Abstract
The Bacillus subtilis murB gene, encoding UDP-N-acetylenolpyruvoylglucosamine reductase, a key enzyme in the peptidoglycan (PG) biosynthetic pathway, is embedded in the dcw (for “division and cell wall”) cluster immediately upstream of divIB. Previous attempts to inactivate murB were unsuccessful, suggesting its essentiality. Here we show that the cell morphology, growth rate, and resistance to cell wall-active antibiotics of murB conditional mutants is a function of the expression level of murB. In one mutant, in which murB was insertionally inactivated in a merodiploid bearing a second xylose-inducible PxylA-murB allele, DivIB levels were reduced and a normal growth rate was achieved only if MurB levels were threefold that of the wild-type strain. However, expression of an extra copy of divIB restored normal growth at wild-type levels of MurB. In contrast, DivIB levels were normal in a second mutant containing an in-frame deletion of murB (ΔmurB) in the presence of the PxylA-murB gene. Furthermore, this strain grew normally with wild-type levels of MurB. During sporulation, the levels of MurB were highest at the time of synthesis of the spore cortex PG. Interestingly, the ΔmurB PxylA-murB mutant did not sporulate efficiently even at high concentrations of inducer. Since high levels of inducer did not interfere with sporulation of a murB+PxylA-murB strain, it appears that ectopic expression of murB fails to support efficient sporulation. These data suggest that coordinate expression of divIB and murB is important for growth and sporulation. The genetic context of the murB gene within the dcw cluster is unique to the Bacillus group and, taken together with our data, suggests that in these species it contributes to the optimal expression of cell division and PG biosynthetic functions during both vegetative growth and spore development.
Peptidoglycan (PG) is a critical component of the eubacterial cell envelope, providing mechanical resistance to withstand osmotic pressure (55) and functioning as a major determinant of cell shape, which ultimately also relies on topological information imposed by the actin cytoskeleton (6, 10, 33, 54). PG is also intimately involved in the cell division process and, in spore-forming bacteria, is additionally required for a cytoskeleton-like role during engulfment of the prespore by the mother cell, as well as for the formation of the spore cortex, a modified layer of PG, essential for spore heat resistance (reviewed in reference 17).
In all eubacteria examined to date, many of the genes involved in PG biosynthesis and cell division are grouped in the highly conserved dcw (for “division and cell wall”) cluster (Fig. 1A). A correlation between bacterial cell shape and the arrangement of genes within the dcw cluster has been suggested (51). Specifically, it appears that rod-shaped bacteria have retained a more conserved and compact dcw cluster, and that transitions to other bacterial shapes have involved rearrangements and loss of gene order conservation within the cluster (51). Other features of the region appear to be associated with the biology of certain groups. Examples are the duplication of the pbpB gene to originate the spoVD gene in spore formers of the genus Bacillus (9, 11, 12) and the presence of a sporulation-specific gene, spoVE, instead of ftsW, which is located in a different chromosomal location. Also, the spoVE-murG-murB-divIB unit (Fig. 1A) appears restricted to this group and, among the Bacillus species whose genomes have been sequenced, is only absent from B. clausii (34, 35). Both spoVE and spoVD are transcribed during sporulation in the mother cell, from σE-dependent promoters located just upstream of their coding regions, and both are dispensable for growth but essential for cortex biogenesis during spore formation (29, 42, 53).
FIG. 1.
Genetic organization of the dcw cluster in B. subtilis. (A) Location, extent, and orientation of the different genes present in the dcw cluster of B. subtilis. Sequences used in the construction of the figure were obtained from the SubtiList server (http://genolist.pasteur.fr/SubtiList/). The figure includes the positions of the transcriptional terminators (stem and loop structures) and main promoters (curved arrows) found in the region, with the regulators involved in their utilization indicated (12, 20, 21, 25, 29, 45). The horizontal bracket indicates the spoVE-murG-murB-divIB unit that appears to be unique to the Bacillus group of spore formers. (B) Region of the dcw cluster cloned to create pGR34 (used to create a murB::neo strain at the indicated position), pGR194 (used to create a ΔmurB in-frame deletion, the boundaries of which are shown in the figure), pGR36, and pGR43 (used to integrate a xylose-inducible murB allele at amyE). The PxylA-murB allele was combined with the murB::neo and ΔmurB alleles to create strains AH1746 and AH3497, respectively, as shown (see Materials and Methods for details).
In B. subtilis the dcw cluster is located in the 133° to 135° region of the chromosome (29). During vegetative growth, several genes in the cluster are cotranscribed in the form of long polycistronic messages, which appear to originate from promoters located just upstream of the murE gene, or even further upstream in the cluster (12, 25, 29). However, individual vegetative promoters have also been identified for specific genes or groups of genes, such as the divIB gene or the pbpB operon (12, 25). The divIB gene codes for a protein required for septum formation during both medial (vegetative) and asymmetric (sporulation) cell division (reference 45 and references therein). divIB is transcribed mainly from a promoter located upstream of murE but also from a weak σA-type promoter located just 93 bp upstream of the divIB coding sequence (25, 45). Cells in which murB is separated from divIB by an integrational plasmid exhibit a defect in nucleoid structure and segregation and fail to activate Spo0A-dependent gene expression at the onset of sporulation (45). Moreover, they show a slight growth defect that is exacerbated upon expression of a second copy of the upstream murB gene (45). Together with the observation that the spoVE-murG-murB-divIB unit is conserved within bacteria of the genus Bacillus (Fig. 1), this observation suggests that coordinated expression of these genes may be important for normal growth. Moreover, because murG and murB appear to be cotranscribed from the σE-dependent promoter of spoVE, this linkage may also be important during sporulation (15).
Here, we have examined whether the chromosomal position of murB is important for its function. The murB gene codes for UDP-N-acetylenolpyruvoylglucosamine reductase, which is required at an early step in the PG biosynthetic pathway. The first step in PG biosynthesis involves the MurA enzyme, which catalyzes the production of UDP-N-acetylenolpyruvylglucosamine (UDPGlcNAcEP) from UDP-N-acetylglucosamine (UDP-GlcNAc) and phosphoenolpiruvate. The enolpyruvyl moiety is then reduced by MurB, yielding UDP-N-acetylmuramic acid (UDPMurNAc). A pentapeptide chain is then added onto UDPMurNAc by the sequential action of MurC, MurD, MurE, and MurF (16, 55). The resultant UDPMurNAc-pentapeptide is transferred to the membrane acceptor (undecaprenylphosphate) by the MraY transferase to form lipid I, and then N-acetylglucosamine (GlcNAc) is added by another transferase, MurG, to form lipid II. The GlcNAc-MurNAc(pentapeptide)-pyrophosphoryl-undecaprenol is next translocated across the membrane, where it is incorporated into nascent PG by the transglycosylase and transpeptidation reactions catalyzed by several penicillin binding proteins (PBPs) (18).
The murB gene is essential in Escherichia coli and Staphylococcus aureus, as demonstrated by the isolation of temperature-sensitive mutants (38, 41, 44). Unsuccessful inactivation attempts also suggested its essentiality in B. subtilis (46). In this work, we have constructed and characterized a murB conditional mutant and have examined the effects of the ectopic expression of murB. We show that the murB gene is essential for normal growth, cell morphology, and resistance to cell wall-active antibiotics. We also show that normal expression of murB is required at the stage of spore cortex synthesis for the formation of heat-resistant spores. The evidence suggests that expression of murB from its normal locus within the dcw cluster is required for normal growth and efficient spore formation.
MATERIALS AND METHODS
Bacterial strains, media, and general methods.
