetic electron transfer, which itself is known to modulate the expression of nuclear genes coding for thylakoid proteins. Light intensity–dependent changes in LHCB1 transcription in the green alga Dunaliella tertiolecta are due to changes in the redox state of the plastoquinone pool and in the transthylakoid membrane potential (Escoubas et al., 1995; Chen et al., 2004), whereas excitation imbalances between the two photosystems induced by changes in light quality control the transcription of certain PSI genes in tobacco (Pfannschmidt et al., 2001). Although prors1 and prpl11 mrpl11 plants displayed very similar transcriptional responses, the redox state of the plastoquinone pool, measured as 1-qP, was increased only by prors1-2, the stronger of the two hypomorphic prors1 alleles. Moreover, the expression of PSAD and PSAF genes, which is upregulated by increased 1-qP values, was repressed in both mutants. This, together with the persistence of downregulation of nuclear photosynthetic genes in dark-adapted prors1 plants (Figure 9), strongly implies that the transcriptional response discussed here is independent of light and photosynthesis. In fact, even in plants with light-independent photomorphogenesis can the accumulation of nuclear photosynthetic transcripts be reduced by treatment with lincomycin or erythromycin (Sullivan and Gray, 1999).
The use of mutants for the dissection of organelle-to-nucleus signaling has often raised the question whether the developmental stage of the organelles might influence nuclear gene expression. A specific and general downregulation of nuclear photosynthetic genes was observed even in prors1-1 leaves, in which the protein composition of mitochondria and plastids was only marginally altered and mutant plants grew like the wild type under low light levels. Therefore, it can be concluded that organelle function, rather than developmental stage, is involved in the observed transcriptional response. Taken together, our data indicate that alterations in rates of translation in plastids and mitochondria are able to modulate synergistically the transcription of nuclear photosynthetic genes independently of light. How these signals are transduced from the organelles to the nucleus and integrated at the molecular levels remains to be revealed.
ROS have been suggested as signaling molecules in the crosstalk between organelles and the nucleus (Apel and Hirt, 2004). In particular, increased H2O2 levels in mitochondria, chloroplasts, or peroxisomes have been associated with changes in the nuclear transcriptome, indicating that information must be transmitted from these organelles to the nucleus. Thus, the YCF3 and AGP genes involved in photosynthesis are downregulated in tobacco leaves that have been acclimatized to oxidative stress (Vranova et al., 2002). Vandenabeele et al. (2003, 2004) showed transcriptional downregulation of a number of photosynthesis-related genes in catalase-deficient tobacco and Arabidopsis plants exposed to high light levels. Similarly, transcriptome analysis of Arabidopsis leaves exposed to high-light stress showed that the set of repressed genes was enriched in photosynthesis-related genes (Desikan et al., 2001). In the case of the prors1 mutants, the progressive reduction in PSII activity during the course of the day (Figure 1B), together with the drastic decrease in PSII-D1 (Figure 2), reflects the accumulation of photooxidative damage, most probably due to the action of ROS in the chloroplast (Niyogi, 1999). Indeed, prors1 leaves show an increase in H2O2 content and, at least in the case of prors1-2, also in the expression of antioxidant genes (Figure 13). This is compatible with the notion that in the prors1 mutants photooxidative stress plays a role in the downregulation of nuclear photosynthetic genes, with ROS acting as a messenger molecule. However, the levels of ROS and the abundance of antioxidant transcripts in prpl11-1 mrpl11-1 leaves, which also show a prors1-like transcriptional response, were identical to the wild type, implying that ROS most probably represent only one of a number of intermediates involved in controlling nuclear photosynthetic gene expression. Interestingly, the marked increase in antioxidant gene expression observed in mrpl11-1 was not associated with a corresponding increase in ROS amounts, which supports that an increase in ROS generation due to mitochondrial dysfunction can be effectively counteracted by scavenging enzymes (Dutilleul et al., 2003b). Moreover, the induction of antioxidant defenses seen in mrpl11-1 was abolished in the prpl11-1 mrpl11-1 double mutant. This implies that concomitant impairment of both organelles results in attenuation of the mrpl11-related oxidative stress and rules out the possibility that increased ROS production in a specific compartment triggers the downregulation of nuclear photosynthetic gene expression (Dutilleul et al., 2003b).
The genome uncoupled mutants of Arabidopsis (gun2, gun3, and gun5) have provided genetic evidence for a role of tetrapyrrole intermediates in plastid signaling (Mochizuki et al., 2001). However, tetrapyrroles appear not to be involved in the transduction of information on the state of organellar translation to the nucleus because treatment of gun2, gun3, and gun5 mutants with lincomycin still downregulates nuclear genes that are sensitive to the rates of organelle protein synthesis, such as LHCB1 (Gray et al., 2002). However, when applied to the gun1 mutant, lincomycin did not repress the same sensitive genes. This strongly suggests that GUN1 might be a component of the signaling pathway activated by the inhibition of organellar protein synthesis (Gray et al., 2002) and might be involved in the general and marked downregulation of nuclear photosynthetic gene expression in prors1 and prpl11 mrpl11 plants (this study).
The data presented here confirm previous reports of preferential downregulation of nuclear photosynthetic genes upon impairment of translation in plastids. However, the earlier studies were limited by the fact that inhibitors of protein synthesis in plastids also inhibit translation in mitochondria. In this work, it was shown that a marked and general downregulation of nuclear photosynthetic genes is the response to simultaneous changes in mitochondrial and chloroplast translation, implying synergistic roles for the two organelles in the regulation of nuclear photosynthetic genes. Such a mechanism that reduces the absorption capacity for light energy would serve to prevent the accumulation of ROS when the capacity for energy transformation is limited.
