Abstract
NtrC-like activators regulate the transcription of a wide variety of adaptive genes in bacteria. Previously, we demonstrated that a mutation in the ntrC-like activator gene nla18 causes defects in fruiting body development in Myxococcus xanthus. In this report, we describe the effect that nla18 inactivation has on gene expression patterns during development and vegetative growth. Gene expression in nla18 mutant cells is altered in the early stages of fruiting body development. Furthermore, nla18 mutant cells are defective for two of the earliest events in development, production of the intracellular starvation signal ppGpp and production of A-signal. Taken together, these results indicate that the developmental program in nla18 mutant cells goes awry very early. Inactivation of nla18 also causes a dramatic decrease in the vegetative growth rate of M. xanthus cells. DNA microarray analysis revealed that the vegetative expression patterns of more than 700 genes are altered in nla18 mutant cells. Genes coding for putative membrane and membrane-associated proteins are among the largest classes of genes whose expression is altered by nla18 inactivation. This result is supported by our findings that the profiles of membrane proteins isolated from vegetative nla18 mutant and wild-type cells are noticeably different. In addition to genes that code for putative membrane proteins, nla18 inactivation affects the expression of many genes that are likely to be important for protein synthesis and gene regulation. Our data are consistent with a model in which Nla18 controls vegetative growth and development by activating the expression of genes involved in gene regulation, translation, and membrane structure.
In nature, biofilms formed by the soil bacterium Myxococcus xanthus feed on prey bacteria to obtain amino acids, which are used as a source of carbon, nitrogen, and energy (1, 7). Upon starvation for amino acids, M. xanthus initiates a complex developmental program that allows large groups of cells to migrate to aggregation centers and begin building multicellular fruiting bodies. Once a fruiting body is molded into its final shape, individual rod-shaped cells within this structure differentiate into dormant, spherically shaped spores that are resistant to many forms of environmental stress (8, 62).
When deprived of amino acids, M. xanthus cells accumulate (p)ppGpp (46, 47, 63), a molecule that serves as an intracellular starvation signal in bacteria (3, 4). After the intracellular pool of (p)ppGpp rises and M. xanthus cells initiate development, a series of cell-cell signals help coordinate large-scale changes in gene expression (6, 19, 38, 39, 43). Of these cell-cell developmental signals, the two that have been studied the most extensively are A-signal and C-signal. A-signal is a diffusible cell density signal that is required in the earliest stages of M. xanthus development, prior to the onset of aggregation (39, 40, 41, 57). In contrast, C-signal is a contact-stimulated signal that is required for the aggregation and sporulation phases of development to proceed normally (33, 34, 35, 38, 44).
Recent findings indicate that M. xanthus uses σ54-like promoters to drive the expression of many developmentally regulated genes (11, 12, 13, 16, 17, 31, 59, 69). Work by Keseler and Kaiser (32) demonstrated that rpoN, which encodes σ54, is essential for vegetative growth in M. xanthus. These results indicate that σ54-like promoters are important for gene expression during vegetative growth and development. Transcriptional activation of σ54-dependent promoters requires an NtrC-like activator, a DNA binding protein that helps σ54- RNA polymerase form a transcriptionally active, open promoter complex (49, 70). Fifteen NtrC-like activators that are required for fruiting body development to proceed normally have been uncovered in the past 10 years (2, 15, 18, 20, 26, 27, 36, 67, 69).
