Abstract
The G1-to-S-phase transition is a key regulatory point in the cell cycle, but the rate-limiting component in plants is unknown. Overexpression of CYCLIN D3;1 (CYCD3;1) in transgenic plants increases mitotic cycles and reduces endocycles, but its effects on cell cycle progression cannot be unambiguously determined. To analyze the cell cycle roles of plant D-type cyclins, we overexpressed CYCD3;1 in Arabidopsis thaliana cell suspension cultures. Changes in cell number and doubling time were insignificant, but cultures exhibited an increased proportion of G2- over G1-phase cells, as well as increased G2 arrest in response to stationary phase and sucrose starvation. Synchronized cultures confirm that CYCD3;1-expressing (but not CYCD2;1-expressing) cells show increased G2-phase length and delayed activation of mitotic genes such as B-type cyclins, suggesting that CYCD3;1 has a specific G1/S role. Analysis of putative cyclin-dependent kinase phosphorylation sites within CYCD3;1 shows that mutating Ser-343 to Ala enhances CYCD3;1 potency without affecting its rate of turnover and results in a fivefold increase in the level of cell death in response to sucrose removal. We conclude that CYCD3;1 dominantly drives the G1/S transition, and in sucrose-depleted cells the decline in CYCD3;1 levels leads to G1 arrest, which is overcome by ectopic CYCD3;1 expression. Ser-343 is likely a key residue in modulating CYCD3;1 activity in response to sucrose depletion.
INTRODUCTION
The cell cycle plays a crucial role in the growth and development of multicellular organisms, including plants (Dewitte and Murray, 2003). In particular, the cyclin D (CYCD)/retinoblastoma (RB) pathway is believed to be involved in controlling both the commitment of cells to the mitotic cell cycle and decisions involving cell growth, cellular differentiation, and cell cycle exit (Ekholm and Reed, 2000; Meijer and Murray, 2001; Boonstra, 2003). The model based on results from mammals suggests that key genes for growth and cell division are regulated by E2F transcription factors, which are inactive when bound by RB. The phosphorylation of RB is initiated by CYCD-containing cyclin-dependent kinases (CDKs) and is completed by cyclin E–CDK2, resulting in the dissociation of RB from E2F factors, triggering the passage of cells from G1- to S-phase. Because CYCDs are unstable proteins in mammals whose expression depends on the continued presence of extracellular mitogenic signals, they link external signals in the cell cycle and trigger the decision of cells to divide (Sherr, 1995). This key role for the G1 exit pathway results in it being the primary and predominant cell cycle control point. However, cyclin E–CDK2 is rate-limiting for entry into S-phase and can trigger S-phase in the absence of RB phosphorylation (Leng et al., 1997; Sherr, 2000).
The key players of the RB pathway are conserved between mammals and plants, albeit under substantially different regulatory mechanisms (Dewitte and Murray, 2003). Accumulating evidence suggests that the CYCD/RB regulatory pathway operates at least in outline in a similar way in plants, but the presence of 10 CYCD genes in seven major groups (Vandepoele et al., 2002), as opposed to 3 in mammals, and the absence of cyclin E in plants suggest additional complexity. In plants, the G1 control point is also of primary importance in responses to nutrient signals, including sucrose removal (Planchais et al., 2004), as well as in developmental arrest, such as that observed in seeds (Barroco et al., 2005; Masubelele et al., 2005).
Of the Arabidopsis thaliana CYCD genes, CYCD2;1 and CYCD3;1 are the best studied examples. Both complement a yeast mutant lacking G1 cyclins (Soni et al., 1995), and expression of their genes is regulated by extrinsic signals, such as sucrose availability (Riou-Khamlichi et al., 2000). CYCD3;1 expression is also regulated by plant hormones, particularly cytokinin (Riou-Khamlichi et al., 1999; Oakenfull et al., 2002) and brassinosteroids (Hu et al., 2000). CYCD3;1 is a highly unstable protein degraded by a proteasome-dependent mechanism and is rapidly lost when the cells enter into resting states, such as stationary phase and sucrose starvation (Healy et al., 2001; Planchais et al., 2004). By contrast, CYCD2;1 is a stable protein that is also present in nondividing stationary phase and sucrose-starved cells (Healy et al., 2001). Both CYCD2;1 and CYCD3;1 bind and activate only the PSTAIRE-containing CDKA and not the plant-specific class of CDKBs (Healy et al., 2001), whereas CYCD4;1 can also bind the plant-specific mitotic kinase CDKB (Kono et al., 2003). This distinct regulation and activity of different CYCDs supports additional complexities to the outline model proposed above, indicating that detailed analysis of the components of this pathway in plants is necessary to understand their role in cell division, growth, and development.
Indeed, previous work has shown that overexpression of CYCDs can have significant effects on plant growth and development. Ectopic expression of Arabidopsis CYCD2;1 in transgenic tobacco (Nicotiana tabacum) plants led to accelerated development and a faster growth rate attributable to a reduction in cell cycle length caused by a reduced G1-phase duration (Cockcroft et al., 2000). No externally visible accompanying effects on morphology were reported, although more detailed analysis has suggested subtle alterations in meristematic zonation (Boucheron et al., 2005).
By contrast, constitutive overexpression of CYCD3;1 in Arabidopsis plants leads to dramatic changes in plant morphology, an expansion of the zone of dividing cells, ectopic cell division, reduced cell size, retardation of cellular differentiation, and a reduction in endoreduplication, the increase in cellular DNA content that occurs in many cells after cessation of mitotic divisions. Leaves of CYCD3;1-overexpressing plants contain a substantially increased number of smaller cells, and cell division replaces cell expansion as the primary mechanism of leaf growth (Dewitte et al., 2003). Explants from these plants also show cytokinin-independent initiation and growth of callus (Riou-Khamlichi et al., 1999). Targeting of CYCD3;1 expression to endoreduplicating trichome cells not only induces DNA replication but also induces cell division, leading to multicellular hairs (Schnittger et al., 2002).
These specific phenotypes resulting from the ectopic expression of particular CYCDs in transgenic plants support distinct biochemical properties of different CYCD proteins. However, their phenotypic effects may be variously attributable to their acting primarily on the cell cycle, or, alternatively, by delaying exit from the cell cycle, or by affecting cellular differentiation or overall development. Thus, although CYCD3;1-overexpressing plants show an increase in cells with a 4C DNA content, this may result from cells that are still engaged in mitotic cycles and accumulating in G2, or from a population of cells that are blocked in the first stage of endoreduplication.
Cell suspension cultures have the advantage that cell cycle processes can be studied in a homogeneous culture independently of cellular differentiation and developmental events. Indeed, overexpression of a tobacco CYCD3;3-Green Fluorescent Protein fusion in tobacco BY-2 cells does not change cellular growth behavior but leads to a change in phase distribution, and these cells exhibit a shorter G1-phase compensated by an increase in the length of S-phase and a reduced overall cell cycle length (Nakagami et al., 2002). Inducible expression of an Antirrhinum CYCD1 gene also in BY-2 cells independently enhances both G1/S entry and progression through S- and G2-phases (Koroleva et al., 2004).
These results obtained in tobacco cells are distinct from those observed in CYCD3;1-overexpressing plants, which do not appear to show an increased S-phase population, perhaps because of either gene-specific effects or the particular responses of tobacco cells. To date, there have been no studies of the effects of ectopic expression of Arabidopsis CYCD genes in cell culture systems, and the effect of CYCD overexpression on cell cycle arrest as a result of stationary phase or nutrient depletion is also unknown.
The Arabidopsis cell suspension cultures MM1 and MM2d can be partially synchronized by sucrose removal, which leads to arrest primarily in G1-phase (Menges and Murray, 2002). Higher synchrony can be achieved by blocking the cells in late G1/early S-phase using aphidicolin, which has been used to study cell cycle progression and the expression of phase-specific genes (Menges and Murray, 2002; Menges et al., 2002, 2003, 2005). These cultures have also been used to investigate the stability of CYCD3;1, and we recently showed that CYCD3;1 is a highly unstable phosphoprotein degraded by a proteasome-dependent mechanism whose presence depends on continued translation (Planchais et al., 2004). Treatment of cells with the proteasome inhibitor MG132 results in the accumulation of a hyperphosphorylated form of CYCD3;1. These results suggested a possible link between phosphorylation and CYCD3;1 turnover, because phosphorylation is frequently linked to ubiquitin-dependent proteolysis (Vierstra, 2003).
Here, we investigate the role of CYCD3;1 in the cell cycle using Arabidopsis cell suspension cultures. We show that CYCD3;1 acts as a dominant driver of the G1/S transition and partially overcomes the G1 arrest induced by stationary phase or sucrose removal. Synchronized CYCD3;1-overexpressing cultures also show an increase in the length of G2-phase, and this extension occurs at a point before the transcriptional activation of B-type cyclins and other mitotic genes, such as CDKB2, in marked contrast with the effect of Antirrhinum CYCD1 in BY-2 cells. We also show that, by contrast, CYCD2;1 does not delay G2-phase, suggesting that CYCD3;1 is a dominant and specific factor in driving Arabidopsis cells through the G1/S transition, potentially acting in a similar manner to cyclin E. We also show that overexpressed CYCD3;1 interacts with its CDK partner CDKA in both dividing and stationary phase cells, suggesting that there are unlikely to be stringent controls over CYCD3-CDKA complex assembly. Mutation of the potential CDK phosphorylation site Ser-343 to Ala apparently increases the potency of CYCD3;1 overexpression, despite there being no change in protein stability, as determined by cycloheximide treatment. Cells expressing the mutant protein respond to sucrose removal by extensive cell death, further supporting a key role for CYCD3;1 in controlling cell cycle progression in response to sucrose availability.