METHODS
Plant Lines and Their Propagation
The prors1-1, prors1-2, and prpl11-1 (Pesaresi et al., 2001) mutants were identified among a collection of Arabidopsis thaliana (ecotype Col-0) lines that had been mutagenized with either AC106 or AC161 T-DNA (see below for details of the screen). The insertion allele prors1-3 was identified in the Cold Spring Harbor Genetrap Collection (http://genetrap.cshl.org), which consists of flank-tagged Ds transposon lines (GT256 line, ecotype Landsberg erecta). The prors1-4 allele was identified by searching the Syngenta Arabidopsis Insertion Library (Sessions et al., 2002), consisting of flank-tagged T-DNA lines (Garlic_420_E02.b.1a.Lb3Fa line, ecotype Col-0). The mrpl11-1 mutant, corresponding to the Salk_090016 line (ecotype Col-0), was obtained by screening the insertion flanking database SIGnAL (http://signal.salk.edu/cgi-bin/tdnaexpress).
Mutant and wild-type plants were grown on Minitray soil (Gebr. Patzer) in a growth chamber (day period of 11 h at 20°C, PFD = 80 μmol m−2 s−1; night period of 13 h at 15°C). Osmocote Plus fertilizer (15% N, 11% P2O5, 13% K2O, and 2% MgO; Scotts Deutschland) was used according to the manufacturer's instructions. Unless stated elsewhere, all the analyses were performed on mutant and wild-type plants at the eight-leaf rosette stage. Leaf material was collected either immediately before (dark-adapted plants) or 8 h after (light-adapted plants) the start of the light period.
Chlorophyll Fluorescence, Pigment, and Leaf Starch Analyses
Mutants that show alterations in ΦII were identified using an automatic pulse amplitude modulation fluorometer system as described previously (Varotto et al., 2000). In vivo chlorophyll a fluorescence of single leaves was measured using the PAM 101/103 fluorometer (Walz). Pulses (800 ms) of white light (6000 μmol m−2 s−1) were used to determine the maximum fluorescence (Fm) and the ratio (Fm − F0)/Fm = Fv/Fm. A 15-min illumination with actinic light (80 μmol m−2 s−1) was supplied to drive electron transport between PSII and PSI before measuring ΦII and qP. Pigment analysis was performed by reverse-phase HPLC as described by Färber et al. (1997). Leaf starch was isolated and assayed as described by Lin et al. (1988).
Two-Dimensional PAGE and Protein Gel Blot Analyses
Leaves were harvested in the middle of the light period, and thylakoids and mitochondria were prepared as described by and Bassi et al. (1985) and Werhahn et al. (2001), respectively. Total proteins were extracted according to Martinez-Garcia et al. (1999).
Mitochondrial proteins, equivalent to 60 g of fresh leaf, were first fractionated by Blue-native PAGE and then by two-dimensional SDS-PAGE as described previously (Schägger, 2001). A gradient gel (4.5 to 16% acrylamide) was used for the first dimension and two-step Tricine-SDS gels (10 and 16% acrylamide) for the second dimension. Detergent was omitted from the cathode buffer for Blue-native gels. Gels were stained with Coomassie-colloidal (Neuhoff et al., 1985).
In the case of two-dimensional SDS-PAGE of thylakoid membranes, samples equivalent to 100 mg of fresh leaf were first fractionated on nondenaturing polyacrylamide gradient gels (4 to 12% acrylamide) according to Peter and Thornber (1991) using lithium dodecyl sulfate instead of lauryl β-d-iminopropionidate in the upper electrophoresis buffer. Fractionation in the second dimension was performed on a denaturing SDS-PAGE gradient gel (10 to 16% acrylamide) as described by Schägger and von Jagow (1987). Densitometric analyses of protein gels after Coomassie Brilliant Blue R 250 staining (Roth) were performed with Lumi Analyst 3.0 (Boehringer Mannheim/Roche).
For protein gel blot analyses, thylakoid proteins or total protein extracts were separated by one-dimensional SDS-PAGE gradient gel (10 to 16% acrylamide) as described by Schägger and von Jagow (1987) and transferred to Immobilon-P membranes (Millipore). Replicate filters were incubated with antibodies specific for the antenna complex of PSI (Lhca1-4), PSI-E, PSI-F, cytochrome b6, cytochrome f, the Rieske protein (PetC), the antenna complex of PSII (Lhcb1, Lhcb3, Lhcb5, and Lhcb6), PSII-D1, the ATPase α+β subunits, mitochondrial COXII, and mitochondrial RPS12. Signals were detected using the Enhanced Chemiluminescence Western Blotting kit (Amersham Biosciences). Densitometric analyses of immunoblot signals were performed using Lumi Analyst 3.0 (Boehringer Mannheim).
Nucleic Acid Analysis
Arabidopsis DNA was isolated as described (Liu et al., 1995), and regions flanking the AC106 and AC161 T-DNA insertions were identified according to Pesaresi et al. (2001). Total RNA was extracted from fresh tissue using the RNeasy plant system (Qiagen), and RNA gel blot analyses were performed according to Sambrook et al. (1989). Probes complementary to the following genes were obtained using gene-specific primer combinations (Table 4). To minimize cross-hybridization, filters were washed at high stringency (0.1× SSPE and 0.1% SDS; 65°C). For QRT-PCR of PRORS1, At3g61190, At5g64870, and At4g02330, first-strand cDNA was synthesized using the SuperScript preamplification system (Invitrogen), and gene-specific 33P-labeled sense primers and unlabeled antisense primers (Table 4) were used to amplify cDNA by PCR for 25 cycles, and the PCR products were separated on a 4.5% (w/v) polyacrylamide gel. Signals were quantified using the Storm 860 PhosphorImager (Molecular Dynamics) and the program Image Quant for Macintosh (version 1.2; Molecular Dynamics).
Table 4.