All of the B. subtilis strains used in this study are congenic derivatives of strain MB24 (trpC2 metC3) (Table 1). The E. coli strain DH5α (BRL) was used for plasmid construction and propagation (Table 1). Growth, selection, and maintenance of drug-resistant transformants of E. coli or B. subtilis was as described previously (27, 28). The high-fidelity Pfu polymerase (Stratagene, La Jolla, Calif.) was used to generate PCR fragments for cloning, which were sequenced to ensure that no mutations were introduced. Sporulation was induced by exhaustion in Difco sporulation medium (DSM) (8, 50), and its frequency was expressed as the percentage of heat-resistant CFU relative to the total cell count (27). Spores were purified on step gradients of Gastrografin (Schering) (26, 27, 47).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype/phenotype | Origin/reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F−lacZΔM15 Δ(lacZYA-argF)U169; wild type | Gibco BRL |
| BL21(DE3)pLysS | F−ompT hsdS (r− m−) gal dcm (DE3) pLysS; Cmr | Stratagene |
| AH1646 | F−ompT hsdS (r− m−) gal dcm (DE3) pLysS pGR28; Ampr Cmr | This work |
| AH3268 | F−ompT hsdS (r− m−) gal dcm (DE3) pLysS pGR144; Ampr Cmr | This work |
| B. subtilis strains | ||
| MB24 | trpC2 metC3; wild type | Laboratory stock |
| SL666 | trpC2 spoVE85 | 26 |
| AH1744 | trpC2 metC3 amyE::PxylA-murB(77-753); Neor | This work |
| AH1745 | trpC2 metC3 amyE::PxylA-murB; Cmr | This work |
| AH1746 | trpC2 metC3 murB::neo amyE::PxylA-murB; Neor Cmr | This work |
| AH3356 | trpC2 metC3 murB::neo amyE::PxylA-murB thrC::Pspac-divIB; Neor Cmr Emr | This work |
| AH3497 | trpC2 metC3 ΔmurB amyE::PxylA-murB; Cmr | This work |
| AH3505 | trpC2 metC3 ΔmurB amyE::PxylA-murB spβ::PgerE-lacZ; Cmr Emr | This work |
| Plasmids | ||
| pBEST501 | bla neo | 32 |
| pMS38 | bla cm | 59 |
| pDG364 | bla amyE::cm | 8 |
| pMLK83 | bla amyE::gus neo | 36 |
| pAH235 | bla amyE::PspoIIIG-gusA neo | 28 |
| pAH256 | bla spec | 28 |
| pMALc2 | bla malE | New England Biolabs |
| pCR2.1-TOPO | bla | Invitrogen, La Jolla, CA |
| pGEM T Easy | bla | Promega |
| pGR28 | bla malE-murB | This work |
| pGR29 | bla PxylA | This work |
| pGR34 | bla murB::neo | This work |
| pGR35 | bla amyE::PxylA-gusA neo | This work |
| pGR36 | bla amyE::PxylA-murB(77-753) neo | This work |
| pGR40 | bla amyE::PxylA neo | This work |
| pGR43 | bla murB(301-1597) cm | This work |
| pGR46 | bla murB(77-753) | This work |
| pGR63 | bla amyE::Pspac-divIB neo | 45 |
| pGR144 | bla malE-divIB | This work |
| pGR163 | bla thrC::Pspac-divIB erm, spec | This work |
| pGR194 | bla ΔmurB | This work |
Purification of MurB and DivIB fusion proteins for antibody production.
The murB gene was PCR amplified from MB24 using primers murB-235D (5′-ATGGAGAAAGTGATACAGG-3′) and murB-1165R (5′-GCTTCAGTCTAGAACTTGAATCAGCG-3′). The 930-bp product was cut with XbaI and inserted between the XmnI and XbaI sites of pMAL-c2 (New England Biolabs) to yield pGR28. The divIB gene (lacking the sequence for its transmembrane domain) was PCR amplified from MB24 using primers divIB-239D (5′-GCGGAGATCTGAGTTTTGGAGTTTGGAC-3′) and divIB-908R (5′-CCAAGCTTCCGTCTCAGCCGCGC-3′). The 669-bp product was digested with BglII and EcoRI and inserted between the BamHI and EcoRI sites of pMAL-c2 to create pGR144. pGR28 or pGR144 was introduced into E. coli BL21(DE3) (pLysS) (Novagen). The transformed cells were grown in 1 liter of LB to an optical density at 600 nm (OD600) of 0.6 and induced with 5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h. The cells were collected and lysed as described previously (48). The lysate was clarified by centrifugation (30,000 × g for 30 min at 4°C) and applied to a 20-ml amylose column (New England Biolabs) equilibrated in lysis buffer. Column washing and elution were as described by the manufacturer. Purified MurB and DivIB fusion proteins were used to raise rabbit polyclonal antibodies (Eurogentec, Herstal, Belgium).
Immunoblot analysis.
Samples (10 ml) of LB or DSM cultures were collected and lysed as described before (48). Samples (20 μg) of total protein were resolved on 12% or 15% polyacrylamide gels containing sodium dodecyl sulfate and subjected to immunoblot analysis as described previously (48). Antibodies were used at the following dilutions: anti-MurB, dilution of 1:1,000; anti-DivIB, 1:100; anti-β′ subunit of RNA polymerase (a gift from Bill Haldenwang), 1:250. A rabbit secondary antibody conjugated to horseradish peroxidase (Sigma) and a mouse secondary antibody were used at dilutions of 1:10,000 and 1:200, respectively. The immunoblots were developed with enhanced chemiluminescence reagents (Amersham Pharmacia Biotech).
Construction of a murB conditional mutant.
For construction of a murB conditional mutant, first a PCR fragment carrying the xylA promoter was produced using primers xylR-61D (5′-CTATTTTAGAATTCCTTTTTCATATGAG-3′) and xylR-1558R (5′-GGAATGAGATCTGAGCCATGTGATTTCCC-3′) and digested with AflIII and BglII, and the 400-bp fragment was inserted into BssHII- and BglII-cut pAH256 (28), yielding pGR29. Then, the PxylA fragment was released from pGR29 with SpeI and XhoI and inserted between the SpeI and SalI sites of pAH235 (28), originating pGR35. Cleavage of pGR35 with EcoRI followed by religation produced pGR40. Next, a PCR fragment containing the first 246 codons of murB, obtained with primers murB-77D (5′-CTGATTGAAGCACTAGTCCGGATTGTAC-3′) and murB-753R (5′-GTAGCTGGGATCCATCTGCTCATTTGTC-3′), was inserted into pCR2.1-TOPO (Invitrogen, La Jolla, CA) to give pGR46, released from this plasmid with SpeI and EcoRI, and introduced between the same sites of pGR40, to yield pGR36 (Fig. 1B). AH1744 (AmyE−) resulted from the transformation of MB24 to neomycin resistance (Nmr) with PstI-linearized pGR36. A plasmid (pGR43, Fig. 1B) containing the 3′ end of murB was made by inserting an NruI- and SphI-cut PCR product obtained with primers murB-213D (5′-GCAAGATCTAATGCGAGGTTACGATGGAG-3′) and murB-1597R (5′-GGTGTTTGCATGCACACAATGATCAGCACC-3′) between the same sites of pDG364 (8). PstI-cut pGR43 was used to transform AH1744 to chloramphenicol resistance (Cmr). This generated AH1745 (Neos AmyE−), in which the presence of murB under PxylA control at amyE was verified by PCR and sequence analysis. A Neor determinant released from pBEST501 (32) with SmaI was introduced into SmaI-cut pGR28 to give pGR34 (Fig. 1B). ScaI-cut pGR34 was used to transform AH1745 to Nmr in the presence of 0.1% xylose. This produced AH1746, in which inactivation of murB within the dcw region was verified by PCR.
Construction of an inducible divIB allele.
The Pspac-divIB-lacI region in pGR90 (45) was PCR amplified with primers pDH45D (5′-GAAGTGGTCAAGAATTCACTAGGCAC-3′) and pDH1685R (5′-GAAGATCTTCACATTAATTGCGTTGCGCTCACTG-3′), and the product was cut with EcoRI and BglII and inserted between the EcoRI and BamHI sites of pDG1664 (24), to yield pGR163. Plasmid pGR163 was used to transform MB24 selecting for Emr, to produce AH3303 (Table 1) in which Pspac-divIB-lacI resides at thrC, as verified by PCR. Chromosomal DNA from AH3303 was used to transform AH1746 to Emr. A transformant was selected and named AH3356 (Table 1).