RESULTS
Constitutive Expression of CYCD3;1 Reduces the Proportion of Cells in G1-Phase
To investigate the role of CYCD3;1 in the plant cell cycle independently of its roles in differentiation and development, CYCD3;1 was expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter in the synchronizable Arabidopsis cell lines MM1 and MM2d. These lines have the same origin but are cultured in either continuous light (MM1) or darkness (MM2d) and have only minor differences in growth characteristics (Menges and Murray, 2002). Two clones of each line expressing CYCD3;1 (35S:CYCD3;1) were selected, and the higher level of CYCD3;1 expression in transgenic lines was confirmed by quantitative real-time RT-PCR and compared with that in wild-type cells (Figure 1A) and by protein blot analysis (Figure 1B).
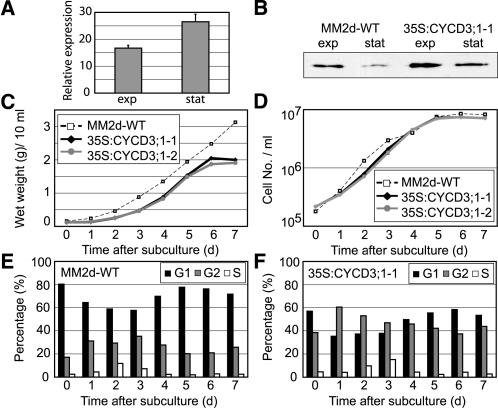
Figure 1.
Characterization of 35S:CYCD3;1 Arabidopsis Cell Lines.
Early stationary phase cells (7 d after subculture) were transferred into fresh medium, and samples were taken for analysis after the days indicated.
(A) Real time RT-PCR analysis to detect increased abundance of CYCD3;1 RNA in cell line 35S:CYCD3;1-1, presented as relative expression to the wild type (MM2d, baseline) during exponential growth (exp; day 3) and early stationary phase (stat; day 7). Error bars indicate sem; n = 4.
(B) Protein gel blot analysis of CYCD3;1 protein during exponential growth (exp; day 3) and early stationary phase (stat; day 7) in the cell lines indicated.
(C) and (D) Determination of biomass (wet weight) (C) and cell density (D) each day after dilution.
(E) and (F) Flow cytometry analysis to monitor changes in DNA distribution during growth in the wild type (E) and transgenic 35S:CYCD3;1-1 (F). The percentages of cells in G1 (black bars), G2 (gray bars), and S (white bars) are shown.
All 35S:CYCD3;1 lines, regardless of the cell line used, showed similar characteristics in all experiments reported in this work. To further confirm that 35S:CYCD3;1 lines do not exhibit significant alterations in growth behavior under different culture conditions (light or dark), a culture of line 35S:CYCD3;1-1 (derived from the dark-grown cell line MM2d) was transferred to continuous light and incubated further for a period of 1 year under these conditions (and designated 35S:CYCD3;1-2). This subline showed no difference in any behavior or results reported here from other 35S:CYCD3;1 lines, regardless of their original parental line (Figures 1C and 1D).
The growth of the untransformed control line MM2d and the 35S:CYCD3;1-1 and 35S:CYCD3;1-2 lines was compared by monitoring wet weight (Figure 1C), cell number (Figure 1D), and doubling times (Table 1) for 7 d after subculture. Differences between lines were small, with a somewhat smaller cell size for CYCD3;1 lines, resulting in a slightly reduced wet weight of the culture (Figure 1C). However, the major difference between 35S:CYCD3;1 lines and the wild type was a large difference in the distribution of cells between cell cycle phases at all times during the growth cycle, from subculture to stationary phase (Figures 1E and 1F). In all cases, the proportion of cells in G1-phase was reduced from 50% (at day 3) to ∼35%, whereas the proportion of cells in G2-phase was increased from 33% in control cells to 50%. This was further confirmed by calculating cell cycle phase durations (Table 1), showing a 25% decrease in the length of G1-phase in 35S:CYCD3;1, little change in the length of S-phase, and a large increase in the duration of G2-phase (Table 1).
Table 1.
Doubling Time and Phase Duration of Wild-Type and 35S:CYCD3;1 Cells during Exponential Growth
| Percentage of Cells in Cell Cycle Phase (Day 3)
|
Duration of Different Phases (h)
|
||||||
|---|---|---|---|---|---|---|---|
| Cell Line | Doubling Time (h) | G1 | S | G2 | G1 | S | G2 |
| Wild type (MM2d) | 19.1 | 57.5 | 7.2 | 35.3 | 10.1 | 3.0 | 6.0 |
| 35S:CYCD3;1-T1 | 17.4 | 37.8 | 15.2 | 47 | 6.3 | 2.1 | 9.0 |
| 35S:CYCD3;1-T2 | 19.3 | 49.2 | 8.6 | 42.1 | 8.8 | 2.1 | 8.4 |
From the midexponential phase of the growth curve (day 2 to day 3) under nonsynchronized conditions, the doubling time was calculated, assuming that all cells are cycling, an assumption supported by the data shown in Figure 3. Flow cytometry was used to determine the proportion of exponentially growing cells (day 3) in G1, S, and G2, and the length of each phase was calculated (Granier and Tardieu, 1998).
As cells enter stationary phase, they normally stop dividing in G1, and for wild-type cells ∼20% are in G2-phase compared with ∼40% for 35S:CYCD3;1. This suggests that cells overexpressing CYCD3;1 may pass a G1/S restriction point more quickly and thereby shorten the G1-phase but exhibit a significant increase in the length of G2. Similar results were consistently obtained with all four 35S:CYCD3;1 lines, regardless of parentage (MM1 or MM2d) or culture conditions (light or dark).
Previous analysis showed that CYCD3;1 is present in exponentially growing MM1 wild-type cells and strongly reduced in stationary phase cells (Planchais et al., 2004). To establish whether the effect on the cell cycle distribution of 35S:CYCD3;1 cells in stationary phase was linked to the continued presence of CYCD3;1 in these cells, we examined protein levels at different stages of culture growth (Figure 1B). Indeed, the relative levels of CYCD3;1 are higher in stationary phase in the 35S:CYCD3;1-1 line because of significantly lower RNA and protein levels for CYCD3;1 in wild-type cells (Figures 1A and 1B). This suggests that the observed effect on the phase distribution in stationary phase could be attributable to a higher expression level of CYCD3;1 in states in which its expression is normally switched off, resulting in a partial overcoming of the preferred tendency of cells to arrest in G1 on reaching stationary phase. The protein and RNA levels of CYCD2;1 and CDKA, the CDK partner of CYCD3;1, were unchanged in 35S:CYCD3;1 cells (data not shown).
35S:CYCD3;1 Cells Have a Higher Frequency of Arrest in G2-Phase after Sucrose Starvation
Removal of sucrose from the growth medium of exponentially growing cells leads to a cessation of S-phase entry and a preferential arrest of cells in G1-phase (70%) within 12 h (Figure 2B) (Menges and Murray, 2002; Planchais et al., 2004). This is correlated with a reduction in CYCD3;1, but not CYCD2;1 or CDKA, protein levels (Planchais et al., 2004). To further investigate the effect of CYCD3;1 on the G1-to-S-phase control point, we examined the response of 35S:CYCD3;1 cells to sucrose removal. We observed that wild-type cells show an increase in G1 cells from ∼55 to ∼70% of the population within 12 h (Figure 2B). By contrast, CYCD3;1 cells show a decrease in G1 cells and an increase in G2 cells over the same period, from ∼45 to ∼65% of the population (Figure 2C). In both wild-type and CYCD3;1 cultures, S-phase cells are substantially reduced or absent within 6 h of sucrose removal. Controls in which cells are placed in normal (3%) or low (0.3%) levels of sucrose show that cycling continues in both cases, with the higher level of G2-phase cells observed in all 35S:CYCD3;1 samples.
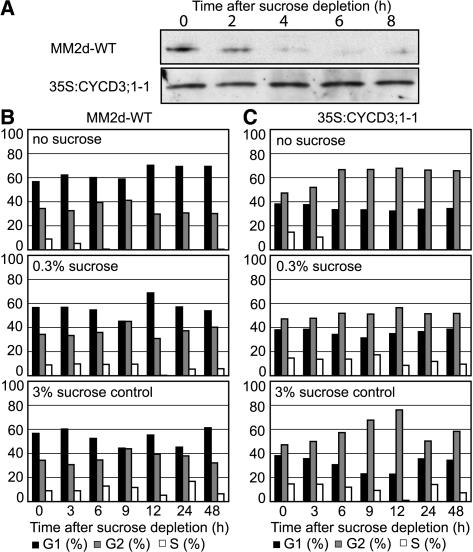
Figure 2.
Effect of Ectopic CYCD3;1 Expression on Cell Cycle Response to Sucrose Depletion.
(A) Midexponentially growing cells (day 3) were washed twice in MS medium (lacking sucrose; see Methods), resuspended, and diluted into MS medium (dilution, 1:5). For protein gel blot analysis of CYCD3;1, samples were taken every 2 h in the cell lines indicated.