Gene-Specific Primers Used for Amplifying RNA Gel Blot Probes or for QRT-PCR
| Gene | Sense Primer | Antisense Primer |
|---|---|---|
| LHCA1 | 5′-GTCAAGCCACTTACTTGGGA-3′ | 5′-GGGATAACAATATCGCCAATG-3′ |
| LHCA2 | 5′-GAGTTCCTAACGAAGATCGG-3′ | 5′-AAGATTGTGGCGTGACCAGG-3′ |
| LHCA3 | 5′-AGGCTGGTCTGATTCCAGCA-3′ | 5′-ACTTGAGGCTGGTCAAGACG-3′ |
| LHCA4 | 5′-TGAGTGGTACGATGCTGGGA-3′ | 5′-GTGTTGTGCCATGGGTCAGA-3′ |
| PSAD1 | 5′-AAGCCGCCGGGATCTTCAAC-3′ | 5′-CTAAGCCTTGTCCCCAAAGC-3′ |
| PSAE1 | 5′-ATGGCGATGACGACAGCATC-3′ | 5′-TGTTGGTCGATATGTTGGCG-3′ |
| PSAF | 5′-GTTCGACAACTACGGGAAGT-3′ | 5′-CTTAGCAATGAGATCACCAT-3′ |
| PSAG | 5′-TCACCAACCA CATTCTCCAC-3′ | 5′-GGGTCGTATCCATTGCTGGA-3′ |
| PSAH1 | 5′-CCAAATCCATCCGGGCTAAT-3′ | 5′-AGGGGAAGAAAAAACGTTTC3′ |
| PSAK | 5′-ATGGTCTTCG AGCCACCAAA-3′ | 5′-CGTTCAGGTGCATGAGAATA-3′ |
| PSAL | 5′-TCAACCAATCAACGGTGATC-3′ | 5′-TGACGAAGTAAGGAAGTTCA-3′ |
| PSAN | 5′-ATGGCGGCCATGAACTCGAG-3′ | 5′-TGCTTGGCAAGATCTTGGCA-3′ |
| PSAO | 5′-ATGGCAGCAACATTTGCAAC-3′ | 5′-GTAATCTTCAGTCCTGCCCT-3 |
| LHCB1.2 | 5′-GACTTTCAGCTGATCCCGAG-3′ | 5′-CGGTCCCTTACCAGTGACAA-3′ |
| LHCB2.2 | 5′-GAGACATTCGCTAAGAACCG-3′ | 5′-CCAGTAACAATGGCTTGGAC-3′ |
| LHCB3 | 5′-GGAGATGGGCAATGTTGGGA-3′ | 5′-TAGTTGCGAAAGCCCACGCA-3′ |
| LHCB4.2 | 5′-AGCTAGTGGATGGATCATCT-3′ | 5′-CAGGAGGAAGAGAAGGTATC-3′ |
| LHCB5 | 5′-CTGGTGCTTTGCTTCTTGATG-3′ | 5′-TCCAGCGATGACGGTAAGCA-3′ |
| LHCB6 | 5′-GCATGGTTTGAAGCTGGAGC-3′ | 5′-ACAAACCAAGAGCACCGAGA-3′ |
| PSBO2 | 5′-AGACGGAAGCGTGAAGTTCA-3′ | 5′-CAATCTGACCGTACCAAACC-3′ |
| PSBP2 | 5′-CTCCTGAACAGTTCCTCTCT-3′ | 5′-ACACTGAAGGACGTAGCAGC-3′ |
| PSBQ2 | 5′-ACAGATAACTCAGACCAAGC-3′ | 5′-GCTTGGCAAGAACATTGTTC-3′ |
| PSBR | 5′-ATGGCTGCTTCAGTGATGCT-3′ | 5′-GCCAAAGCACTTGTGTTGTA-3′ |
| PSBT2 | 5′-ATGGCGTCAATGACCATGAC-3′ | 5′-CAGTTACGGCATATCTTGGC-3′ |
| PSBW | 5′-ATGGCTAGCTTTACTGCCTC-3′ | 5′-TTCTTCATCCTCCTCGAGAG-3′ |
| PSBX | 5′-ATGGCTTCTACCTCCGCGAT-3′ | 5′-TAGGTTCTCTTGACAGGGTC-3′ |
| PSBY | 5′-ATGGCAGCAGCTATGGCAAC-3′ | 5′-CTCCGGAGGTGGAGTCAAAA-3′ |
| AOX1 | 5′-GGTTCTGAATGGAAGTGGAAC-3′ | 5′-GGAGCTGGAGCTTCCTTTAGT-3′ |
| CAT1 | 5′-CTTCTTTGACTGTCGGAACTC-3′ | 5′-CCAGTATCCTCCAGTTCTCC-3′ |
| CAT2 | 5′-AACTCTGGTGCTCCTGTATGG-3′ | 5′-CTCCAGTTCTCTTGGATGTG-3′ |
| cAPX1 | 5′-GGAGGAAGCTCAGAGGTTTG-3′ | 5′-TGTCCATGCACCTTCGAAGC-3′ |
| FERRITIN1 | 5′-ATGGCCTCAAACGCACTCTC-3′ | 5′-ATGCCCTCTCTCTTCCTCAC-3′ |
| ACTIN1 | 5′-TGCGACAATGGAACTGGAATG-3′ | 5′-GGATAGCATGTGGAAGTGCATACC-3′ |
| PRORS1 | 5′-GCAACATGTACTTCCCACAG-3′ | 5′-GCAAATCTGGTGTAGATCTCG-3′ |
| At3g61190 | 5′-CATGGGAAGGTATTCACCTG-3′ | 5′-ACCGGAACTCCGGTAACAGT-3′ |
| At5g64870 | 5′-CTTGCGATTCAAAAGGCTGC-3′ | 5′-GCTTGCTTCTGCTTCTCGTA-3′ |
| At4g02330 | 5′-CATGAAAGTCAAGGGAAACG-3′ | 5′-ACTCAGCGGTTTGTGCAGGT-3′ |
| APT1 | 5′-TCCCAGAATCGCTAAGATTGCC-3′ | 5′-CCTTTCCCTTAAGCTCTG-3′ |
Complementation of the mrpl11-1 Mutation
The mrpl11-1 gene was ligated into the plant expression vector pJAN33 under the control of the 35S promoter of Cauliflower mosaic virus. Flowers of mrpl11-1 mutant plants were transformed according to Clough and Bent (1998). The plants were then transferred into the greenhouse, and seeds were collected 3 weeks later. Twenty-five independent transgenic plants were selected on the basis of their resistance to kanamycin. Successful complementation was confirmed by wild-type leaf coloration and growth. In addition, the presence and expression of the transgene was confirmed by PCR and RT-PCR.