In-frame deletion of murB.
First, two PCR products of 600 bp encompassing the 5′ and 3′ regions of murB were produced using primers murB-77D (see above) and murBfaseR(BglII) (5′-GCTCATATCAGATCTGTGAGCGCCGGC-3′) and primers murBfaseD(BglII) (5′-CGTGAAAGATCTGAGATTGATATGCACACAGAG-3′) and divIB430R(EcoRI) (5′-CTGAGCCGAATTCAAGCACTTCATAGTAC-3′). The two products were digested with BglII, ligated together, and inserted into pGEM-Teasy (Promega). This created pGR194 (Fig. 1B), which was used with pDG1731 (24) to cotransform AH1745 to spectinomycin resistance (Spr). One erythromycin-susceptible transformant (AH3497) was dependent on xylose for growth and had a murB 447-bp in-frame deletion, as verified by PCR and sequence analysis.
Microscopy.
Culture samples were collected and stained with the membrane dye FM4-64 (Molecular Probes), as described previously (45). Phase-contrast or fluorescence images were acquired in a Leica DMRA2 microscope coupled to a CoolSNAP HQ Photometrics camera (Roper Scientific).
Antimicrobial susceptibility.
Susceptibility to antibiotics was determined by measuring bacterial growth on square gradient plates (12 cm by 12 cm) as described previously (2, 7). Briefly, each plate contained LB agar supplemented with 0.001%, 0.005%, 0.01%, or 0.1% xylose (50 ml in each of the top and bottom layers). Antibiotics used for the top layer were as follows: vancomycin, 0.25 μg ml−1; oxacillin, 0.2 μg ml−1; fosfomycin, 20 μg ml−1; d-cycloserine, 10 μg ml−1; cephalosporin, 10 μg ml−1. Strains were grown in LB (supplemented with 0.0025% xylose when required) for ∼16 h, diluted 100-fold into LB supplemented with 0.01% xylose, and incubated at 37°C to an OD600 of 0.2. Cell plating was as described previously (1, 22, 23). Confluent growth along the gradient was scored after 18 h at 37°C.
RESULTS
murB is essential in B. subtilis.
Previous unsuccessful attempts to insertionally inactivate the murB gene in B. subtilis were interpreted as indicating its essentiality (46). Here, we constructed a murB conditional mutant by first inserting a copy of murB under the control of the xylose-inducible PxylA promoter at the nonessential amyE locus and then by using the murB merodiploid (AH1745) to disrupt the murB gene at its normal locus, by insertion of a Nmr cassette within the gene's coding sequence. Attempts to obtain Nmr transformants of AH1745 in the absence of xylose failed, but those readily arose when the medium was supplemented with 0.1% xylose. One, in which the murB gene was shown to be inactivated, was designated AH1746 (Table 1). To test the effect of varying the expression level of murB on cell growth, overnight cultures of AH1746 grown in the presence of 0.05% xylose were centrifuged, washed with LB medium, resuspended in xylose-free medium, and diluted to an OD600 of 0.02 in prewarmed LB medium without xylose or supplemented with increasing concentrations of inducer. The growth rate of the cultures was determined by monitoring the rate of increase in the OD600. In parallel, we monitored the levels of MurB in cells of the conditional mutant using an anti-MurB antibody (see Materials and Methods).
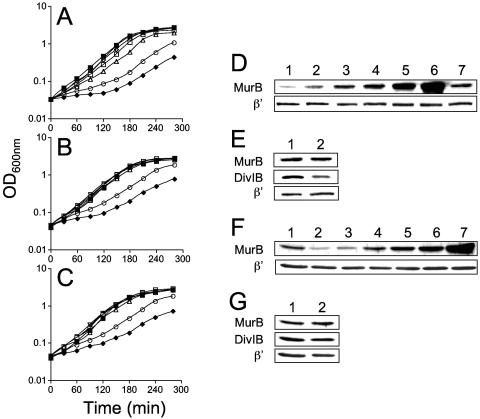
As shown in Fig. 2A, the growth rate of the murB conditional mutant (AH1746) was proportional to the concentration of xylose in the medium. The wild-type strain exhibited a doubling time of about 30 min in LB medium at 37°C in the absence or in the presence of 1% xylose, whereas the doubling time of the murB conditional mutant in the absence of inducer was of 93.1 min (Table 2). Residual growth of the mutant in the absence of xylose is presumably caused by leakiness of the PxylA promoter, allowing MurB to accumulate to about 30% of the level in the wild-type strain (Fig. 2D; compare lanes 1 and 7). Increasing the concentration of xylose in the medium led to an increment in the growth rate, in parallel with an increase in the cellular level of MurB (Fig. 2D). Note that as a control for these experiments, the same membranes were probed with an antibody against the β′ subunit of RNA polymerase, whose levels remained constant regardless of the xylose concentration and similar to those in the wild-type strain MB24 (Fig. 2D). As shown in Fig. 2D, at a xylose concentration of 0.0025%, MurB accumulated to wild-type levels (compare lanes 3 and 7), but surprisingly the growth rate of this culture was lower than that of wild-type cells, with a doubling time of 37.3 min compared to 30.1 min for the wild type (Table 2). A xylose concentration of 0.01% restored a wild-type growth rate (doubling time of about 30 min) to the conditional mutant (Fig. 2A and Table 2). However, under these conditions MurB accumulated to levels about three times higher than in the wild-type strain (Fig. 2D; compare lanes 5 and 7).
FIG. 2.
Inducer-dependent growth and levels of MurB in PxylA-murB conditional mutants. (A) Growth of wild-type strain MB24 (filled squares) or AH1746 (ΔmurB::neo amyE::PxylA-murB) in the absence of xylose (filled diamonds) or in the presence of increasing xylose concentrations, as follows: 0.001%, open circles; 0.0025%, open triangles; 0.005%, open squares; 0.01%, open diamonds; 0.1%, open inverted triangles. (B) Growth of MB24 (filled squares) or AH3356 (murB::neo amyE::PxylA-murB thrC::Pspac-divIB) in the presence of 0.5 μM IPTG and in the absence of xylose (filled diamonds) or in the presence of xylose concentrations, as follows: 0.001%, open circles; 0.0025%, open triangles; 0.005%, open squares; 0.01%, open diamonds; 0.1%, open inverted triangles. (C) Growth of strain MB24 (filled squares) or AH3497 (ΔmurB amyE::PxylA-murB) in the absence of xylose (filled diamonds) or in the presence of increasing xylose concentrations, as follows: 0.001%, open circles; 0.0025%, open triangles; 0.005%, open squares; 0.01%, open diamonds; 0.1%, open inverted triangles. Xylose (at all concentrations tested) or IPTG did not influence the growth rate of the wild-type strain, whose growth is represented only in the absence of either inducer. (D and F) Immunoblot analysis using anti-MurB and anti-β′ subunit of the RNA polymerase antisera (Material and Methods) of mid-log-phase samples of cultures of MB24 (lane 7 in panel D and lane 1 in panel F) and AH1746 (panel D, lanes 1 to 6) or AH3497 (panel F, lanes 2 to 7), as represented in panels A and C. Lanes in panels D are as follows: lane 1, strain AH1746 grown in the absence of xylose; lane 2, 0.001% xylose; lane 3, 0.0025% xylose; lane 4, 0.005% xylose; lane 5, 0.01% xylose; lane 6, 0.1% xylose; lane 7, MB24 (no xylose). Lanes in panel F are as follows: lane 1, strain MB24 grown in the absence xylose; lanes 2 to 7, strain AH3497 grown in the absence or presence of xylose (lane 2, no xylose; lane 3, 0.001% xylose; lane 4, 0.0025% xylose; lane 5, 0.005% xylose; lane 6, 0.01% xylose; lane 7, 0.1% xylose). (E and G) Samples of the above cultures of MB24, AH1746, or AH3497 were subject to immunoblot analysis using anti-MurB, anti-DivIB, and anti-β′ RNA polymerase subunit antisera (see Materials and Methods). (E) Lane 1, MB24 (wild type); lane 2, AH1746 grown in the presence of 0.0025% xylose. (G) Lane 1, MB24 (wild type); lane 2, AH3497 grown in the presence of 0.0025% xylose.