(B) and (C) Midexponentially growing cells (day 3) were washed twice in MS medium (lacking sucrose), resuspended, and diluted in MS medium containing no sucrose, 0.3% sucrose, or 3% sucrose (dilution, 1:5). The arrest of cell cycle activity on sucrose depletion and change in DNA distribution was monitored by flow cytometry in wild-type (B) and transgenic 35S:CYCD3;1-1 (C) cells. The percentages of cells in G1 (black bars), G2 (gray bars), and S (white bars) are shown.
The continued increase in the G2-phase proportion on sucrose removal shows that CYCD3;1 cells preferentially arrest in G2, whereas wild-type cells preferentially arrest in G1-phase. Protein gel blots confirmed that CYCD3;1 is present in sucrose-deprived 35S:CYCD3;1 cells (Figure 2A, bottom) but not in wild-type cells, in which degradation of CYCD3;1 is observed 4 h after depletion (Figure 2A, top) (Planchais et al., 2004). Hence, ectopic expression of CYCD3;1 is associated with a reduced tendency to arrest in G1-phase, suggesting that it partially overcomes the normal control mechanism at the G1/S restriction point but does not remove the signal for cessation of cycling in response to sucrose removal. Therefore, cells arrest at the subsequent control point in G2-phase. Consistent with this interpretation, 35S:CYCD3;1 cultures show a higher level of S-phase cells than wild-type cultures in low (0.3%) or absent sucrose (Figures 2B and 2C).
35S:CYCD3;1 Cells Have an Extended G2-Phase
A higher proportion of cells have a G2-phase DNA content in both 35S:CYCD3;1 cultures and transgenic plants (Dewitte et al., 2003). One possible explanation is that a proportion of cells are permanently arrested in G2 and thus no longer cycling, perhaps as a result of their initiation of a blocked endocycle program. Alternatively, most cells may continue to cycle, but with an extended duration of G2-phase. To distinguish between these possibilities, we used aphidicolin block–release experiments to generate cultures with a high degree of synchrony from S- to M-phases (Menges and Murray, 2002). Synchronization of wild-type and 35S:CYCD3;1 cells was performed and is shown in Figures 3A and 3B. Both wild-type and 35S:CYCD3;1 cells were synchronized to a high degree, with ∼85% of cells arrested at the G1/S boundary. Release of the aphidicolin block resulted in a high degree of synchrony of both cultures. Importantly, >90% of 35S:CYCD3;1 cells were found in S-phase at 1 to 2 h after release, demonstrating that all cells in the culture are cycling and that there is no significant population of noncycling cells blocked in G2-phase (Figure 3B). Interestingly, S-phase is completed in 35S:CYCD3;1 cells up to 1 h earlier than in control cells, although S-phase length calculation in unsynchronized cultures showed little difference (Table 1). This finding suggests that 35S:CYCD3;1 cells are able to progress further into S-phase in the presence of aphidicolin than are control cells; indeed, no G1-phase cells remained 1 h after aphidicolin removal.
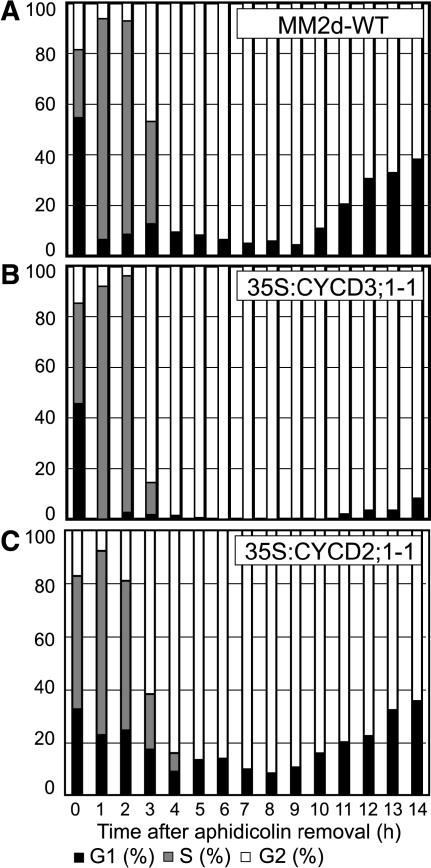
Figure 3.
Cell Cycle Progression in Arabidopsis Cell Lines after Aphidicolin-Induced Synchrony.
Cells were treated for 21 h with aphidicolin, and after release of the block, the synchronous progression of cells was monitored by flow cytometry in wild-type (A), 35S:CYCD3;1 (B), and 35S:CYCD2;1 (C) cells. The percentages of cells in G1 (black bars), G2 (white bars), and S (gray bars) are represented as DNA histograms.
Consistent with the prevalence of G2-phase cells observed in 35S:CYCD3;1 cultures, these cultures show a significantly longer period before the proportion of G1-phase cells increases after the completion of mitosis. Control cultures show increasing numbers of G1 cells after 10 h and reach 40% G1-phase cells after 14 h. 35S:CYCD3;1 cultures exhibit only 10% G1 cells after 14 h. A similar delay was also observed in all 35S:CYCD3;1 lines, independent of the cell line from which they were derived.
To establish whether the observed consequences of ectopic CYCD3;1 expression on S-phase and G2 progression are a general effect of CYCD or even an artifact of the transformation process, we repeated this analysis in a transgenic Arabidopsis line constitutively expressing CYCD2;1 (35S:CYCD2;1). No delay was observed in these cells in the progression through G2-phase (Figure 3C), nor was there any difference from wild-type cells in the distribution of cells between phases in nonsynchronized cultures (Menges and Murray, 2004). We conclude that the effect of CYCD3;1 expression on promoting the G1-to-S-phase transition and on provoking G2-phase delay is not observed with CYCD2;1.
35S:CYCD3;1 Delays the Activation of Cyclin B and G2/M Gene Expression
We recently showed that cyclin B and most cyclin A genes are coactivated in late G2 and show an expression peak 10 h after release of the aphidicolin block. Eighty-two genes share this common regulatory pattern (Menges et al., 2005), including two genes of the plant-specific CDKB class, CDKB2;1 and CDKB2;2. Expression of the related CDKB1 gene peaks ∼4 h before the CDKB2/CYCB cluster in mid-G2-phase. Because synchronization experiments demonstrated that CYCD3;1 overexpression leads to an extended G2-phase, we sought to investigate the timing of this delay; therefore, we examined the expression of the marker genes characteristic for S- and G2-phases using RT-PCR (Figure 4).
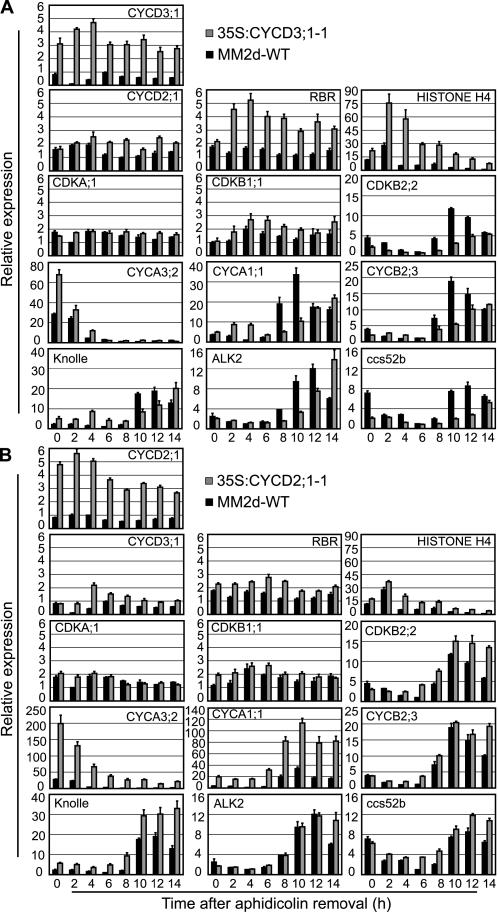
Figure 4.
35S:CYCD3;1 Delays the Activation of G2/M-Phase Genes.
Expression of the genes indicated using RNA prepared from the synchronized cell samples in Figure 3 was monitored by quantitative RT-PCR using Actin-2 as an internal control. Error bars indicate se; n = 4.
(A) Relative expression of genes in line 35S:CYCD3;1-1 compared with the wild type (MM2d) relative to the lowest expression observed in the experiment, except CYCD3;1, which is presented relative to the maximum wild-type expression (wild type, black bars; 35S:CYCD3;1-1, gray bars).
(B) Relative expression of genes in line 35S:CYCD2;1-1 compared with the wild type (MM2d) as for (A) (wild-type, black bars; 35S:CYCD2;1-1, gray bars).
As expected, 35S:CYCD3;1 lines show higher levels of CYCD3;1 mRNA (Figure 4A); interestingly, they also show a normal pattern of expression but strongly increased levels of the S-phase genes HISTONE H4 and CYCA3;2, as well as RBR (Menges et al., 2005). This is likely attributable to the resulting increase in E2F activity in 35S:CYCD3;1 lines, because these genes are also increased as a result of increased E2F expression (S.M. de Jager and J.A.H. Murray, unpublished data). Increased RBR levels were also reported in plants overexpressing CYCD3;1 (Dewitte et al., 2003). Both wild-type and 35S:CYCD3;1 cells show a similar pattern of expression and levels of the B-type CDK CDKB1;1, which is activated from the start of S-phase. However, when we examined the timing of expression of genes of the CDKB2/CYCB regulatory cluster, we found a significant delay in the activation of a sample of these genes (Figure 5), including CYCA1;1, CYCB2;3, CDKB2;2, the mitotic syntaxin KNOLLE, the aurora-like kinase ALK2 (Menges et al., 2005), and the APC regulatory subunit CCS52b (Tarayre et al., 2004). In wild-type cells, as well as in 35S:CYCD2;1 cells (Figures 4A and 4B), these genes show peak expression after 10 to 12 h, whereas in 35S:CYCD3;1 cells, expression is still increasing at 14 h. We conclude that 35S:CYCD3;1 induces a G2 delay before the activation of expression of CYCB and coregulated genes (Figure 5).