Expression Profiling and Analysis
Generation and use of the nylon array carrying sequence tags for 3292 genes, including 2661 nuclear genes for chloroplast proteins and 631 genes coding for nonchloroplast proteins, has been described before (Richly et al., 2003). Total RNA was isolated from 5-g samples of leaf material harvested from dark- or light-adapted plants (see above). Three independent experiments were performed with different filters and independent cDNA probes derived from plant material corresponding to pools of at least 50 wild-type, prors1-1, or prors1-2 plants, thus minimizing the effects of variation between individual plants, filters, or probes. cDNA probes were synthesized using as a primer a mixture of oligonucleotides matching the 3292 genes in the antisense orientation and hybridized to the array as described (Kurth et al., 2002; Richly et al., 2003). Images were read using the Storm 860 PhosphorImager. Hybridization images were imported into the ArrayVision program (version 6; Imaging Research), which removes artifacts, corrects for background, and normalizes the resulting values with reference to the intensity of all spots on the array (Kurth et al., 2002). In the next step, these data were imported into the ArrayStat program (version 1.0, rev. 2.0; Imaging Research), and a z-test (nominal α set to 0.05) was performed employing false discovery rate correction (Benjamini and Hochberg, 1995) to identify statistically significant differential expression values. Only differential expression values fulfilling the criteria of this statistical procedure were considered for further analysis. Classification of genes, extending the previous grouping of Pesaresi et al. (2003), resulted in 15 functional classes: amino acid metabolism, carbohydrate metabolism, secondary metabolism, other metabolic functions, photosynthesis (dark reaction), photosynthesis (light reaction), protein modification and fate, protein phosphorylation, sensing, signaling and cellular communication, stress response, transport, transcription, protein synthesis, other functions, and unknown function.
In Vitro Import Assay
PRORS1, PRPL11, and MRPL11 precursors were synthesized using the in vitro transcription/translation coupled reticulocyte lysate TNT system (SDS-Promega) in the presence of [35S]Met (Amersham). The precursor proteins were then incubated together with both chloroplasts and mitochondria as described by Rudhe et al. (2002). After allowing for protein import, the organelles were separated on a 4% Percoll gradient, imported products were analyzed by SDS-PAGE, and images were acquired using the Storm 860 PhosphorImager.
Microscopy
Scanning electron microscopy was used to analyze freshly dissected siliques mounted on aluminum specimen stubs using Tissue-Tek OCT compound (Sakura Finetek) and immediately shock frozen in liquid nitrogen. The samples were subsequently transferred to a Zeiss DSM 940 electron microscope equipped with a cryo-chamber (Oxford Instruments). After sublimation of any ice on their surfaces, the samples were sputter-coated with gold and examined at an accelerating voltage of 5 kV.
To analyze defects in seed and embryo development, seeds from siliques of heterozygous prors1-3 or -4 plants were cleared overnight using a solution comprising 160 g of chloral hydrate (C-8383; Sigma-Aldrich), 100 mL of water, and 50 mL of glycerol. After clearing, the seeds were mounted on slides and covered with cover slips.
Ovules were dissected from premature pistils, which were staged according to Schneitz et al. (1995). Clearing and staining were performed as described by Schneitz et al. (1995). Ovules were immersed in the methyl benzoate/Spurr's resin mixture, mounted on slides, covered with cover slips, and sealed with Canada Balsam (Sigma-Aldrich).
Both seeds and ovules were analyzed using the Zeiss Axiophot microscope equipped with differential interface contrast optics. Photographs were taken using a video imaging system mounted on the microscope that consisted of a Hitachi charge-coupled device video camera operated by the DISKUS software package (Technisches Büro Hilgers).
Determination of H2O2 Content
The H2O2 content of leaves was determined according to the method of Veljovic-Jovanovic et al. (2002). Light-adapted leaves (200 mg) were ground to a fine powder in liquid nitrogen, and the powder was extracted with 2 mL of 1 M HClO4. The homogenates were centrifuged at 12,000g for 10 min, and the supernatant was neutralized, pH 5.6, with 5 M K2CO3 in 50 μL of 0.3 M phosphate buffer, pH 5.6. The homogenate was centrifuged at 12,000g for 2 min to remove KClO4. Before the assay, samples were incubated with 1 unit of ascorbate oxidase (Sigma-Aldrich). The reaction mixture consisted of 0.1 M phosphate buffer, pH 6.5, 3.3 mM 3-(dimethylamino) benzoic acid, 0.07 mM 3-methyl-2-benzothiazoline hydrazone, and 50 ng of horseradish peroxidase (Sigma-Aldrich). The change in absorbance induced by adding 50 μL of sample was then monitored at 590 nm. For each assay, the H2O2 content in the extract was quantified with reference to an internal standard (1.5 nmol H2O2 added to the reaction mixture upon completion of the absorbance change due to the sample).
In Vivo Translation Assay
For radioactive labeling of thylakoid proteins, leaf discs of Arabidopsis plants at the eight-leaf rosette stage were pressed extremely gently against coarse sandpaper and then vacuum-infiltrated in a syringe containing 1 mCi of [35S]Met in 10 mL of 1 mM KH2PO4, pH 6.3, and 0.1% Tween 20. Directly after infiltration, three leaf discs were frozen in nitrogen (t0). The remaining leaves were transferred into light (200 μmol m−2 s−1), and three leaf discs were collected at each of the four time points (t = 15, 30, 45, and 60 min). Subsequently, thylakoid proteins were prepared and fractionated as described by Pesaresi et al. (2002). Signals were detected and quantified using the Storm 860 PhosphorImager and the program Image Quant for Macintosh (version 1.2).