TABLE 2.
Growth rates of the murB conditional mutant in LB medium
| Strain | Relevant genotype | % Xylosea | Doubling time (min)b |
|---|---|---|---|
| MB24 | Wild type | 0 | 30.1 ± 2.2 |
| 1 | 30.9 ± 1.7 | ||
| AH1746 | ΔmurB::neo amyE::PxylA-murB | 0 | 93.1 ± 5.4 |
| 0.001 | 69.2 ± 3.1 | ||
| 0.0025 | 37.3 ± 3.2 | ||
| 0.005 | 35.2 ± 2.7 | ||
| 0.01 | 30.0 ± 3.3 | ||
| 0.1 | 30.9 ± 1.2 | ||
| 1 | 29.0 ± 2.4 | ||
| AH3356 | ΔmurB::neo amyE::PxylA-murB thrC::Pspac-divIB(+) | 0 | 89.0 ± 4.2 |
| 0.001 | 63.0 ± 4.7 | ||
| 0.0025 | 30.6 ± 3.4 | ||
| 0.005 | 30.1 ± 3.1 | ||
| 0.01 | 30.8 ± 4.1 | ||
| 0.1 | 30.9 ± 2.2 | ||
| 1 | 28.6 ± 1.7 | ||
| AH3497 | ΔmurB amyE::PxylA-murB | 0 | 88.1 ± 6.2 |
| 0.001 | 53.1 ± 6.8 | ||
| 0.0025 | 30.1 ± 2.3 | ||
| 0.005 | 29.1 ± 1.3 | ||
| 0.01 | 30.1 ± 1.6 | ||
| 0.1 | 30.5 ± 1.2 | ||
| 1 | 30.5 ± 1.7 |
The numbers in bold are the minimum xylose concentration at which a wild-type doubling time is observed.
Data are averages from three independent experiments. IPTG, for induction of divIB, was added at a concentration of 0.5 μM.
A link between murB and divIB.
The separation of the murB and divIB genes by an integrational plasmid produced a strain with a slow growth phenotype, which could be corrected by a copy of divIB in trans (45). In that mutant, divIB was transcribed only from a weak σA-type promoter located just upstream of the gene (45). To test whether the Neor cassette inserted into murB affected the levels of DivIB, we conducted immunoblot analysis with an anti-DivIB antibody (see Materials and Methods). We examined the levels of DivIB in strain AH1746 during the exponential phase of growth in the presence of 0.0025% xylose, which permits MurB to accumulate to wild-type levels (see above). The results in Fig. 2E indicate that the levels of DivIB in AH1746 were slightly reduced (by about 10%) relative to MB24 (wild type), while the levels of MurB or of the β′ subunit of RNA polymerase remained unchanged in the two strains (Fig. 2E). The reduction in the levels of DivIB was not sufficient to cause any discernible effect on cell division, as the cell length distribution was similar to that of the wild-type strain (data not shown; see also reference 45).
To test whether the requirement for higher than wild-type levels of MurB for normal growth of AH1746 was due to reduced levels of DivIB, we placed a second copy of divIB under the control of the IPTG-inducible Pspac promoter atthenonessential thrC locus. The resulting strain, AH3356 (ΔmurB::neo amyE::PxylA-murB thrC::Pspac-divIB), was, like its parent, xylose dependent for growth (not shown). However, in the presence of 0.5 μM IPTG and 0.001% xylose, the doubling time of AH3356 was 63 min, compared to 93 min for AH1746 (Fig. 2B and Table 2). Moreover, in the presence of 0.5 μM IPTG, a wild-type doubling time was observed for AH3356 at a xylose concentration of only 0.0025%; that is, four times lower than for the parental strain AH1746 (Table 2; see also above). Thus, the need for increased levels of MurB to sustain a normal growth rate in AH1746 results from the reduction in DivIB levels; conversely, increased expression of divIB reduces the requirement for MurB.
A nonpolar murB in-frame deletion mutant.
Because of the effect of the murB::neo allele on the levels of DivIB, we constructed an in-frame deletion of the murB gene (see Materials and Methods) and transferred the new allele into strain AH1745. The new conditional mutant, AH3497, exhibited xylose dependency for growth (Table 2). AH3497 grew slowly, with a doubling time of 88.1 min in the absence of inducer (Table 2 and Fig. 2C). However, in contrast to AH1746 (murB::neo), which grew normally at a xylose concentration of 0.01%, AH3497 showed the same doubling time as the wild type in the presence of only 0.0025% xylose (Table 2 and Fig. 2C), a concentration at which AH3356 (murB::neo Pspac-divIB) also exhibited normal growth. As for AH1746, the level of MurB, but not that of the β′ subunit of RNA polymerase, increased with the concentration of xylose in the growth medium (Fig. 2F). For AH3497 growing in the presence of 0.0025% xylose, MurB accumulated to wild-type levels (Fig. 2F, lanes 1 and 4, and Fig. 2G), and the doubling time was 30.6 min, similar to that of the wild type (30.1 min; Fig. 2C and Table 2). Moreover, under these conditions, DivIB accumulated to wild-type levels, as shown by the immunoblot analysis documented in Fig. 2G, where probing of the same membranes with an anti-MurB and with an anti-RNA polymerase β′ subunit antibody confirmed equivalent loadings. The results show that the in-frame deletion of murB has no impact on the levels of DivIB, and accordingly the conditional mutant exhibited a normal growth rate at wild-type levels of MurB.
Morphology of the murB conditional mutant.
To examine whether growth of AH3497 in the absence of xylose or in the presence of low levels of inducer caused alterations in cell shape, cells in mid-log-phase cultures of the conditional mutant were examined by fluorescence and phase-contrast microscopy. We found that in the absence of inducer, approximately 30% of the cells were slightly elongated and showed bulges, often at one end of the cell, close to a division septum (Fig. 3A and a, arrowheads). Occasionally, what looked like empty vesicles bulged out from the cells, and many cells were lysed (data not shown). Induction of the xylose promoter with 0.001% xylose reduced the frequency of bulge formation to 17%, and when these appeared (Fig. 3B and b, arrowheads), they were not as pronounced as in the absence of inducer. Cell swelling has been observed for other B. subtilis mutants with lesions affecting the expression of genes essential for PG biosynthesis. For example, low levels of expression of the rodA morphogene, essential for cell elongation, causes swelling and lysis (30), as does inactivation of the genes coding for PBP2a and PBPH, which are partially redundant functional homologues of the E. coli elongation-specific PBP2 (11, 57, 58).
FIG. 3.
MurB depletion causes a defect in cell morphology. The figure depicts the morphology of cells in LB cultures of the wild-type (wt) strain MB24 and AH3497 (ΔmurB amyE::PxylA-murB). (A/a through D/d) Cells from strain AH3497 grown in the absence of xylose (A/a) or in the presence of 0.001% xylose (B/b), 0.01% xylose (C/c), or 0.1% xylose (D/d). (E/e) MB24 cells. The cells were harvested during the exponential phase of growth and stained with the membrane dye FM4-64 to allow visualization of membranes and septa for observation by phase-contrast microscopy (A to E) and by fluorescence microscopy (a to e). Arrowheads point to bulges close to division septa. Scale bars, 2 μm.