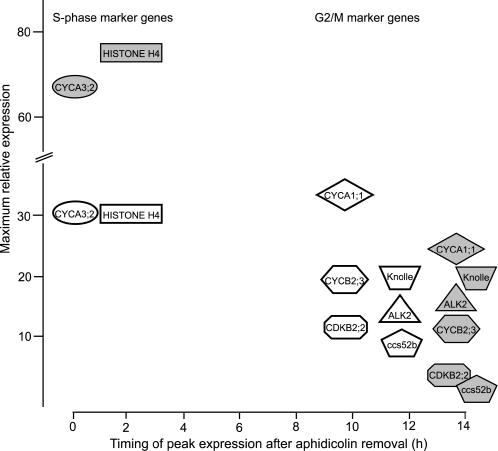
Figure 5.
Summary of Data from Figure 4.
Expression of selected cell cycle marker genes in cell line 35S:CYCD3;1-1 (gray symbols) compared with the wild type (white symbols). The genes are indicated at the time of their peak expression and positioned according to their fold increase in expression over the minimum level observed in wild-type cells. In particular, the higher expression of the S-phase genes HISTONE H4 and CYCA3;2 and the later expression of G2/M genes in 35S:CYCD3;1 cells is apparent.
By contrast, 35S:CYCD2;1 cells, although showing a similar level of overexpression to that of CYCD3;1 in 35S:CYCD3;1 cells, show little increase in the expression of S-phase genes and no delay in the activation of G2/M-phase genes. Indeed, these cells show higher levels and/or more sustained expression of mRNA for certain genes, such as CYCA1;1 and KNOLLE (Figure 4B), although this was not reflected in any detectable delay in mitotic exit, as determined by the rate of accumulation of G1-phase cells (Figure 3C).
Constitutively Expressed CYCD3;1 Binds CDKA
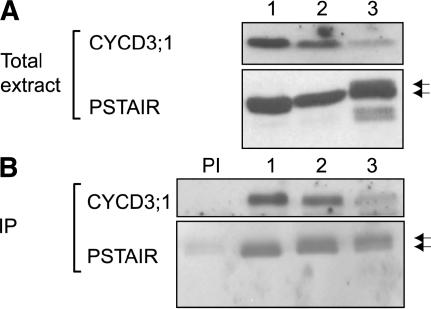
CYCD3;1 binds CDKA to form functional complexes but does not bind CDKB1 in vivo (Healy et al., 2001), but CYCD3;1 abundance normally decreases during stationary phase and is absent after sucrose starvation. We investigated whether overexpressed CYCD3;1 in the 35S:CYCD3;1 lines interacts with CDKA in all growth states with the same specificity. Using specific anti-CYCD3;1 antisera (Healy et al., 2001) and a monoclonal antibody against the conserved PSTAIRE epitope of CDKA, protein gel blot analysis showed that CYCD3 and CDKA are present in exponential, stationary phase, and sucrose-starved cells, although CYCD3;1 abundance is significantly reduced after sucrose starvation, suggesting a higher turnover of the protein under these conditions (Figure 6). Immunoprecipitation using anti-CYCD3;1 antisera showed that CYCD3;1 and CDKA coprecipitate in all conditions tested (Figure 6). Because CYCD3;1 is only detectable in exponentially growing MM1 wild-type cells, no CDKA;1 could be detected in immunoprecipitates from other growth states (data not shown). No CDKB1 was detectable in any immunoprecipitates, as reported previously (Healy et al., 2001).
Figure 6.
Constitutively Expressed CYCD3;1 Interacts with CDKA;1.
Total protein extracted from 35S:CYCD3;1-3 was analyzed by protein gel blot analysis (A) and immunoprecipitation (B).
(A) Protein gel blot analysis using anti-CYCD3;1 antibody (Healy et al., 2001) and CDKA (using an anti-PSTAIRE monoclonal antibody). Lane 1, exponentially growing cells (day 3); lane 2, early stationary phase cells (day 7); lane 3, 24 h after sucrose starvation of exponentially growing cells (day 3).
(B) Immunoprecipitation (IP) of CYCD3;1 followed by protein gel blot detection of CYCD3;1 and CDKA (PSTAIRE). PI, preimmune serum with protein extracts of sample 1. Lanes 1 to 3 are as in (A).
Interestingly after sucrose starvation, we observed additional bands with the anti-PSTAIRE CDKA antibody, including two smaller proteins, although only the two larger bands coimmunoprecipitated with CYCD3;1. These higher molecular weight bands may represent alternative phosphorylated forms of CDKA (Figure 6A, lane 3).
Mutation of Ser-343 to Ala Alters Functional Properties of CYCD3;1
Investigations in mammalian cells (Diehl and Sherr, 1997) revealed that the stability and localization of D-type cyclins are regulated by phosphorylation of the cyclin, and Nakagami et al. (2002) showed that phosphorylation of Thr-191 in the tobacco D-type cyclin NictaCYCD3;3 may be necessary for full kinase activity and localization.
We found recently that Arabidopsis CYCD3;1 is phosphorylated in sucrose-starved cells and accumulates in a hyperphosphorylated form when the proteasome degradation pathway is blocked by the inhibitor MG132 (Planchais et al., 2004). To investigate whether CYCD3;1 is phosphorylated in normally cycling cells, protein extracts from exponentially growing wild-type cells were treated with λ protein phosphatase, which dephosphorylates phosphoserines, phosphothreonines, and phosphotyrosines, followed by protein gel blot analysis. A shift of CYCD3;1 migration to a faster-migrating form was observed between untreated and treated samples (Figure 7A, lanes 1 and 2), indicating that a phosphorylated form of the protein exhibits retarded migration. No band was observed in the untreated sample at the equivalent migration to phosphatase-treated CYCD3;1, indicating that all, or a significant majority, of CYCD3;1 present in exponentially growing cells is phosphorylated, as was also reported by Planchais et al. (2004) in sucrose-starved cells. This apparent complete and constitutive phosphorylation of CYCD3;1 was also observed in 35S:CYCD3;1 cells (Figure 7A), in which the level of CYCD3;1 was increased severalfold, whether exponentially growing (lanes 3 and 4), in stationary phase (lanes 5 and 6), or after sucrose starvation (lanes 7 and 8). CYCD3;1, therefore, is a constitutive phosphoprotein in suspension-cultured cells. Note that this constitutive phosphorylation is distinct from the hyperphosphorylation detected after MG132 treatment (Planchais et al., 2004) (Figure 8C).
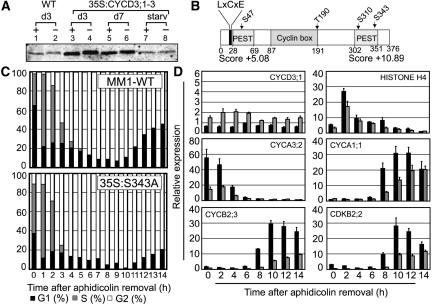
Figure 7.
CYCD3;1 Is a Constitutive Phosphoprotein.
(A) Protein extracts of wild-type and 35S:CYCD3;1 Arabidopsis cell lines were treated with λ protein phosphatase. CYCD3;1 was detected by protein gel blot analysis in treated (+) and untreated (−) samples of exponentially growing cells (day 3 [d3]; lanes 1 to 4), early stationary phase cells (day 7 [d7]; lanes 5 and 6), and after 24 h of sucrose removal from the medium of exponentially growing cells (starv; lanes 7 and 8).
(B) Scheme of the CYCD3;1 sequence. Important domains and positions of mutated residues are highlighted. The LxCxE motif at the N terminus is necessary for binding to RB (Huntley et al., 1998), and PEST sequences with PESTfind scores are indicated.
(C) Cell cycle progression in Arabidopsis wild type and mutant cell line CYCD3-S343A (35S:S343A) after aphidicolin-induced synchrony. Cells were treated for 21 h with aphidicolin, and after release of the block, the synchronous progression of cells was monitored by flow cytometry. The percentages of cells in G1 (black bars), G2 (white bars), and S (gray bars) are represented as DNA histograms.
(D) Mutation of CYCD3-S343A (35S:S343A) results in a delay of activation of G2/M genes. Expression of the genes indicated was monitored by quantitative RT-PCR using Actin-2 as an internal control, shown as described in the legend to Figure 4A. Relative expression of mutant cell line 35S:S343A compared with the wild type (MM1) is shown relative to the lowest expression observed, except for CYCD3;1, which is presented relative to the maximum wild-type expression (wild type, black bars; 35S:S343A, gray bars). Error bars indicate se; n = 4.