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource database (http://www.arabidopsis.org) with Arabidopsis Genome Initiative locus identifiers At5g52520 (PRORS1), At4g35490 (MRPL11), and At1g32990 (PRPL11). Sequence data of ProRS sequences from species other than Arabidopsis can be found in the National Center for Biotechnology Information database: Oryza sativa (gi 22202767), Giardia intestinalis (gi 10800405), Thermus thermophilus (gi 14423353), Aeropyrum pernix (gi 14601997), Chlorobium tepidum (gi 21647484), and Clostridium sticklandii (gi 6899998). Original data for the four expression profiles analyzed are available at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE1232 (light-adapted prors1-1 versus the wild type), GSE1233 (light-adapted prors1-2 versus the wild type), GSE1495 (dark-adapted prors1-1 versus the wild type), and GSE1496 (dark-adapted prors1-2 versus the wild type).
Supplemental Data
The following materials are available in the online version of the article.
Supplemental Figure 1. T-DNA Tagging and mRNA Expression of the MRPL11 Gene.
Supplemental Figure 2. Protein Sequence Comparison of the L11 Subunit of Ribosomes (RPL11) in Organelles of Higher Plants and in Prokaryotes.
Supplemental Figure 3. In Vitro Dual-Import of MRPL11 and PRPL11.
Supplementary Material
Acknowledgments
We thank the SALK Institute, Syngenta, and the Cold Spring Harbor Laboratory for making T-DNA insertion lines publicly available, Kay Schneitz and Christine Foyer for valuable advice and practical help, and Paul Hardy for critical comments on the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Dario Leister (leister@lrz.uni-muenchen.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.039073.
References
- Adamska, I. (1995). Regulation of early light-inducible protein gene expression by blue and red light in etiolated seedlings involves nuclear and plastid factors. Plant Physiol. 107 1167–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, K., Grandjean, O., and Small, I. (1998). Potential dual targeting of an Arabidopsis archaebacterial-like histidyl-tRNA synthetase to mitochondria and chloroplasts. FEBS Lett. 431 39–44. [DOI] [PubMed] [Google Scholar]
- Apel, K., and Hirt, H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. [DOI] [PubMed] [Google Scholar]
- Aro, E.M., and Ohad, I. (2003). Redox regulation of thylakoid protein phosphorylation. Antioxid. Redox. Signal 5 55–67. [DOI] [PubMed] [Google Scholar]
- Aronsson, H., and Jarvis, P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529 215–220. [DOI] [PubMed] [Google Scholar]
- Bannai, H., Tamada, Y., Maruyama, O., Nakai, K., and Miyano, S. (2002). Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18 298–305. [DOI] [PubMed] [Google Scholar]
- Bassi, R., Peruffo, A.D., Barbato, R., and Ghisi, R. (1985). Differences in chlorophyll-protein complexes and composition of polypeptides between thylakoids from bundle sheaths and mesophyll cells in maize. Eur. J. Biochem. 146 589–595. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 57 289–300. [Google Scholar]
- Bradbeer, J.W., Atkinson, Y.E., Borner, T., and Hagemann, R. (1979). Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesised RNA. Nature 279 816–817. [Google Scholar]
- Briat, J.F., and Lobreaux, S. (1998). Iron storage and ferritin in plants. Met. Ions Biol. Syst. 35 563–584. [PubMed] [Google Scholar]
- Buchanan, B.B., Schurmann, P., and Jacquot, J.P. (1994). Thioredoxin and metabolic regulation. Semin. Cell Biol. 5 285–293. [DOI] [PubMed] [Google Scholar]
- Bunjun, S., Stathopoulos, C., Graham, D., Min, B., Kitabatake, M., Wang, A.L., Wang, C.C., Vivares, C.P., Weiss, L.M., and Soll, D. (2000). A dual-specificity aminoacyl-tRNA synthetase in the deep-rooted eukaryote Giardia lamblia. Proc. Natl. Acad. Sci. USA 97 12997–13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.B., Durnford, D.G., Koblizek, M., and Falkowski, P.G. (2004). Plastid regulation of Lhcb1 transcription in the chlorophyte alga Dunaliella tertiolecta. Plant Physiol. 136 3737–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet, Y., and Wollman, F.A. (2002). Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 529 39–42. [DOI] [PubMed] [Google Scholar]
- Christensen, A.C., Lyznik, A., Mohammed, S., Elowsky, C.G., Elo, A., Yule, R., and Mackenzie, S.A. (2005). Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell 17 2805–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros, M.G., and Vincens, P. (1996). Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241 779–786. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Constant, S., Perewoska, I., Alfonso, M., and Kirilovsky, D. (1997). Expression of the psbA gene during photoinhibition and recovery in Synechocystis PCC 6714: Inhibition and damage of transcriptional and translational machinery prevent the restoration of photosystem II activity. Plant Mol. Biol. 34 1–13. [DOI] [PubMed] [Google Scholar]
- Cusack, S., Yaremchuk, A., Krikliviy, I., and Tukalo, M. (1998). tRNAPro anticodon recognition by Thermus thermophilus prolyl-tRNA synthetase. Structure 6 101–108. [DOI] [PubMed] [Google Scholar]
- Danon, A. (1997). Translational regulation in the chloroplast. Plant Physiol. 115 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X.W., and Gruissem, W. (1987). Control of plastid gene expression during development: The limited role of transcriptional regulation. Cell 49 379–387. [DOI] [PubMed] [Google Scholar]
- Desikan, R., A-H-Mackerness, S., Hancock, J.T., and Neill, S.J. (2001). Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchene, A.M., Giritch, A., Hoffmann, B., Cognat, V., Lancelin, D., Peeters, N.M., Zaepfel, M., Marechal-Drouard, L., and Small, I.D. (2005). Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102 16484–16489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchene, A.M., Peeters, N., Dietrich, A., Cosset, A., Small, I.D., and Wintz, H. (2001). Overlapping destinations for two dual targeted glycyl-tRNA synthetases in Arabidopsis thaliana and Phaseolus vulgaris. J. Biol. Chem. 276 15275–15283. [DOI] [PubMed] [Google Scholar]