Xylose concentrations of 0.0025% (not shown) or higher restored normal length and cell shape to the murB conditional mutant AH3497 (Fig. 3C through d). Addition of 0.01% xylose to cells growing in the absence of inducer restored normal cell morphology, suggesting that the cell envelope deficiency could be overcome if expression of murB was restored (Fig. 3C through d). The average cell length of the population in the presence of 0.01% (3.12 μm ± 0.75) or 0.1% (3.26 μm ± 0.65) xylose, concentrations at which no alteration of cell shape was noticed, did not significantly differ from that of a wild-type population (3.55 μm ± 0.88). We infer from these results that expression of murB is required for normal cell shape. Also, in agreement with earlier work (10, 30, 33, 56, 57), the results also suggest a correlation between a normal growth rate and the acquisition of rod-shaped morphology by B. subtilis cells.
Cells depleted for MurB show increased susceptibility to antibiotics.
Strains engineered for low-level expression of single essential genes, and thus rendered hypersensitive to inhibitors of the corresponding gene product, have been used in screens for new antimicrobial compounds (13). No MurB inhibitors with antimicrobial activity were available to us (49), but we tested whether MurB depletion could cause hypersensitivity to several other cell wall-active antibiotics. The tests were performed on gradient plates (2, 7) containing the antibiotics fosfomycin, d-cycloserine, oxacillin, vancomycin, and cephalosporin, supplemented with increasing concentrations of xylose (from 0.001 to 0.1%; see Materials and Methods). Under these conditions, the extent of the zone of growth is a function of the resistance level of the strain (Fig. 4A), and the MIC is defined as the lowest concentration of antibiotic that prevents growth (37). The results in Fig. 4B show that while xylose did not influence the resistance of the wild-type strain MB24, the murB conditional mutant AH3497 was more susceptible to the antibiotics fosfomycin, oxacillin, d-cycloserine, and cephalosporin at all the concentrations of xylose tested. However, while the resistance to oxacillin and cephalosporin clearly augmented in parallel with the increase in xylose concentration in the medium, the resistance of AH3497 to fosfomycin and d-cycloserine was much less pronounced. Also, even at a xylose concentration of 0.1%, which permits the accumulation of MurB to levels almost 15 times higher than in the wild type (Fig. 2E; also see above), wild-type levels of resistance were never achieved (Fig. 4A and B). In contrast, the murB conditional mutant was found to be less susceptible to vancomycin, chloramphenicol, or tetracycline at any xylose concentration tested (data not shown). Fosfomycin is an inhibitor of MurA, the UDP-GlcNAc enolpyruvyl transferase that catalyzes the first committed step in the synthesis of PG, and acts just upstream of MurB in the pathway (16, 49). MurA and MurB act sequentially to convert UDP-GlcNac into UDP-MurNAc. d-Cycloserine is a competitive inhibitor of alanine racemase (Alr) and of d-alanine:d-alanine ligase (Ddl) (16, 49). Alr and Ddl act sequentially to produce d-ala-d-ala, which is then added to UDP-MurNAc tripeptide by the MurF lyase (16, 49). Oxacillin and cephalosporin are β-lactam antibiotics that interfere with PG synthesis by inhibiting the final transpeptidation needed for the cross-linking of the PG molecules (49) Thus, a reduction in the level of expression of the murB gene does not cause a generalized hypersensitivity of the cells to antibiotics. Rather, the enzymatic steps catalyzed by MurA, and by Alr or Ddl in the cytoplasm, and the transpeptidation step that takes place at the cell wall become more sensitive to inhibition by specific antibiotics, when the level of expression of murB is altered.
FIG. 4.
The murB conditional mutant shows increased susceptibility to cell wall-active antibiotics. (A) Susceptibility of MB24 (wild type) and AH3497 (ΔmurB amyE::PxylA-murB) to the antibiotic cephalosporin when grown in increasing concentrations of xylose. (B) Quantification of the susceptibility of strains MB24 (black columns) and AH3497 (white columns) to the indicated antibiotics as a function of the xylose concentration, as determined on antibiotic gradient plates (as depicted in panel A). Susceptibility is shown as the MIC of antibiotic (as defined in Materials and Methods) relative to the level of xylose present in the gradient plates. The graphs shown represent averages of three independent experiments; error bars show standard deviations.
MurB levels throughout sporulation.
Formation of heat-resistant spores involves the synthesis of a layer of modified PG known as the spore cortex (3, 18, 43). Synthesis of the spore cortex is likely to involve most of the vegetative components of the PG-synthesizing machinery, although it also requires the expression of several sporulation-specific genes (3, 18). Here, we used an anti-MurB antibody to examine the profile of accumulation of MurB during sporulation. For that we grew the wild-type strain MB24 in DSM medium, in which sporulation is induced by nutrient exhaustion, and took samples during growth, at the onset of the stationary phase of growth (defined as the initiation of sporulation), and at various intervals thereafter for immunoblot analysis. The results in Fig. 5B show that the levels of MurB appeared to decrease as cells approach the end of the exponential phase of growth in DSM and that the decline in MurB levels persisted until about 1 h after the onset of sporulation (Fig. 5B). The levels of MurB then appear to steadily increase until at least hour 6 of sporulation (Fig. 5B). Synthesis of the spore cortex follows the activation of the late mother cell regulator σK, which takes place around hour 4 of sporulation (17, 18, 43). Thus, the levels of MurB appear to coincide with the time of synthesis of the spore cortex. The increase in the level of MurB during sporulation is consistent with the results of a recent study suggesting that the transcription of both murG and murB is enhanced during sporulation, presumably from the sporulation-specific spoVE promoter (14, 15) (Fig. 1). Presumably, increased transcription of murB is required to increase the levels of MurB at the time of cortex formation.
FIG. 5.
Effect of MurB depletion during growth and sporulation in DSM medium. (A) Overnight cultures of the wild-type strain MB24 (filled squares) and of AH3497 (ΔmurB amyE::PxylA-murB) in LB medium, later grown in the presence of 0.0025% xylose, were centrifuged to collect the cells. These were washed with DSM medium, resuspended in xylose-free DSM, and used to inoculate prewarmed DSM to an OD600 of 0.02 in the absence of xylose (filled diamonds) or in the presence of xylose at the following concentrations: 0.0005%, open circles; 0.001%, open inverted triangles; 0.0025%, filled circles; 0.005%, open triangles; 0.01%, open diamonds; 0.1%, open squares. Growth of the cultures was followed by monitoring of the rate of increase in OD600 through time (in minutes) following inoculation. Note that some cultures reach stationary phase, defined as the initiation of sporulation (T0), at different times. (B through F) Cells of the wild type (wt) (MB24), AH1745 (amyE::PxylA-murB), or AH3497 (ΔmurB amyE::PxylA-murB) were grown as described above and collected at different times during growth and sporulation to monitor the levels of MurB using an anti-MurB antibody. Panel B shows the levels of MurB in the wild-type strain MB24. Panels C to F compare the levels of MurB in MB24, AH1745, and AH3497 grown in the absence of xylose (−) or in the presence of 0.1% xylose (+). The numbers above each lane represent time (in hours) before or after the onset of sporulation (T0).
We then examined the impact of varying the expression level of murB from the PxylA promoter on the efficiency of sporulation in DSM. Under the conditions used, the Spo+ strain MB24 produced 6.7 × 108 to 7.0 × 108 spores per ml of culture, independently of the level of xylose present in the sporulation medium (Table 3). In the absence of xylose, AH3497 (ΔmurB PxylA-murB) was still able to grow, albeit slower than the wild type, entered stationary phase at an OD600 slightly lower than a culture of the wild type (Fig. 5A), and formed only about 104 spores per ml of culture (Table 3). At xylose concentrations of 0.0005% or 0.001%, AH3497 grew slower than the wild type in DSM, but at xylose concentrations of 0.0025% or higher (up to 0.1%), AH3497 showed a wild-type growth rate in DSM (Fig. 5A). However, at all xylose concentrations tested, AH3497 was impaired in the formation of heat-resistant spores, with titers in the range of 104 per ml of culture (Table 3). The reduced sporulation efficiency was not caused by addition of xylose, because a murB+ strain (AH1745) harboring an extra copy of murB under the control of PxylA at amyE sporulated at wild-type levels regardless of the xylose concentration in the medium (Table 3).