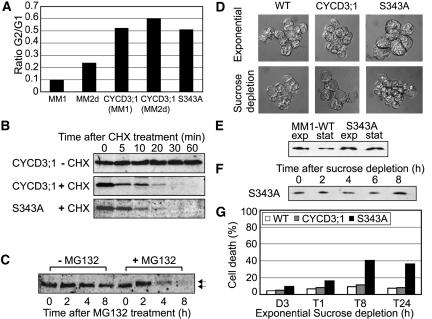
Figure 8.
Characterization of CYCD3;1 Mutant S343A.
(A) Flow cytometry analysis of early stationary phase cells (day 7) in the cell lines indicated, presented as the relative distribution of G1:G2 cells in the population and showing the greater propensity of CYCD3;1– and CYCD3;1-S343A–expressing cells to exit division in G2-phase.
(B) Cells expressing CYCD3;1 or CYCD3;1-S343A were incubated with (+) or without (−) cycloheximide (CHX) for the times indicated, and extracts were analyzed by immunoblotting with anti-CYCD3;1 antibody.
(C) Exponentially growing cells from line CYCD3-S343A (day 4) were washed twice with MS medium to remove sucrose, resuspended, and treated with (+MG132) or in the absence of (−MG132) 100 μM MG132 for the times indicated. After protein extraction, CYCD3;1 was detected by protein gel blot analysis. Normal and hyperphosphorylated bands were detected for CYCD3-S343A, as observed previously with similar kinetics for wild-type CYCD3;1 (Planchais et al., 2004).
(D) Cell morphology in exponentially growing cells (day 3; top) and after 24 h of sucrose depletion (bottom) in the wild type (MM1), 35S:CYCD3;1, and CYCD3-S343A.
(E) Protein gel blot analysis of CYCD3;1 during exponential growth (exp; day 3) and early stationary phase (stat; day 7) in the cell lines indicated.
(F) Midexponentially growing cells of line 35S:S343A (day 3) were washed twice in MS medium (lacking sucrose), resuspended, and diluted in MS medium (dilution, 1:5) as described for Figure 2A. For protein gel blot analysis of CYCD3;1, samples were taken every 2 h.
(G) Effect of ectopic CYCD3;1 expression on viability in response to sucrose depletion. Midexponentially growing cells (day 3) were washed twice in MS medium (lacking sucrose), resuspended, and diluted in medium containing no sucrose (dilution, 1:5). Samples were taken as indicated and subjected to trypan blue staining to determine the percentage of dead or dying cells.
In Arabidopsis CYCD3;1, there are four putative phosphorylation sites for CDKs and other Pro-directed kinases, based on the conserved motif S/TP. Three of these sites lie within PEST sequences predicted by the program PESTfind (www.at.embnet.org/embnet/tools/bio/PESTfind/) and proposed to be involved in protein turnover (Rechsteiner and Rogers, 1996), whereas the fourth (Thr-190) is located at the C-terminal end of the cyclin box (Figure 7B). All four sites were separately mutated to Ala as a residue not susceptible to phosphorylation and expressed in tobacco BY2 cells under the control of the CaMV 35S promoter. The wild-type CYCD3;1 protein and the S47A, T190A, and S310A mutant proteins all exhibited constitutive phosphorylation in exponential BY2 cells, whereas CYCD3;1-S343A did not show a shift on phosphatase treatment, indicating that unlike wild-type CYCD3;1, the mutant protein may not be phosphorylated during normal growth conditions (data not shown).
To establish whether mutation of Ser-343 has effects on CYCD3;1 activity in Arabidopsis cells, the construct was stably transformed into MM1 cells. We observed that the resulting cell line, 35S:CYCD3-S343A (35S:S343A), showed effects on cell cycle similar to wild-type CYCD3;1 (35S:CYCD3;1), including a significant shift to G2-phase cells similar to that observed with 35S:CYCD3;1 lines in both MM1 and MM2d backgrounds (Figure 8A), and in synchronized cultures a more rapid progression through S-phase from the arrest point, a lengthened G2-phase (Figure 7C), and delayed activation of the G2/M cluster of coregulated genes, such as CYCA1;1, CYCB2;3, and CDKB2;2 (Figure 7D).
Examining the expression levels of CYCD3;1 in synchronized 35S:S343A cells, we noted that the overall level of CYCD3;1 detected was only approximately twofold the wild-type levels (Figure 7D), implying that mRNA encoding the S343A variant was approximately equal to that of endogenous CYCD3;1 in dividing cells, compared with the 4- to 10-fold increase in the 35S:CYCD3;1-1 line (Figure 4A). Because the effects on cell cycle progression were similar to those of 35S:CYCD3;1 lines despite the only marginally increased overall CYCD3;1 level, the S343A mutant of CYCD3;1 may have a more potent effect. Whereas the expression of other G2/M-phase marker genes was similarly delayed in 35S:S343A cells as in 35S:CYCD3;1 cells (Figure 7D), the increased level of the S-phase genes HISTONE H4 and CYCA3;2 observed in 35S:CYCD3;1 cells was not seen; rather, expression of the latter gene was significantly lower in 35S:S343A cells (Figure 7D). Reduced expression of the S-phase marker gene HISTONE H4 was confirmed in nonsynchronized cells (see Supplemental Figure 1B online). These results suggest that the effects of CYCD3;1 on S-phase gene expression may be independent of the effects on G2 delay and G2/M-phase gene expression.
We also noted that 35S:S343A cells showed morphological differences, which were particularly apparent during sucrose depletion of exponentially growing cells (Figure 8D). Whereas wild-type and 35S:CYCD3;1 cells respond to deprivation of the carbon source by vacuolization (Figure 8D, bottom left and middle), only small or no vacuoles are observed in 35S:S343A cells 24 h after sucrose depletion (Figure 8D, bottom right). To establish the effect of this failure of vacuolization, we monitored cell death during sucrose depletion in wild-type, 35S:CYCD3;1, and 35S:S343A cells using trypan blue staining. We noted that even in exponential cultures, a higher level of dead cells was present in 35S:S343A cultures (Figure 8G), and this increased to ∼40% of the population after 8 h of sucrose depletion. Wild-type and 35S:CYCD3;1 cells both exhibited ∼10% cell death at this time. These data suggest that the S343A mutant of CYCD3;1 prevents the normal cellular response of vacuolization to sucrose removal, resulting in a high level of cell death, and implicate Ser-343 as a potential regulatory residue.
The effect of CYCD3;1-S343A during sucrose removal suggested a possible effect of the S343A mutation on protein level during sucrose starvation or on protein stability. The protein level was monitored in 35S:S343A and wild-type cells (Figure 8E) and after sucrose removal and showed no difference from 35S;CYCD3;1 cells (cf. Figures 8F and 2A).
We have previously established that CYCD3;1 has a short half-life, as determined in cells in which protein synthesis has been blocked using cycloheximide, and is destroyed by a proteasome-dependent mechanism. Because proteasome degradation often depends on prior phosphorylation, and indeed treatment of MM1 cells with the proteasome inhibitor MG132 resulted in the accumulation of CYCD3;1 in a hyperphosphorylated form, we speculated that Ser-343 might determine the stability of CYCD3;1, particularly given that it is located within the C-terminal PEST sequence (Figure 7B) (Soni et al., 1995). However, no significant difference was observed between the stability of CYCD3;1 and CYCD3-S343A when de novo protein synthesis was blocked by treating cells with cycloheximide (Figure 8B), and both showed similar stability to endogenous CYCD3;1, which was calculated to have a half-life of 7 min under these conditions (Planchais et al., 2004). Despite the apparent absence of constitutive phosphorylation of CYCD3;1-S343A during normal growth conditions, on MG132 treatment CYCD3-S343A accumulates in a hyperphosphorylated form, as does the wild-type protein (Planchais et al., 2004), indicating that Ser-343 is neither required for degradation nor phosphorylated as part of the degradation process (Figure 8C). Therefore, additional and/or multiple phosphorylation(s) is presumably involved in the degradation of CYCD3;1.
DISCUSSION
CYCD3;1 Is a Dominant and Specific Driver of the G1-to-S-Phase Transition
The cell cycle is regulated by control points of which the most significant is the G1 restriction point, at which cells must integrate a variety of nutritional, hormonal, and developmental signals in making the irreversible decision to commit to the cycle and hence progress from G1- to S-phase and initiate DNA replication. Therefore, understanding of the G1 control point and the G1-to-S-phase transition is of primary importance in plant cell cycle studies.
Microarray expression analysis shows that CYCD3;1 is probably the highest expressed cyclin in Arabidopsis suspension cultured cells (Menges et al., 2005). Here, we show that CYCD3;1 is a rate-limiting regulator of the G1/S transition in cycling Arabidopsis cells. Overexpression of CYCD3;1 results in a reduction of G1-phase length and reduces the stringency of the G1 control point, such that cells show a reduced arrest at the stringent G1 control point in response to stationary phase or sucrose removal. Previous studies have shown that CYCD3;1 expression is regulated by nutrient and hormonal signals, including sucrose (Riou-Khamlichi et al., 2000), cytokinin (Riou-Khamlichi et al., 1999), and brassinosteroids and other hormones (Hu et al., 2000; Oakenfull et al., 2002). The data presented here are consistent with CYCD3;1 levels playing a key role in controlling cell progression through the G1/S-phase transition in response to the continued presence of such external signals. However, 35S:CYCD3;1 cells stop dividing and start to enter stationary phase at day 5 (Figure 1D), as do wild-type cells, showing that CYCD3;1 overexpression does not override the cessation of division as cells enter stationary phase but rather alters the relative stringency of the G1 and G2 control points. We did not observe these effects in cells expressing CYCD2;1 to a similar level.