- Dutilleul, C., Driscoll, S., Cornic, G., De Paepe, R., Foyer, C.H., and Noctor, G. (2003. a). Functional mitochondrial complex I is required by tobacco leaves for optimal photosynthetic performance in photorespiratory conditions and during transients. Plant Physiol. 131 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul, C., Garmier, M., Noctor, G., Mathieu, C., Chetrit, P., Foyer, C.H., and de Paepe, R. (2003. b). Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and Von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani, G., Delarue, M., Poch, O., Gangloff, J., and Moras, D. (1990). Partition of transfer-RNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347 203–206. [DOI] [PubMed] [Google Scholar]
- Escoubas, J.M., Lomas, M., LaRoche, J., and Falkowski, P.G. (1995). Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc. Natl. Acad. Sci. USA 92 10237–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Färber, A., Young, A.J., Ruban, A.V., Horton, P., and Jahns, P. (1997). Dynamics of xanthophyll-cycle activity in different antenna subcomplexes in the photosynthetic membranes of higher plants (The relationship between zeaxanthin conversion and nonphotochemical fluorescence quenching). Plant Physiol. 115 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiler, H.S., and Newton, K.J. (1987). Altered mitochondrial gene expression in the nonchromosomal stripe 2 mutant of maize. EMBO J. 6 1535–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosch, S., Jabben, M., Bergfeld, R., Kleinig, H., and Mohr, H. (1979). Inhibition of carotenoid biosynthesis by the herbicide SAN9789 and its consequences for the action of phytochrome on plastidogenesis. Planta 145 497–505. [DOI] [PubMed] [Google Scholar]
- Ge, S.J., Yao, X.L., Yang, Z.X., and Zhu, Z.P. (1998). An Arabidopsis embryonic lethal mutant with reduced expression of alanyl-tRNA synthetase gene. Cell Res. 8 119–134. [DOI] [PubMed] [Google Scholar]
- Gilmore, A.M., and Bjorkman, O. (1995). Temperature-sensitive coupling and uncoupling of ATPase-mediated, nonradiative energy-dissipation - Similarities between chloroplasts and leaves. Planta 197 646–654. [Google Scholar]
- Gray, J.C., Sornarajah, R., Zabron, A.A., Duckett, C.M., and Khan, M.S. (1995). Chloroplast control of nuclear gene expression. In Photosynthesis, from Light to Biosphere, P. Mathis, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 543–550.
- Gray, J.C., Sullivan, J.A., Wang, J.H., Jerome, C.A., and MacLean, D. (2002). Coordination of plastid and nuclear gene expression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineke, D., Bykova, N., Gardestrom, P., and Bauwe, H. (2001). Metabolic response of potato plants to an antisense reduction of the P-protein of glycine decarboxylase. Planta 212 880–887. [DOI] [PubMed] [Google Scholar]
- Hess, W.R., Müller, A., Nagy, F., and Börner, T. (1994). Ribosome-deficient plastids affect transcription of light induced nuclear genes: Genetic evidence for a plastid derived signal. Mol. Gen. Genet. 242 505–512. [DOI] [PubMed] [Google Scholar]
- Hess, W.R., Schendel, R., Börner, T., and Rüdiger, W. (1991). Reduction of mRNA levels for two nuclear encoded light regulated genes in the barley mutant albostrians is not correlated with phytochrome content and activity. J. Plant Physiol. 138 292–298. [Google Scholar]
- Hoefnagel, M.H.N., Atkin, O.K., and Wiskich, J.T. (1998). Interdependence between chloroplasts and mitochondria in the light and the dark. Biochim. Biophys. Acta 1366 235–255. [Google Scholar]
- Hunt, M.D., and Newton, K.J. (1991). The NCS3 mutation: Genetic evidence for the expression of ribosomal protein genes in Zea mays mitochondria. EMBO J. 10 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba, M., and Soll, D. (2000). Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69 617–650. [DOI] [PubMed] [Google Scholar]
- Igamberdiev, A.U., Bykova, N.V., Lea, P.J., and Gardestrom, P. (2001). The role of photorespiration in redox and energy balance of photosynthetic plant cells: A study with a barley mutant deficient in glycine decarboxylase. Physiol. Plant 111 427–438. [DOI] [PubMed] [Google Scholar]
- Karpinski, S., Escobar, C., Karpinska, B., Creissen, G., and Mullineaux, P.M. (1997). Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen, R., Pursiheimo, S., Rintamaki, E., Van Wijk, K.J., and Aro, E.M. (1997). Transcriptional and translational adjustments of psbA gene expression in mature chloroplasts during photoinhibition and subsequent repair of photosystem II. Eur. J. Biochem. 247 441–448. [DOI] [PubMed] [Google Scholar]
- Kromer, S. (1995). Respiration during photosynthesis. Annu. Rev. Plant Physiol. 46 45–70. [Google Scholar]
- Kurth, J., Varotto, C., Pesaresi, P., Biehl, A., Richly, E., Salamini, F., and Leister, D. (2002). Gene-sequence-tag expression analyses of 1,800 genes related to chloroplast functions. Planta 215 101–109. [DOI] [PubMed] [Google Scholar]
- Leister, D. (2005). Genomics-based dissection of the cross-talk of chloroplasts with the nucleus and mitochondria in Arabidopsis. Gene 354 110–116. [DOI] [PubMed] [Google Scholar]
- Lin, T.P., Caspar, T., Somerville, C., and Preiss, J. (1988). Isolation and characterization of a starchless mutant of Arabidopsis thaliana (L) Heynh lacking ADP-glucose pyrophosphorylase activity. Plant Physiol. 86 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8 457–463. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Monte, E., and Quail, P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20 251–257. [DOI] [PubMed] [Google Scholar]
- Menand, B., Marechal-Drouard, L., Sakamoto, W., Dietrich, A., and Wintz, H. (1998). A single gene of chloroplast origin codes for mitochondrial and chloroplastic methionyl-tRNA synthetase in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95 11014–11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mireau, H., Lancelin, D., and Small, I.D. (1996). The same Arabidopsis gene encodes both cytosolic and mitochondrial alanyl-tRNA synthetases. Plant Cell 8 1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7 405–410. [DOI] [PubMed] [Google Scholar]