TABLE 3.
Sporulation of a murB conditional mutant in DSM medium
| Strain | Relevant genotype | % Xylose | Viable cell count | Heat-resistant cell count | % Sporulation |
|---|---|---|---|---|---|
| MB24 | Wild type | 0 | 6.7 × 108 | 7.0 × 108 | 100 |
| 0.001 | 6.8 × 108 | 6.7 × 108 | 97.8 | ||
| 0.01 | 6.9 × 108 | 6.8 × 108 | 98.5 | ||
| 1 | 7.1 × 108 | 6.9 × 108 | 97.1 | ||
| AH1745 | amyE::PxylA-murB | 0 | 5.6 × 108 | 5.4 × 108 | 96.4 |
| 0.001 | 5.5 × 108 | 5.3 × 108 | 96.3 | ||
| 0.01 | 6.7 × 108 | 6.4 × 108 | 95.5 | ||
| 1 | 6.9 × 108 | 6.4 × 108 | 92.7 | ||
| AH3497 | ΔmurB amyE::PxylA-murB | 0 | 7.7 × 107 | 1.6 × 104 | 0.02 |
| 0.001 | 5.5 × 107 | 1.7 × 104 | 0.03 | ||
| 0.01 | 6.3 × 108 | 1.4 × 104 | 0.002 | ||
| 1 | 6.4 × 108 | 1.3 × 104 | 0.002 |
To test whether the failure of AH3497 to sporulate efficiently could be due to insufficient levels of MurB, samples of DSM cultures in the absence or in the presence of 0.1% xylose (the highest concentration tested) were collected throughout sporulation, and the levels of MurB were analyzed by immunoblot analysis. The results in Fig. 5E show that no MurB could be detected throughout sporulation of AH3497 in DSM in the absence of inducer. In the presence of 0.1% xylose, however, the levels of MurB at the onset of sporulation (T0) were much higher than for the wild type and remained higher than for the wild type throughout sporulation (Fig. 5F). Therefore, the failure of AH3497 to sporulate efficiently in the presence of 0.1% xylose is not due to reduced levels of MurB during sporulation. Conversely, the sporulation phenotype of AH3497 also does not appear to be due to increased levels of MurB. We base this inference on the observation that in AH1745 (murB+ PxylA-murB) grown in the presence of 0.1% xylose (a condition under which the strain sporulates efficiently; Table 3), the levels of MurB during sporulation were also elevated relative to the wild type (Fig. 5D), at least to the same extent of the increase in AH3497 (Fig. 5F). We propose that the sporulation phenotype of AH3497 is due to the ectopic expression of murB in this strain.
Sporulating cells of the murB conditional mutant are blocked at the stage of cortex formation.
In an attempt to determine the stage in the sporulation pathway at which AH3497 was blocked, spores from cultures of the murB conditional mutant growing in the absence or in the presence of increasing concentrations of xylose were purified 24 h after the onset of sporulation and observed under phase-contrast microscopy. Approximately 97% of the spores produced by the wild-type strain (Fig. 6A) or by AH1745 (murB+ PxylA-murB) (not shown) were bright phase, independently of the amount of xylose present in the medium. In contrast, almost 99% of spores of the murB conditional mutant grown in the absence and presence of increasing concentrations of inducer were dark phase (Fig. 6B through F). Dark-phase immature spores were seen inside the mother cell or as free “sporelets” in the culture medium. The production of dark-phase, immature spores is a characteristic of mutants with lesions in genes involved in the synthesis of the spore cortex PG (29). These results suggest that the ectopic expression of murB fails to promote efficient synthesis of the spore cortex and hence to support formation of heat-resistant spores.
FIG. 6.
MurB depletion causes the production of dark-phase spores. The figure depicts spores produced by the wild-type strain MB24 (A) and the murB conditional mutant AH3497 (ΔmurB amyE::PxylA-murB) (B through F) in DSM medium in the absence of xylose (A and B) or in the presence of the following xylose concentrations: 0.001% (C), 0.005% (D), 0.01% (E), 0.1% (F). The spores were collected and purified as described in Material and Methods and visualized under phase-contrast microscopy. Open and filled arrowheads point to free spores or to spores still inside the mother cell, respectively. Bars, 2 μm.
DISCUSSION
In an extension of earlier work (46), the results herein presented indicate that expression of the murB gene is essential for normal growth and cell shape in B. subtilis. We base this conclusion on the analysis of two different murB conditional mutants, in which the only functional copy of the gene was placed under the control of the xylose-inducible PxylA promoter at the nonessential amyE locus. In one mutant (AH1746), the murB gene present in the dcw cluster was insertionally inactivated. In the second (AH3497), an in-frame deletion was introduced into murB in the dcw cluster. The two strains exhibited slow growth in the absence of inducer and greatly altered cell morphology, and their growth rate was proportional to the level of murB expression.
However, the two strains showed different requirements for MurB for normal growth in liquid medium. The murB::neo insertional mutant AH1746 required a level of MurB at least three times higher than a congenic wild-type strain for normal growth. The mutant also showed a slight reduction in the level of DivIB, encoded by the downstream divIB gene, although not sufficient to perturb normal cell division. However, a second copy of the divIB gene permitted normal growth at wild-type levels of MurB, suggesting that DivIB somehow contributes to the normal growth of the murB conditional mutant. In agreement with this suggestion, the conditional mutant AH3497, in which the murB gene at the dcw cluster was deleted in frame, showed normal levels of DivIB and a normal growth rate at wild-type levels of MurB. Separation of murB from divIB by an integrational plasmid resulted in a strain with reduced levels of DivIB but essentially normal cell division and with a slow growth phenotype (45). The reason for this slow growth phenotype was not clear, but it was not due to a polar effect on genes located downstream of divIB, and while a copy of divIB in trans corrected the slow growth phenotype of the mutant, an extra copy of the murB gene aggravated the phenotype (45). This suggested that at least in this strain, a reduction in the levels of DivIB may not be accompanied by a reduction in the levels of MurB. In any case, these observations suggested that murB and divIB have to be coordinately expressed. Cell division mutants have been reported to show a normal growth rate before cell lysis occurs. However, the function of divIB may not be restricted to cell division (45). The suggestion that coordinated expression of murB and divIB is important for normal growth is in agreement with the presence of the two cistrons in long polycistronic messages that transverse most of the dcw cluster of B. subtilis during growth (25) and with the observation that the spoVE-murG-murB-divIB unit within the dcw cluster appears to be unique to spore formers of the genus Bacillus. It could be that in Bacillus species, the coordination between synthesis of cell wall precursors and cell division is exerted at least in part at the level of transcription of the murG-murB-divIB unit (see also below).
Synthesis of the spore cortex PG is essential for spore heat resistance (3, 17, 18). The spore cortex is formed between the two membranes that surround the prespore at an intermediate stage of development, from precursors that may be produced mainly in the mother cell (3, 17, 18). The activity of the sporulation-specific σE factor is confined to the mother cell and is required for transcription of several genes involved in synthesis of the spore cortex PG (e.g., see references 9 and 53). However, synthesis of the spore cortex commences only following the complete engulfment of the prespore by the mother cell, with the concomitant activation of the late mother cell regulator σK (3, 17, 18, 43). Activation of σK takes place around hour 4 of sporulation, and synthesis of the spore cortex ensues, in parallel with the appearance of bright-phase spores (39). It seems that expression of murB during sporulation is required for synthesis of the spore cortex. Increased transcription of the murG and murB genes is seen during sporulation (15), presumably from the σE-dependent promoter of the upstream spoVE gene (53), and the levels of MurB increase coincidentally with the period of cortex formation (this work).