More recently, Caberoy et al. (2) demonstrated that an insertion in the ntrC-like activator gene nla18 (MXAN_3692; see http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?orgsearch=my&org=gmx) causes defects in aggregation and sporulation. In this paper, we show that the nla18 mutation reduces or abolishes the expression of genes that are activated throughout the course of fruiting body development. In addition, cells carrying the nla18 insertion are defective for ppGpp accumulation and A-signal production, indicating that Nla18 is required in the earliest stages of fruiting body development. We also found that in nutrient broth the doubling time of the nla18 mutant is about two- to threefold longer than that of wild-type cells, indicating that the nla18 mutant has a vegetative growth defect. By DNA microarray analysis, we show that the nla18 insertion alters the normal expression patterns of vegetative genes, including a large number of genes whose products are likely to be involved in gene regulation and protein synthesis, and genes that encode membrane and membrane-associated proteins. Taken together, our findings indicate that Nla18 is an important regulator of both vegetative and developmental gene expression.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are shown in Table 1. DNA sequence analysis of the nla18 locus revealed that the mx887 gene is located immediately downstream of nla18, suggesting that nla18 and mx887 may be part of the same operon. Two lines of evidence indicate that the nla18 insertion does not impact mx887 transcription. (i) A plasmid (pFOR18) carrying a wild-type copy of nla18 and the upstream nla18 promoter element rescues the defects of nla18 mutant strain AG343 when it is integrated into the Mx8 phage attachment site (attB) in the chromosome. (ii) Reverse transcription-PCR and DNA microarray analysis confirm that mx887 is expressed in the nla18 mutant.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristic(s) or sequence | Source, reference, or amplicon size |
|---|---|---|
| Strains | ||
| AG318 | DK1622 pNBC18::nla18 | 2 |
| AG339 | DK101 pNBC18::nla18 | This study |
| AG343 | DK1622 pNBC31::nla18 | 2 |
| AG390 | pNBC31::nla18 sdeK::Ω4408 Tn5lacZ | This study |
| AG391 | pNBC31::nla18 spi::Ω4521 Tn5lacZ | This study |
| AG392 | pNBC31::nla18 devR::Ω4414 Tn5lacZ | This study |
| AG393 | pNBC31::nla18 Ω4403 Tn5lacZ | This study |
| AG395 | pNBC31::nla18 Ω4435 Tn5lacZ | This study |
| AG407 | pNBC31::nla18 exo::Ω7536 Tn5lacZ | This study |
| AG1001 | AG343 Mx8attB::pFOR18 | This study |
| DK101 | pilQ1 (wild-type development) | 24 |
| DK476 | pilQ1 asgA476 | 19 |
| DK1622 | Wild-type development | 29 |
| DK4323 | pilQ1 asgA476 spi::Ω4521 Tn5lacZ | 39 |
| DK4300 | sdeK::Ω4408 Tn5lacZ | 37 |
| DK4368 | Ω4403 Tn5lacZ | 37 |
| DK4521 | spi::Ω4521 Tn5lacZ | 37 |
| DK5204 | Ω4435 Tn5lacZ | 37 |
| DK5208 | csgA::Tn5-132 ΩLS205 | 38 |
| DK5508 | devR::Ω4414 Tn5lacZ | 37 |
| DK7536 | exo::Ω7536 Tn5lacZ | 45 |
| DK11063 | fruA::Ω7540 Tn5lacZ | 64 |
| MS1000 | DK101ΔrelA1 | This study |
| Plasmids | ||
| pBGS18 | Kanr | 66 |
| pCR2.1-TOPO | Kanr | Invitrogen |
| pSWU19 | KanrattP | S. S. Wu and D. Kaiser |
| pSWU22 | Tetr | S. S. Wu and D. Kaiser |
| pBJ114 | Kanr, galK vector | 28 |
| pMS302 | 4.8 PstI fragment containing relA in pBGS18 | 23 |
| pMS302R | 4.8 PstI fragment containing relA (opposite orientation) in pBGS18 | This study |
| pMS330 | pMS302 with 1.3 kb of relA deleted | This study |
| pMS331 | pBJ114 carrying the 3.5-kb PstI fragment from pMS330 | This study |
| pNBC18 | 477-bp internal fragment of nla18 in pCR2.1-TOPO | 2 |
| pNBC31 | 477-bp internal fragment of nla18 in pSWU22 | 2 |
| pFOR18 | Kanr, pSWU19 containing 1.8 kb of nla18 on a BamHI-HindIII fragment | This study |
| Primers | ||
| relAfwd | 5′-TCATCGCCTTTCGCATCATCGC-3′ | 129 bp |
| relArev | 5′-ACATGTTGGGCTTCGGAATCGC-3′ |
Strain MS1000 carries a 1,311-bp in-frame deletion in the relA gene. To construct the relA deletion mutant, we used pMS302, a plasmid that contains the relA gene and flanking DNA on a PstI fragment. Plasmid pMS302 was digested with SacI, and the large DNA fragment was gel purified and then self-ligated to generate plasmid pMS330. Plasmid pMS330 carries a 1,311-bp internal deletion of relA due to the removal of adjacent 87-bp and 1,224-bp SacI fragments. The PstI fragment carrying the internal deletion of relA was isolated from plasmid pMS330 and ligated into the PstI site of galK vector pBJ114, yielding plasmid pMS331. Plasmid pMS331 was introduced into DK101 cells by electroporation as described by Plamann et al. (58). Kanr transformants with a single copy of pMS331 integrated into the relA locus were identified by Southern blot analysis (60) and used to isolate Galr Kans strains as previously described (11, 68). Galr Kans strains carrying the relA deletion were identified by Southern blot analysis (60). The presence of the relA deletion was confirmed by screening for (p)ppGpp accumulation in response to nutrient downshift. Strains AG390, AG391 AG392, AG393, AG395, and AG407 were constructed by introducing plasmid pNBC31 into strains DK4300, DK4521, DK5508, DK4368, DK5204, and DK7536, respectively.