- Mochizuki, N., Brusslan, J.A., Larkin, R., Nagatani, A., and Chory, J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 98 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux, P., and Karpinski, S. (2002). Signal transduction in response to excess light: Getting out of the chloroplast. Curr. Opin. Plant Biol. 5 43–48. [DOI] [PubMed] [Google Scholar]
- Nakai, K., and Kanehisa, M. (1992). A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhoff, V., Stamm, R., and Eibl, H. (1985). Clear background and highly sensitive protein staining with Coomassie blue dyes in polyacrylamide gels - A systematic analysis. Electrophoresis 6 427–448. [Google Scholar]
- Newton, K.J., and Coe, E.H. (1986). Mitochondrial DNA changes in abnormal growth mutants of maize. Proc. Natl. Acad. Sci. USA 83 7363–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, K.J., Coe, E.H., Gabay-Laughnan, S., and Laughnan, J.R. (1989). Abnormal growth phenotypes and mitochondrial mutants in maize. Maydica 34 291–296. [Google Scholar]
- Niyogi, K.K. (1999). PHOTOPROTECTION REVISITED. Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 333–359. [DOI] [PubMed] [Google Scholar]
- Noctor, G., Dutilleul, C., De Paepe, R., and Foyer, C.H. (2004). Use of mitochondrial electron transport mutants to evaluate the effects of redox state on photosynthesis, stress tolerance and the integration of carbon/nitrogen metabolism. J. Exp. Bot. 55 49–57. [DOI] [PubMed] [Google Scholar]
- Oelmüller, R., Levitan, I., Bergfeld, R., Rajasekhar, V.K., and Mohr, H. (1986). Expression of nuclear genes as affected by treatments acting on plastids. Planta 168 482–492. [DOI] [PubMed] [Google Scholar]
- op den Camp, R.G., Przybyla, D., Ochsenbein, C., Laloi, C., Kim, C., Danon, A., Wagner, D., Hideg, E., Gobel, C., Feussner, I., Nater, M., and Apel, K. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15 2320–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmond, B., Badger, M., Maxwell, K., Bjorkman, O., and Leegood, R. (1997). Too many photos: Photorespiration, photoinhibition and photooxidation. Trends Plant Sci. 2 119–121. [Google Scholar]
- Padmasree, K., Padmavathi, L., and Raghavendra, A.S. (2002). Essentiality of mitochondrial oxidative metabolism for photosynthesis: Optimization of carbon assimilation and protection against photoinhibition. Crit. Rev. Biochem. Mol. Biol. 37 71–119. [DOI] [PubMed] [Google Scholar]
- Padmasree, K., and Raghavendra, A.S. (1999). Prolongation of photosynthetic induction as a consequence of interference with mitochondrial oxidative metabolism in mesophyll protoplasts of the pea (Pisum sativum L.). Plant Sci. 142 29–36. [Google Scholar]
- Padmasree, K., and Raghavendra, A.S. (2001). Restriction of mitochondrial oxidative metabolism leads to suppression of photosynthetic carbon assimilation but not of photochemical electron transport in pea mesophyll protoplasts. Curr. Sci. 81 680–684. [Google Scholar]
- Paul, M.J., and Pellny, T.K. (2003). Carbon metabolite feedback regulation of leaf photosynthesis and development. J. Exp. Bot. 54 539–547. [DOI] [PubMed] [Google Scholar]
- Peeters, N., and Small, I. (2001). Dual targeting to mitochondria and chloroplasts. Biochim. Biophys. Acta 1541 54–63. [DOI] [PubMed] [Google Scholar]
- Peeters, N.M., Chapron, A., Giritch, A., Grandjean, O., Lancelin, D., Lhomme, T., Vivrel, A., and Small, I. (2000). Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J. Mol. Evol. 50 413–423. [DOI] [PubMed] [Google Scholar]
- Pesaresi, P., Gardner, N.A., Masiero, S., Dietzmann, A., Eichacker, L., Wickner, R., Salamini, F., and Leister, D. (2003). Cytoplasmic N-terminal protein acetylation is required for efficient photosynthesis in Arabidopsis. Plant Cell 15 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi, P., Lunde, C., Jahns, P., Tarantino, D., Meurer, J., Varotto, C., Hirtz, R.D., Soave, C., Scheller, H.V., Salamini, F., and Leister, D. (2002). A stable LHCII-PSI aggregate and suppression of photosynthetic state transitions in the psae1-1 mutant of Arabidopsis thaliana. Planta 215 940–948. [DOI] [PubMed] [Google Scholar]
- Pesaresi, P., Varotto, C., Meurer, J., Jahns, P., Salamini, F., and Leister, D. (2001). Knock-out of the plastid ribosomal protein L11 in Arabidopsis: Effects on mRNA translation and photosynthesis. Plant J. 27 179–189. [DOI] [PubMed] [Google Scholar]
- Peter, G.F., and Thornber, J.P. (1991). Biochemical composition and organisation of higher plant photosystem II light-harvesting pigment-proteins. J. Biol. Chem. 266 16745–16754. [PubMed] [Google Scholar]
- Pfannschmidt, T. (2003). Chloroplast redox signals: How photosynthesis controls its own genes. Trends Plant Sci. 8 33–41. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt, T., and Liere, K. (2005). Redox regulation and modification of proteins controlling chloroplast gene expression. Antioxid. Redox. Signal 7 607–618. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt, T., Nilsson, A., and Allen, J.F. (1999). Photosynthetic control of chloroplast gene expression. Nature 397 625–628. [Google Scholar]
- Pfannschmidt, T., Schutze, K., Brost, M., and Oelmuller, R. (2001). A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment. J. Biol. Chem. 276 36125–36130. [DOI] [PubMed] [Google Scholar]
- Porse, B.T., Cundliffe, E., and Garrett, R.A. (1999). The antibiotic micrococcin acts on protein L11 at the ribosomal GTPase centre. J. Mol. Biol. 287 33–45. [DOI] [PubMed] [Google Scholar]
- Raghavendra, A.S., and Padmasree, K. (2003). Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 8 546–553. [DOI] [PubMed] [Google Scholar]
- Reiss, T., Bergfeld, R., Link, G., Thien, W., and Mohr, H. (1983). Photooxidative destruction of chloroplasts and its consequences to cytosol enzyme levels and plant development. Planta 159 518–528. [DOI] [PubMed] [Google Scholar]