Two previous studies have examined the role of D-type cyclins in tobacco BY-2 cells, but important differences were seen in the results reported. Koroleva et al. (2004) expressed the heterologous Antirrhinum CYCD1 gene and showed that it could accelerate both the G1-to-S and G2-to-M transitions of BY-2 cells. Nakagami et al. (2002) showed that a tobacco CYCD3;3-Green Fluorescent Protein fusion protein also reduced the length of G1-phase, but this was compensated by an increase in S-phase length. We show here that CYCD3;1 overexpression in Arabidopsis results in an increase in the proportion of G2-phase cells, and we further show that this is attributable to the extension of G2-phase before the induction of B-type cyclins (CYCB) and coregulated genes, and not to the permanent arrest of a proportion of cells in G2-phase. A similar striking increase in cells with a 4C (G2) DNA content (26% instead of 16%) and a reduced proportion of cells in G1-phase (31% instead of 45%) were also observed in meristems of CYCD3-overexpressing plants compared with wild-type plants (Dewitte et al., 2003), and the results obtained in cell culture show that this is also likely attributable to an extended G2-phase. However, because CYCD3-overexpressing plants also show a striking reduction in endoreduplication, it was not possible to draw this conclusion from in planta studies.
The results we present here clearly demonstrate that CYCD3;1 does not promote the G2-to-M-phase transition, in contrast with previous suggestions from studies of the same gene in trichomes (Schnittger et al., 2002) and of the action of the Antirrhinum CYCD1 gene in tobacco BY-2 cells. Therefore, we believe that the promotion of mitotic cycles in trichomes by CYCD3;1 is attributable to its inhibitory effect on endocycles, as also observed in leaves (Dewitte et al., 2003), rather than to its ability to promote the G2/M-phase transition per se. The ability of CYCD1 to promote mitosis in tobacco cells is thus presumably linked to its different characteristics, or to differences between tobacco and Arabidopsis cells.
We cannot determine the mechanism or cause of the extended G2-phase in response to CYCD3;1 overexpression, but we clearly show that the G2-phase is extended before the activation of the coordinately regulated set of genes in late G2 (Figure 4A). This set comprises at least 82 genes, including most A- and B-type cyclins and both CDKB2 genes (Menges et al., 2005), all of which contain mitosis-specific activator sequences in their promoters and probably respond to activation by a specific three-repeat MYB (Ito et al., 2001; Araki et al., 2004). Analysis of a number of these genes demonstrates that their coordinate activation is delayed in 35S:CYCD3;1, and because Araki et al. (2004) have shown that MYB activation requires CYCB-containing kinase activation, this is likely delayed in 35S:CYCD3;1. This may be connected with the abbreviated G1-phase, such that G2-phase is extended to allow cells to reach the critical size that cells would normally attain during G1-phase. No activation of DNA damage genes was seen in CYCD3;1-expressing cells, suggesting that this is not the cause of the G2 delay.
In mammalian cells, acceleration of G1-phase by overexpression of cyclin D and/or cyclin E has been reported to lead either to an abbreviation of the overall cell cycle in rodent fibroblasts (Jiang et al., 1993; Ohtsubo and Roberts, 1993; Quelle et al., 1993; Imoto et al., 1997) or to a compensatory extension of S/G2-phases (Resnitzky et al., 1994). Interestingly, in these cases, acceleration of cell cycle reentry (G0-G1-S), as well as G1 transit (G1-S) by cycling cells, were observed. However, cyclin E, which is induced later in G1 than cyclin D, is less effective at shortening the G1-phase in cells reentering the cell cycle (Jiang et al., 1993).
In the case of cells overexpressing CYCD3;1, we did not observe a reduction in the time taken for sucrose-starved or stationary phase cells to reenter the cycle and progress to S-phase (see Supplemental Figure 1A online). This suggests that CYCD3;1, although rate-limiting for transit through G1-phase in cycling cells, is not able to increase the rate at which cells transit G1 as cells reenter the cycle. In this regard, it is interesting that the normal timing of induction of CYCD3;1 in cells reentering the cell cycle is rather late in G1-phase, after 4 to 5 h (Menges and Murray, 2002), whereas the D-type cyclins CYCD3;3 and CYCD5;1 have a peak of mRNA abundance 2 h after sucrose readdition and hence are activated much earlier in cell cycle reentry (Menges et al., 2005). Therefore, these cyclins, or CYCD2;1, which shows modest but marked upregulation and activation in response to sucrose addition (Riou-Khamlichi et al., 2000), may be rate-limiting during sucrose-induced cell cycle reentry.
The absence of a cyclin E homolog in plants leads to the question of whether other cyclins fulfill this role. CYCD3;1 has a number of characteristics of cyclin E, including its ability to drive cells through the G1-to-S-phase transition. However, cyclin E expression is characterized by cyclical expression with a peak in late G1-phase. Although CYCD3;1 expression increases rapidly during the time in which cells are reentering the cycle, subsequent transcript levels do not show strong oscillations (Menges et al., 2005). Moreover, CYCD3;1 expression depends on external signals (Riou-Khamlichi et al., 1999, 2000; Oakenfull et al., 2002), characteristic of cyclin D expression. Another candidate for playing the role of cyclin E in plants is CYCA3, whose cyclical expression initiates in late G1/early S-phases (Menges et al., 2005).
Posttranslational Regulation of CYCD3;1
Using protein gel blot analysis and immunodetection, we show here that the expression of CYCD3;1 under the control of the CaMV 35S promoter is constitutive. Hence, unlike the situation in wild-type cells, CYCD3;1 is expressed in stationary and sucrose-starved cells, resulting in the overriding of the preferential G1 arrest point. Furthermore, we show that CYCD3;1 is a phosphoprotein under all three growth states examined. Our previous analysis showed that CYCD3;1 is an unstable protein that accumulates in a hyperphosphorylated form in the presence of the proteasome inhibitor MG132 (Planchais et al., 2004). However, we show here that Ser-343 phosphorylation does not appear to affect protein stability.
In mammalian cells, phosphorylation of D-type cyclins on Thr-286 is important for their full ability to activate CDKs and for intracellular localization, stability, and degradation (Diehl and Sherr, 1997; Diehl et al., 1997, 1998; Germain et al., 2000). In plants, Nakagami et al. (2002) showed that for tobacco CYCD3;3, Thr-191 is a putative phosphorylation site important for full kinase activity and nuclear import in tobacco BY-2 cells.
In contrast with these roles identified for cyclin D function in mammalian cells, we found no strong evidence that Ser-343 phosphorylation is involved in protein turnover or cell cycle activity, on the basis of observed phenotypes obtained by expression of the S343A mutant of CYCD3;1. However, some differences were observed in cellular phenotype, with an increase in cell size and, in some cultures, in division pattern. Cell growth has often been linked to G1 processes in yeast and mammalian cells, and indeed, yeast G1 cyclins link cell growth to division rate (Schneider et al., 2004). However, most striking was the effect of sucrose depletion on cells expressing CYCD3-S343A, which resulted in a failure to develop a vacuole and a high level of cell death (Figure 8G).
We have extensively used the removal of sucrose from the growth medium of Arabidopsis cells as a method to promote the cessation of cell cycling, allowing the readdition of sucrose to be used to generate partially synchronous cultures for cell cycle reentry (Menges and Murray, 2002; Menges et al., 2003, 2005). The cessation of division, development of the vacuole, and cytoplasmic reduction is a normal response to sucrose depletion in plant cell suspension cultures, which allows cells to survive without loss of viability for >24 h (Moriyasu and Ohsumi, 1996; Contento et al., 2004). Although the ectopic expression of CYCD3;1 does not significantly affect the ability of cells to undergo this response, even relatively low-level expression of the S343A mutant of CYCD3;1 leads to extensive cell death associated with a failure of the vacuole to develop. It is also notable that cells expressing S343A showed the same tendency to override the G1 checkpoint and to delay the G2-phase, consistent with the possibility that Ser-343 is a negative regulatory residue. Therefore, we suggest that Ser-343 is a key regulatory residue in CYCD3;1 function and that phosphorylation of this site is likely to be of particular importance for the inactivation of CYCD3;1 activity in response to signals such as sucrose starvation, which normally arrest the cell cycle. In this model, ectopic CYCD3;1 expression can be mitigated by phosphorylation during sucrose starvation, potentially explaining why 35S:CYCD3;1 cells are still able to cease division in a timely manner in response to such signals and cannot be accelerated through G1-phase during cell cycle reentry.
Although we observed similar effects on cell cycle distribution and G2-phase delay in both 35S:CYCD3;1 and 35S:S343A cells, there are differences in the effects on S-phase gene expression. In particular, whereas the expression of HISTONE H4 and CYCA3;2 is strongly increased in 35S:CYCD3;1 cells, probably because of increased E2F activity, HISTONE H4 is not increased and CYCA3;2 is decreased relative to the wild type in 35S:S343A cells. This result shows that the G2 delay, which is common to both lines, is probably not provoked by increased S-phase gene expression.
Overexpression of CYCD3;1 in transgenic plants was found to promote progression of the cell cycle into S-phase, resulting in the accumulation of cells with a 4C DNA content (Dewitte et al., 2003). This was accompanied by a retardation of differentiation, as cells do not exit from the cell cycle but continue to divide, resulting in extensive hyperplasia of poorly differentiated cells and reduced endoreduplication. Therefore, we could not conclude from this analysis whether CYCD3;1 acts to promote the G1/S transition. Here, we confirm that CYCD3;1 is rate-limiting for entry into S-phase, providing a cell cycle explanation for the previously reported data.