Surprisingly, in cells with murB absent from the dcw cluster, expression of the PxylA-murB inducible allele from an ectopic position drastically reduced the frequency of heat-resistant spores and resulted in the accumulation of over 99% dark-phase spores, a sign of incomplete cortex formation (29, 43). Remarkably, ectopic expression of PxylA-murB in cells that kept murB at its normal position within the dcw cluster did not interfere in any detectable way with the frequency of sporulation of the formation of refractile spores. Hence, displacement of the murB gene from its normal location in the dcw cluster appears detrimental for cortex formation during sporulation. This is in keeping with the idea that the spoVE-murG-murB-divIB unit within the dcw region, which is confined to Bacillus spore formers (Fig. 1), is of biological importance for this group of organisms. Both spoVE and murG appear to be involved in the membrane-linked steps of PG synthesis (4, 31). It is tempting to speculate that the clustering of murG, murB, and divIB would contribute to the coordination between synthesis of PG precursors and cell division during the vegetative life cycle and that linkage of at least the murG-murB unit to spoVE would coordinate PG precursor synthesis to their translocation across the spore membrane for the formation of the spore cortex.
Assembly of the PG molecule involves the sequential addition of l-alanine, d-glutamic acid, diaminopimelic acid, and d-alanyl-d-alanine to the UDP-MurNAc. The observation that the murB conditional mutant (AH3497) is more sensitive to d-cycloserine, an inhibitor of the d-alanine racemase (Alr) and of d-alanyl-d-alanine ligase (Ddl), suggests that a reduction in the efficiency of the MurB-catalyzed step of the PG biosynthetic pathway will lead to a rapid depletion of downstream precursors, and as a consequence it will make these cells more sensitive to any antibiotic acting downstream of that step. At suboptimal concentrations of inducer, the increased susceptibility of the mutant to d-cycloserine and to the β-lactamic antibiotics oxacillin and cephalosporin suggests that its cell wall may accumulate tripeptide muropeptides. If so, a further decrease in cross-linking caused by β-lactam antibiotics could compromise the structural stability of the cell wall. That resistance to the β-lactams cephalosporin and oxacillin increases with increased expression of murB is reminiscent of the situation with a Pspac-murE conditional mutant of S. aureus (19). At suboptimal levels of inducer, the mutant shows increased susceptibility to oxacillin (19).
In B. subtilis, cell wall-active antibiotics induce the extracytoplasmic sigma factors σM and σW possibly via a chemical or physical signal related to a defect in the structure or composition of the cell wall (5, 52). Significantly, vancomycin, which acts as a potent inducer of sigW, and of a large number of σW-dependent genes, results in induction of the contiguous murG, murB, divIB, and ylxW genes (5). We note that the murB conditional mutant is not more susceptible to vancomycin. However, a sigW mutant is not significantly affected in its sensitivity to vancomycin (5). It is possible that in B. subtilis, increased expression of murB in the conditional mutant somehow induces expression of the genes for specific PBPs. However, the pool of soluble PG precursors, or the composition of the cell wall in the murB conditional mutant, has not yet been examined, and it is unknown whether σW is induced in the PxylA-murB mutant.
The murB conditional mutant is also more sensitive to fosfomycin, an inhibitor of MurA (16, 49). MurA catalizes the step immediately upstream of MurB, the production of UDPGlcNAcEP. In vitro experiments have shown that the activity of MurA is inhibited by its reaction product and indicated that this reaction is an important control step in the PG biosynthetic pathway (40). Increased susceptibility to fosfomycin under conditions where MurB levels are low could be caused by accumulation of UDPGlcNAcEP and inhibition of MurA, or simply to the combined effect of inhibition of two consecutive steps in the pathway. In any event, because different steps in the PG biosynthetic pathway appear to be more sensitive to known antibiotics in a way that depends on the expression of murB, we suggest that the murB conditional mutant could be used as a tool in screening for new antibacterial compounds that act at the level of cell wall biogenesis (13).
Acknowledgments
We thank Mónica Serrano, Teresa M. Barbosa, and Sérgio R. Filipe for helpful discussions. We thank Hermínia de Lencastre for the gift of some of the antibiotics used in this study, Bill Haldenwang for the gift of the antibody against the β′ subunit of RNA polymerase, and John Helmann and Anne Moir for the gift of strains. We also thank an anonymous reviewer for helpful suggestions.
This work was supported by grant POCTI/BCI/48647/2002 from Fundação para a Ciência e a Tecnologia (FCT) to A.O.H. G.R. was the recipient of a Ph.D. fellowship (PRAXIS XXI/BD/21560/99) from the FCT.
REFERENCES
- 1.Amabile-Cuevas, C. F., and B. Demple. 1991. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 19:4479-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ariza, R. R., S. P. Cohen, N. Bachhawat, S. B. Levy, and B. Demple. 1994. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 176:143-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchanan, C. E., A. O. Henriques, and P. J. Piggot. 1993. Wall metabolism during sporulation, p. 167-186. In J.-M. Ghuysen and R. Hakenback (ed.), Bacterial cell wall, vol. 8. Elsevier Science Publishers, New York, N.Y. [Google Scholar]
- 4.Bupp, K., and J. van Heijenoort. 1993. The final step of peptidoglycan subunit assembly in Escherichia coli occurs in the cytoplasm. J. Bacteriol. 175:1841-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, M., T. Wang, R. Ye, and J. D. Helmann. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons. Mol. Microbiol. 45:1267-1276. [DOI] [PubMed] [Google Scholar]
- 6.Carballido-Lopez, R., and J. Errington. 2003. A dynamic bacterial cytoskeleton. Trends Cell Biol. 13:577-583. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham, R. P., S. M. Saporito, S. G. Spitzer, and B. Weiss. 1986. Endonuclease IV (nfo) mutant of Escherichia coli. J. Bacteriol. 168:1120-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biology methods for Bacillus. John Wiley and Sons, Ltd., New York, N.Y.