- Richly, E., Dietzmann, A., Biehl, A., Kurth, J., Laloi, C., Apel, K., Salamini, F., and Leister, D. (2003). Covariations in the nuclear chloroplast transcriptome reveal a regulatory master-switch. EMBO Rep. 4 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix, J.D. (2001). Posttranscriptional control of chloroplast gene expression. From RNA to photosynthetic complex. Plant Physiol. 125 142–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodermel, S., and Park, S. (2003). Pathways of intracellular communication: Tetrapyrroles and plastid-to-nucleus signaling. Bioessays 25 631–636. [DOI] [PubMed] [Google Scholar]
- Rolland, F., Moore, B., and Sheen, J. (2002). Sugar sensing and signaling in plants. Plant Cell 14(Suppl.), 185–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudhe, C., Chew, O., Whelan, J., and Glaser, E. (2002). A novel in vitro system for simultaneous import of precursor proteins into mitochondria and chloroplasts. Plant J. 30 213–220. [DOI] [PubMed] [Google Scholar]
- Sabar, M., De Paepe, R., and de Kouchkovsky, Y. (2000). Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris. Plant Physiol. 124 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schägger, H. (2001). Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 65 231–244. [DOI] [PubMed] [Google Scholar]
- Schägger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166 368–379. [DOI] [PubMed] [Google Scholar]
- Schein, A.I., Kissinger, J.C., and Ungar, L.H. (2001). Chloroplast transit peptide prediction: A peek inside the black box. Nucleic Acids Res. 29 E82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz, K., Hulskamp, M., and Pruitt, R.E. (1995). Wild-type ovule development in Arabidopsis thaliana: A light-microscope study of cleared whole-mount tissue. Plant J. 7 731–749. [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Filho, M.C. (2003). One ticket for multiple destinations: Dual targeting of proteins to distinct subcellular locations. Curr. Opin. Plant Biol. 6 589–595. [DOI] [PubMed] [Google Scholar]
- Stehlin, C., Burke, B., Yang, F., Liu, H., Shiba, K., and Musier-Forsyth, K. (1998). Species-specific differences in the operational RNA code for aminoacylation of tRNAPro. Biochemistry 37 8605–8613. [DOI] [PubMed] [Google Scholar]
- Strand, A., Asami, T., Alonso, J., Ecker, J.R., and Chory, J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421 79–83. [DOI] [PubMed] [Google Scholar]
- Sullivan, J.A., and Gray, J.C. (1999). Plastid translation is required for the expression of nuclear photosynthesis genes in the dark and in the roots of the pea lip 1 mutant. Plant Cell 11 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkyn, J.C., Deng, X.W., and Gruissem, W. (1992). Regulation of plastid gene expression during photooxidative stress. Plant Physiol. 99 1406–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwer, U., Willmitzer, L., and Altmann, T. (1998). Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. Plant Cell 10 1277–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele, S., Van Der Kelen, K., Dat, J., Gadjev, I., Boonefaes, T., Morsa, S., Rottiers, P., Slooten, L., Van Montagu, M., Zabeau, M., Inze, D., and Van Breusegem, F. (2003). A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc. Natl. Acad. Sci. USA 100 16113–16118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele, S., Vanderauwera, S., Vuylsteke, M., Rombauts, S., Langebartels, C., Seidlitz, H.K., Zabeau, M., Van Montagu, M., Inze, D., and Van Breusegem, F. (2004). Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 39 45–58. [DOI] [PubMed] [Google Scholar]
- Varotto, C., Pesaresi, P., Maiwald, D., Kurth, J., Salamini, F., and Leister, D. (2000). Identification of photosynthetic mutants of Arabidopsis by automatic screening for altered effective quantum yield of photosystem 2. Photosynthetica 38 497–504. [Google Scholar]
- Veljovic-Jovanovic, S., Noctor, G., and Christine, H.F. (2002). Are leaf hydrogen peroxide concentrations commonly overestimated? The potential influence of artefactual interference by tissue phenolics and ascorbate. Plant Physiol. Biochem. 40 501–507. [Google Scholar]
- Vranova, E., Atichartpongkul, S., Villarroel, R., Van Montagu, M., Inze, D., and Van Camp, W. (2002). Comprehensive analysis of gene expression in Nicotiana tabacum leaves acclimated to oxidative stress. Proc. Natl. Acad. Sci. USA 99 10870–10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werhahn, W., Niemeyer, A., Jansch, L., Kruft, V., Schmitz, U.K., and Braun, H. (2001). Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis. Identification of multiple forms of TOM20. Plant Physiol. 125 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman, F.A. (2001). State transitions reveal the dynamics and flexibility of the photosynthetic apparatus. EMBO J. 20 3623–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, K., and Subramanian, A.R. (2000). The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 275 28466–28482. [DOI] [PubMed] [Google Scholar]
- Yamamoto, Y. (2001). Quality control of photosystem II. Plant Cell Physiol. 42 121–128. [DOI] [PubMed] [Google Scholar]
- Yaremchuk, A., Tukalo, M., Grotli, M., and Cusack, S. (2001). A succession of substrate induced conformational changes ensures the amino acid specificity of Thermus thermophilus prolyl-tRNA synthetase: Comparison with histidyl-tRNA synthetase. J. Mol. Biol. 309 989–1002. [DOI] [PubMed] [Google Scholar]
- Yokozawa, J., Okamoto, K., Kawarabayasi, Y., Kuno, A., and Hasegawa, T. (2003). Molecular recognition of proline tRNA by prolyl-tRNA synthetase from hyperthermophilic archaeon, Aeropyrum pernix K1. Nucleic Acids Res. (suppl.), 247–248. [DOI] [PubMed]
- Yoshida, R., Sato, T., Kanno, A., and Kameya, T. (1998). Streptomycin mimics the cool temperature response in rice plants. J. Exp. Bot. 49 221–227. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.