METHODS
Plant Material and Culture Conditions
Suspension cultures of Arabidopsis thaliana cell lines MM1 and MM2d (Menges and Murray, 2002) were used, both of which were selected from a cell suspension originally produced from Landsberg erecta stem explants by May and Leaver (1993). For routine subculture, cell lines MM1, MM2d, and transgenic derivatives were maintained as described previously in Murashige and Skoog salts (MSS) medium (ICN Biomedicals) with addition of 3% sucrose, 0.5 mg/L napthalene acetic acid, and 0.05 mg/L kinetin (Menges and Murray, 2002).
Transformation of Arabidopsis MM1 and MM2d Cell Lines
MM1 and MM2d cells were transformed with constructs of CYCD3;1 (Riou-Khamlichi et al., 1999) and CYCD2;1 (Cockcroft et al., 2000; Masubelele et al., 2005), as described previously (Menges and Murray, 2004). Transgenic cell cultures 35S:CYCD3;1-1 and 35S:CYCD2;1 were derived from line MM2d. A culture of line 35S:CYCD3;1-1 was transferred to continuous light (designated 35S:CYCD3;1-2) and incubated for a period of 1 year under these conditions before further analysis. 35S:CYCD3;1-3/4 and 35S:S343A were derived from line MM1.
Characterization of Transgenic Cell Cultures
For growth analysis, 5 mL of an early stationary phase culture (7 d after subculture) was inoculated into 100 mL of fresh MSS and incubated. Samples were taken at ∼24-h intervals, and cells were observed microscopically and counted in triplicate in a counting chamber (0.2-mm depth). To determine nuclear DNA content, a sample of frozen cell pellets was treated to release nuclei and analyzed by flow cytometry as described (Menges and Murray, 2002). Calculation for cell doubling time and phase duration were as described by Granier and Tardieu (1998). Trypan blue staining of dead or dying cells was as described by Menges and Murray (2004).
Site-Directed Mutagenesis
Mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) (Table 2). The CYCD3;1 open reading frame cloned into the pART7 vector (Gleave, 1992) was used as a template. All mutations were confirmed by sequencing. Mutated CYCD3;1 was subsequently cloned into the binary transformation vector pART27 (Gleave, 1992) and transformed into bacterial strain GV3101. These constructs were used to transform Arabidopsis MM1 and tobacco (Nicotiana tabacum) BY-2 cell cultures.
Table 2.
Primers Used for Site-Directed Mutagenesis of CYCD3;1
| S47A | 5′-CTCTTCCTTGAGCTCTTCTTCTGCTCCATTCGTTG-3′ |
| 5′-CAACGAATGGAGCAGAAGAAGAGCTCAAGGAAGAG-3′ | |
| T190A | 5′-GGAAGATGCACCTCATTGCTCCAATTTCGTTTG-3′ |
| 5′-CAAACGAAATTGGAGCAATGAGGTGCATCTTCC-3′ | |
| S310A | 5′-CATCATCGTTGAACGCTCCAAGCTGCGTGATCGATGCAAACC-3′ |
| 5′-GGTTTGCATCGATCACGCAGCTTGGAGCGTTCAACGATGATG-3′ | |
| S343A | 5′-CCCACCAACGAGCTCGTCGGCCCCGCAGCAACAACCTCC-3′ |
| 5′-GGAGGTTGTTGCTGCGGGGCCGACGAGCTCGTTGGTGGG-3′ |
Mutagenesis was carried out using the QuikChange site-directed mutagenesis kit (Stratagene). Changes in the primers are indicated as underlined and in boldface.
Sucrose Depletion of Cell Cultures
For sucrose depletion experiments, an aliquot of 3-d-old cells was gently washed twice by centrifugation with MS medium (composition the same as MSS, but lacking sucrose), as described previously (Menges and Murray, 2002), resuspended, and diluted 1:5 in fresh MS medium (i.e., lacking sucrose) or MS medium supplemented with 0.3% sucrose or 3% sucrose. Samples were taken every 3 h over a 12-h range and after 24 and 48 h and analyzed by flow cytometry.
Aphidicolin-Induced Synchronization in Wild-Type and Transgenic Cell Cultures
Arabidopsis MM1, MM2d, and transgenic derivative cell lines 35S:CYCD3;1, 35S:CYCD2;1, and 35S:S343A were reversibly blocked in late G1/early S-phase with aphidicolin, as described previously (Menges and Murray, 2002). After removal of aphidicolin by washing, samples were taken hourly for analysis of DNA distribution by flow cytometry. RNA was extracted (Verwoerd et al., 1989), and the expression of marker genes was analyzed by RT-PCR, as described previously (Menges et al., 2005) (Table 3). Relative expression was calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001), in which the observed threshold cycle (CT) value of the RNA sample for the transgenic line was compared with an equivalent wild-type RNA sample. CT values of both samples were normalized using Actin-2 as an internal control.
Table 3.
Accession Numbers of Genes and Primer Pairs Used for RT-PCR
| Gene Identifier | Gene | Primer Pair |
|---|---|---|
| At4g34160 | CYCD3;1 | 5′-GCAAGTTGATCCCTTTGACC-3′ |
| 5′-CAGCTTGGACTGTTCAACGA-3′ | ||
| At2g22490 | CYCD2;1 | 5′-GCTGCTGCAGTGTCTGTTTC-3′ |
| 5′-ACAGCTCTTACCGCAACTCG-3′ | ||
| At3g12280 | RBR | 5′-CAAGTGGCTCAGGACTGTCA-3′ |
| 5′-TCCATCAGGTCAACAGCTTG-3′ | ||
| At2g28740 | HISTONE H4 | 5′-TTAGGCAAAGGAGGAGCAAA-3′ |
| 5′-CTCCTCGCATGCTCAGTGTA-3′ | ||
| At3g48750 | CDKA;1 | 5′-CCGAGCACCAGAGATACTCC-3′ |
| 5′-GTTACCCCACGCCATGTATC-3′ | ||
| At3g54180 | CDKB1;1 | 5′-CGATTACTCTGCGTCGAACA-3′ |
| 5′-TATGACAATGCGCAACACCT-3′ | ||
| At1g20930 | CDKB2;2 | 5′-AGCCTTCACTCTCCCAATGA-3′ |
| 5′-TCAGAGTCTCCCGCAAAGAT-3′ | ||
| At1g47210 | CYCA3;2 | 5′-GTCCGAAACAACATCCTTGG-3′ |
| 5′-AAAAGGTAACCGGCAGCTCT-3′ | ||
| At1g44110 | CYCA1;1 | 5′-GGCTAAGAAGCGACCTGATG-3′ |
| 5′-TACAAGCCACACCAAGCAAC-3′ | ||
| At1g20610 | CYCB2;3 | 5′-TAAACCACCTGTGCATCGAC-3′ |
| 5′-ATCTCCTCCAGCATTGCTTC-3′ | ||
| At1g08560 | Knolle | 5′-CGCTGGAGGTGAAGAGTTTC-3′ |
| 5′-TCGATCTCGTCCATTGTTCA-3′ | ||
| At4g32830 | ALK2 | 5′-CTCCTTTTGAAGCCATGGAG-3′ |
| 5′-TCCAGAAGGATCAGCGTTTT-3′ | ||
| At5g13840 | ccs52b | 5′-TGCTCTGGAAGTACCCATCC-3′ |
| 5′-CCCAATGACCAGAGACCTGT-3′ |
Protein Analysis
Procedures for protein extraction, immunoprecipitation, peptide competition, and protein gel blot analysis were as described previously (Cockcroft et al., 2000; Healy et al., 2001). For detection of the conserved PSTAIRE sequence of CDKA, a mouse monoclonal anti-PSTAIRE antibody was used (Sigma-Aldrich; catalog number P7962; dilution, 1:1000). Procedures for λ protein phosphatase, cycloheximide, and MG132 treatment were as described by Planchais et al. (2004).
Accession Numbers
The accession numbers of genes referred to in this article are listed in Table 3.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Expression of HISTONE H4 RNA to Monitor S-Phase Progression.
Supplementary Material
Acknowledgments
We thank Graham Armstrong for work on the figures and manuscript and Susan Howroyd for technical support. This work was supported by Biotechnology and Biological Science Research Council Grant 8/C15792 and by a European Community Marie Curie Fellowship (HPMF-CT-2000 00957) to A.K.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: James A.H. Murray (j.murray@biotech.cam.ac.uk).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.039636.