- 9.Daniel, R. A., S. Drake, C. E. Buchanan, R. Scholle, and J. Errington. 1994. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J. Mol. Biol. 235:209-220. [DOI] [PubMed] [Google Scholar]
- 10.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767-776. [DOI] [PubMed] [Google Scholar]
- 11.Daniel, R. A., E. J. Harry, and J. Errington. 2000. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol. Microbiol. 35:299-311. [DOI] [PubMed] [Google Scholar]
- 12.Daniel, R. A., A. M. Williams, and J. Errington. 1996. A complex four-gene operon containing essential cell division gene pbpB in Bacillus subtilis. J. Bacteriol. 178:2343-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeVito, J. A., J. A. Mills, V. G. Liu, A. Agarwal, C. F. Sizemore, Z. Yao, D. M. Stoughton, M. G. Cappiello, M. D. Barbosa, L. A. Foster, and D. L. Pompliano. 2002. An array of target-specific screening strains for antibacterial discovery. Nat. Biotechnol. 20:478-483. [DOI] [PubMed] [Google Scholar]
- 14.Eichenberger, P., M. Fujita, S. T. Jensen, E. M. Conlon, D. Z. Rudner, S. T. Wang, C. Ferguson, K. Haga, T. Sato, J. S. Liu, and R. Losick. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 16.El Zoeiby, A., F. Sanschagrin, and R. C. Levesque. 2003. Structure and function of the Mur enzymes: development of novel inhibitors. Mol. Microbiol. 47:1-12. [DOI] [PubMed] [Google Scholar]
- 17.Errington, J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117-126. [DOI] [PubMed] [Google Scholar]
- 18.Foster, S. J., and D. L. Popham. 2002. Structure and synthesis of cell wall, spore cortex, teichoic acids, S-layers, and capsules, p. 21-42. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 19.Gardete, S., A. M. Ludovice, R. G. Sobral, S. R. Filipe, H. de Lencastre, and A. Tomasz. 2004. Role of murE in the expression of beta-lactam antibiotic resistance in Staphylococcus aureus. J. Bacteriol. 186:1705-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gholamhoseinian, A., Z. Shen, J. J. Wu, and P. Piggot. 1992. Regulation of transcription of the cell division gene ftsA during sporulation of Bacillus subtilis. J. Bacteriol. 174:4647-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzy-Treboul, G., C. Karmazyn-Campelli, and P. Stragier. 1992. Developmental regulation of transcription of the Bacillus subtilis ftsAZ operon. J. Mol. Biol. 224:967-979. [DOI] [PubMed] [Google Scholar]
- 22.Greenberg, J. T., J. H. Chou, P. A. Monach, and B. Demple. 1991. Activation of oxidative stress genes by mutations at the soxQ/cfxB/marA locus of Escherichia coli. J. Bacteriol. 173:4433-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenberg, J. T., P. Monach, J. H. Chou, P. D. Josephy, and B. Demple. 1990. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:6181-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57-61. [DOI] [PubMed] [Google Scholar]
- 25.Harry, E. J., S. L. Rowland, M. S. Malo, and R. G. Wake. 1994. Expression of divIB of Bacillus subtilis during vegetative growth. J. Bacteriol. 176:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henriques, A. O., B. W. Beall, and C. P. Moran, Jr. 1997. CotM of Bacillus subtilis, a member of the alpha-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J. Bacteriol. 179:1887-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henriques, A. O., B. W. Beall, K. Roland, and C. P. Moran, Jr. 1995. Characterization of cotJ, a σE-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilis spores. J. Bacteriol. 177:3394-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henriques, A. O., E. M. Bryan, B. W. Beall, and C. P. Moran, Jr. 1997. cse15, cse60, and csk22 are new members of mother-cell-specific sporulation regulons in Bacillus subtilis. J. Bacteriol. 179:389-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henriques, A. O., H. de Lencastre, and P. J. Piggot. 1992. A Bacillus subtilis morphogene cluster that includes spoVE is homologous to the mra region of Escherichia coli. Biochimie 74:735-748. [DOI] [PubMed] [Google Scholar]
- 30.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda, M., T. Sato, M. Wachi, H. K. Jung, F. Ishino, Y. Kobayashi, and M. Matsuhashi. 1989. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J. Bacteriol. 171:6375-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itaya, M., K. Kondo, and T. Tanaka. 1989. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 17:4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 34.Kanehisa, M. 1997. Linking databases and organisms: GenomeNet resources in Japan. Trends Biochem. Sci. 22:442-444. [DOI] [PubMed] [Google Scholar]
- 35.Kanehisa, M., S. Goto, S. Kawashima, and A. Nakaya. 2002. The KEGG databases at GenomeNet. Nucleic Acids Res. 30:42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karow, M. L., and P. J. Piggot. 1995. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene 163:69-74. [DOI] [PubMed] [Google Scholar]
- 37.Marty, N., L. Agueda, V. Carratala, and G. Chabanon. 1994. Evaluation of the Spiral method to determine the MIC of various antibiotics. Pathol. Biol. (Paris) 42:448-453. [PubMed] [Google Scholar]
- 38.Matsuo, M., K. Kurokawa, S. Nishida, Y. Li, H. Takimura, C. Kaito, N. Fukuhara, H. Maki, K. Miura, K. Murakami, and K. Sekimizu. 2003. Isolation and mutation site determination of the temperature-sensitive murB mutants of Staphylococcus aureus. FEMS Microbiol. Lett. 222:107-113. [DOI] [PubMed] [Google Scholar]
- 39.Meador-Parton, J., and D. L. Popham. 2000. Structural analysis of Bacillus subtilis spore peptidoglycan during sporulation. J. Bacteriol. 182:4491-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mengin-Lecreulx, D., B. Flouret, and J. van Heijenoort. 1983. Pool levels of UDP N-acetylglucosamine and UDP N-acetylglucosamine-enolpyruvate in Escherichia coli and correlation with peptidoglycan synthesis. J. Bacteriol. 154:1284-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyakawa, T., H. Matsuzawa, M. Matsuhashi, and Y. Sugino. 1972. Cell wall peptidoglycan mutants of Escherichia coli K-12: existence of two clusters of genes, mra and mrb, for cell wall peptidoglycan biosynthesis. J. Bacteriol. 112:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyao, A., G. Theeragool, M. Takeuchi, and Y. Kobayashi. 1993. Bacillus subtilis spoVE gene is transcribed by σE-associated RNA polymerase. J. Bacteriol. 175:4081-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piggot, P. J., and J. G. Coote. 1976. Genetic aspects of bacterial endospore formation. Bacteriol. Rev. 40:908-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pucci, M. J., L. F. Discotto, and T. J. Dougherty. 1992. Cloning and identification of the Escherichia coli murB DNA sequence, which encodes UDP-N-acetylenolpyruvoylglucosamine reductase. J. Bacteriol. 174:1690-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Real, G., S. Autret, E. J. Harry, J. Errington, and A. O. Henriques. 2005. Cell division protein DivIB influences the Spo0J/Soj system of chromosome segregation in Bacillus subtilis. Mol. Microbiol. 55:349-367. [DOI] [PubMed] [Google Scholar]
- 46.Rowland, S. L., J. Errington, and R. G. Wake. 1995. The Bacillus subtilis cell-division 135-137 degree region contains an essential orf with significant similarity to murB and a dispensable sbp gene. Gene 164:113-116. [DOI] [PubMed] [Google Scholar]
- 47.Serrano, M., R. Zilhao, E. Ricca, A. J. Ozin, C. P. Moran, Jr., and A. O. Henriques. 1999. A Bacillus subtilis secreted protein with a role in endospore coat assembly and function. J. Bacteriol. 181:3632-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seyler, R. W., Jr., A. O. Henriques, A. J. Ozin, and C. P. Moran, Jr. 1997. Assembly and interactions of cotJ-encoded proteins, constituents of the inner layers of the Bacillus subtilis spore coat. Mol. Microbiol. 25:955-966. [DOI] [PubMed] [Google Scholar]
- 49.Silver, L. L. 2003. Novel inhibitors of bacterial cell wall synthesis. Curr. Opin. Microbiol. 6:431-438. [DOI] [PubMed] [Google Scholar]
- 50.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamames, J., M. Gonzalez-Moreno, J. Mingorance, A. Valencia, and M. Vicente. 2001. Bringing gene order into bacterial shape. Trends Genet. 17:124-126. [DOI] [PubMed] [Google Scholar]
- 52.Thackray, P. D., and A. Moir. 2003. SigM, an extracytoplasmic function sigma factor of Bacillus subtilis, is activated in response to cell wall antibiotics, ethanol, heat, acid, and superoxide stress. J. Bacteriol. 185:3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Theeragool, G., A. Miyao, K. Yamada, T. Sato, and Y. Kobayashi. 1993. In vivo expression of the Bacillus subtilis spoVE gene. J. Bacteriol. 175:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van den Ent, F., L. A. Amos, and J. Lowe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39-44. [DOI] [PubMed] [Google Scholar]
- 55.van Heijenoort, J. 1996. Murein synthesis, p. 1025-1034. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaecter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 56.Varley, A. W., and G. C. Stewart. 1992. The divIVB region of the Bacillus subtilis chromosome encodes homologs of Escherichia coli septum placement (minCD) and cell shape (mreBCD) determinants. J. Bacteriol. 174:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei, Y., T. Havasy, D. C. McPherson, and D. L. Popham. 2003. Rod shape determination by the Bacillus subtilis class B penicillin-binding proteins encoded by pbpA and pbpH. J. Bacteriol. 185:4717-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yanouri, A., R. A. Daniel, J. Errington, and C. E. Buchanan. 1993. Cloning and sequencing of the cell division gene pbpB, which encodes penicillin-binding protein 2B in Bacillus subtilis. J. Bacteriol. 175:7604-7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zilhao, R., G. Naclerio, A. O. Henriques, L. Baccigalupi, C. P. Moran, Jr., and E. Ricca. 1999. Assembly requirements and role of CotH during spore coat formation in Bacillus subtilis. J. Bacteriol. 181:2631-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]