References
- Araki, S., Ito, M., Soyano, T., Nishihama, R., and Machida, Y. (2004). Mitotic cyclins stimulate the activity of c-Myb-like factors for transactivation of G(2)/M-phase-specific genes in tobacco. J. Biol. Chem. 279 32979–32988. [DOI] [PubMed] [Google Scholar]
- Barroco, R.M., Van Poucke, K., Bergervoet, J.H.W., De Veylder, L., Groot, S.P.C., Inze, D., and Engler, G. (2005). The role of the cell cycle machinery in resumption of postembryonic development. Plant Physiol. 137 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonstra, J. (2003). Progression through the G(1)-phase of the on-going cell cycle. J. Cell. Biochem. 90 244–252. [DOI] [PubMed] [Google Scholar]
- Boucheron, E., Healy, J.M.S., Bajon, C., Sauvanet, A., Rembur, J., Noin, M., Sekine, M., Riou-Khamlichi, C., Murray, J.A.H., Van Onckelen, H., and Chriqui, D. (2005). Ectopic expression of Arabidopsis CycD2 and CycD3 in tobacco has distinct effects on the structural organization of the shoot apical meristem. J. Exp. Bot. 56 123–134. [DOI] [PubMed] [Google Scholar]
- Cockcroft, C.E., den Boer, B.G., Healy, J.M., and Murray, J.A.H. (2000). Cyclin D control of growth rate in plants. Nature 405 575–579. [DOI] [PubMed] [Google Scholar]
- Contento, A.L., Kim, S.-J., Diane, C., and Bassham, D.C. (2004). Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 135 2330–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte, W., and Murray, J.A.H. (2003). The plant cell cycle. Annu. Rev. Plant Biol. 54 235–264. [DOI] [PubMed] [Google Scholar]
- Dewitte, W., Riou-Khamlichi, C., Scofield, S., Healy, J.M., Jacqmard, A., Kilby, N.J., and Murray, J.A.H. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl, J.A., Cheng, M.G., Roussel, M.F., and Sherr, C.J. (1998). Glycogen synthase kinase 3 beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12 3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl, J.A., and Sherr, C.J. (1997). A dominant-negative cyclin D1 mutant prevents nuclear import of cyclin-dependent kinase 4 (Cdk4) and its phosphorylation by Cdk-activating kinase. Mol. Cell. Biol. 17 7362–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl, J.A., Zindy, F., and Sherr, C.J. (1997). Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 11 957–972. [DOI] [PubMed] [Google Scholar]
- Ekholm, S.V., and Reed, S.I. (2000). Regulation of G(1) cyclin dependent kinases in the mammalian cell cycle. Curr. Opin. Cell Biol. 12 676–684. [DOI] [PubMed] [Google Scholar]
- Germain, D., Russell, A., Thompson, A., and Hendley, J. (2000). Ubiquitination of free cyclin D1 is independent of phosphorylation on threonine 286. J. Biol. Chem. 275 12074–12079. [DOI] [PubMed] [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20 1203–1207. [DOI] [PubMed] [Google Scholar]
- Granier, C., and Tardieu, F. (1998). Spatial and temporal analyses of expansion and cell cycle in sunflower leaves. A common pattern of development for all zones of a leaf and different leaves of a plant. Plant Physiol. 116 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy, J.M.S., Menges, M., Doonan, J.H., and Murray, J.A.H. (2001). The Arabidopsis D-type cyclins CycD2 and CycD3 both interact in vivo with the PSTAIRE cyclin-dependent kinase Cdc2a but are differentially controlled. J. Biol. Chem. 276 7041–7047. [DOI] [PubMed] [Google Scholar]
- Hu, Y., Bao, F., and Li, J. (2000). Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J. 24 693–701. [DOI] [PubMed] [Google Scholar]
- Huntley, R., et al. (1998). The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol. Biol. 37 155–169. [DOI] [PubMed] [Google Scholar]
- Imoto, M., Doki, Y., Jiang, W., Han, E.K.H., and Weinstein, I.B. (1997). Effects of cyclin D1 overexpression on G1 progression-related events. Exp. Cell Res. 236 173–180. [DOI] [PubMed] [Google Scholar]
- Ito, M., Araki, S., Matsunaga, S., Itoh, T., Nishihama, R., Machida, Y., Doonan, J.H., and Watanabe, A. (2001). G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13 1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W., Kahn, S.M., Zhou, P., Zhang, Y.J., Cacace, A.M., Infante, A.S., Doi, S., Santella, R.M., and Weinstein, I.B. (1993). Overexpression of cyclin D1 in rat fibroblasts causes abnormalities in growth control, cell cycle progression and gene expression. Oncogene 8 3447–3457. [PubMed] [Google Scholar]
- Kono, A., Umeda-Hara, C., Lee, J., Ito, M., Uchimiya, H., and Umeda, M. (2003). Arabidopsis D-type cyclin CycD4;1 is a novel cyclin partner of B2-type cyclin-dependent kinase. Plant Physiol. 132 1315–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva, O.A., Tomlinson, M., Parinyapong, P., Sakvarelidze, L., Leader, D., Shaw, P., and Doonan, J.H. (2004). CycD1, a putative G1 cyclin from Antirrhinum majus, accelerates the cell cycle in cultured tobacco BY-2 cells by enhancing both G1/S entry and progression through S and G2 phases. Plant Cell 16 2364–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng, X., Connell-Crowley, L., Goodrich, D., and Harper, J.W. (1997). S-phase entry upon ectopic expression of G1 cyclin-dependent kinases in the absence of retinoblastoma protein phosphorylation. Curr. Biol. 7 709–712. [DOI] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Masubelele, N.H., Dewitte, W., Menges, M., Maughan, S., Collins, C., Huntley, R., Nieuwland, J., Scofield, S., and Murray, J.A.H. (2005). D-type cyclins activate division in the root apex to promote seed germination in Arabidopsis. Proc. Natl. Acad. Sci. USA 102 15694–15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, M.J., and Leaver, C.J. (1993). Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 103 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, M., and Murray, J.A.H. (2001). Cell cycle controls and the development of plant form. Curr. Opin. Plant Biol. 4 44–49. [DOI] [PubMed] [Google Scholar]
- Menges, M., De Jager, S.M., Gruissem, W., and Murray, J.A.H. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41 546–566. [DOI] [PubMed] [Google Scholar]
- Menges, M., Hennig, L., Gruissem, W., and Murray, J.A.H. (2002). Cell cycle-regulated gene expression in Arabidopsis. J. Biol. Chem. 277 41987–42002. [DOI] [PubMed] [Google Scholar]
- Menges, M., Hennig, L., Gruissem, W., and Murray, J.A.H. (2003). Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 53 423–442. [DOI] [PubMed] [Google Scholar]
- Menges, M., and Murray, J.A.H. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30 203–212. [DOI] [PubMed] [Google Scholar]
- Menges, M., and Murray, J.A.H. (2004). Cryopreservation of transformed and wild-type Arabidopsis and tobacco cell suspension cultures. Plant J. 37 635–644. [DOI] [PubMed] [Google Scholar]
- Moriyasu, Y., and Ohsumi, Y. (1996). Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol. 111 1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami, H., Kawamura, K., Sugisaka, K., Sekine, M., and Shinmyo, A. (2002). Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakenfull, E.A., Riou-Khamlichi, C., and Murray, J.A.H. (2002). Plant D-type cyclins and the control of G1 progression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo, M., and Roberts, J.M. (1993). Cyclin-dependent regulation of G(1) in mammalian fibroblasts. Science 259 1908–1912. [DOI] [PubMed] [Google Scholar]
- Planchais, S., Samland, A.K., and Murray, J.A.H. (2004). Differential stability of Arabidopsis D-type cyclins: CycD3;1 is a highly unstable protein degraded by a proteasome-dependent mechanism. Plant J. 38 616–625. [DOI] [PubMed] [Google Scholar]
- Quelle, D.E., Ashmun, R.A., Shurtleff, S.A., Kato, J.Y., Barsagi, D., Roussel, M.F., and Sherr, C.J. (1993). Overexpression of mouse D-type cyclins accelerates G(1) phase in rodent fibroblasts. Genes Dev. 7 1559–1571. [DOI] [PubMed] [Google Scholar]
- Rechsteiner, M., and Rogers, S.W. (1996). PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21 267–271. [PubMed] [Google Scholar]
- Resnitzky, D., Gossen, M., Bujard, H., and Reed, S.I. (1994). Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol. 14 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi, C., Huntley, R., Jacqmard, A., and Murray, J.A.H. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283 1541–1544. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi, C., Menges, M., Healy, J.M., and Murray, J.A.H. (2000). Sugar control of the plant cell cycle: Differential regulation of Arabidopsis D-type cyclin gene expression. Mol. Cell. Biol. 20 4513–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, B.L., Zhang, J., Markwardt, J., Tokiwa, G., Volpe, T., Honey, S., and Futcher, B. (2004). Growth rate and cell size modulate the synthesis of, and requirement for, G(1)-phase cyclins at start. Mol. Cell. Biol. 24 10802–10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger, A., Schobinger, U., Bouyer, D., Weinl, C., Stierhof, Y.D., and Hulskamp, M. (2002). Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc. Natl. Acad. Sci. USA 99 6410–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr, C.J. (1995). D-type cyclins. Trends Biochem. Sci. 20 187–190. [DOI] [PubMed] [Google Scholar]
- Sherr, C.J. (2000). The Pezcoller Lecture. Cancer cell cycles revisited. Cancer Res. 60 3689–3695. [PubMed] [Google Scholar]
- Soni, R., Carmichael, J.P., Shah, Z.H., and Murray, J.A.H. (1995). A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell 7 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarayre, S., Vinardell, J.M., Cebolla, A., Kondorosi, A., and Kondorosi, E. (2004). Two classes of the CDH1-type activators of the anaphase-promoting complex in plants: Novel functional domains and distinct regulation. Plant Cell 16 422–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele, K., Raes, J., De Veylder, L., Rouze, P., Rombauts, S., and Inze, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd, T.C., Dekker, B.M.M., and Hoekema, A. (1989). A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra, R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8 135–142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.