Media used for growth and development.
M. xanthus strains were grown at 28οC or 32οC in CTT broth (1.0% Casitone [Difco], 10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4, 8 mM MgSO4), on CTTYE broth (CTT containing 0.5% yeast extract [Difco]), or on solid support plates containing CTTYE broth and 1.5% Difco Bacto Agar. CTTYE broth and plates were supplemented with 40 μg of kanamycin sulfate/ml or 10 μg of oxytetracycline/ml as needed. Fruiting body development was carried out at 32οC on plates containing TPM buffer (10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4, 8 mM MgSO4) and 1.5% Difco Bacto Agar. A-factor assays were performed with microtiter plates containing MC7 starvation buffer (10 mM MOPS, 1 mM CaCl2, final pH 7.0). CTT soft agar contains CTT broth and 0.7% Difco Bacto Agar.
Escherichia coli strains were grown at 37οC in Luria broth (LB) containing 1.0% tryptone (Difco), 0.5% yeast extract (Difco), and 0.5% NaCl or in plates containing LB and 1.5% Difco Bacto Agar. LB and LB plates were supplemented with 40 μg of kanamycin sulfate/ml or 10 μg of oxytetracycline/ml as needed.
M. xanthus development.
M. xanthus strains were inoculated into flasks containing CTTYE broth, and the cultures were incubated at 28οC or 32οC with vigorous swirling. After the cultures reached a density of 5 × 108 cells/ml, the cells were pelleted, the supernatants were removed, and the cells were resuspended in TPM buffer to a density of 5 × 109 cells/ml. Aliquots (20 μl) of the cell suspensions were spotted onto TPM agar plates and incubated at 32οC. M. xanthus cells were harvested at various times during development on TPM agar and used for RNA slot blot hybridization studies, quantitative PCR (QPCR), β-galactosidase assays, or Western blot analysis as described below.
β-Galactosidase assays.
Cells were harvested at different times during development on TPM and quick-frozen in liquid nitrogen as described previously (11). β-Galactosidase assays were performed on quick-frozen cell extracts by the technique of Kaplan et al. (30). β-Galactosidase specific activity is defined as nanomoles of o-nitrophenol produced per minute per milligram of protein.
A-factor assays.
DK101, DK476, and AG339 cells were inoculated into flasks containing CTTYE broth, and the cultures were incubated at 32°C with vigorous swirling. After the cultures reached a density of 5 × 108 cells/ml, the cells were pelleted, washed with MC7 buffer, and resuspended in MC7 buffer to a density of 2.5 × 1010 cells/ml. The cell suspensions were placed in flasks and shaken at 32°C. After 3 h, the cells were pelleted and the conditioned MC7 buffer was removed. A-factor assays were performed with M. xanthus test strain DK4323 and aliquots of conditioned MC7 buffer as described previously (23, 57).
Western blot assays.
Approximately 109 M. xanthus cells/ml were harvested from TPM agar plates, placed in sodium dodecyl sulfate (SDS) lysis buffer, and boiled for 10 min. Protein samples were separated by electrophoresis through a 12% polyacrylamide gel and transferred to an Immobilon P membrane (Millipore) with a semidry blotting apparatus. The blots were probed with anti-FruA antibody, followed by incubation with peroxidase-conjugated goat anti-rabbit immunoglobulin G (Boehringer Mannheim). The blots were developed with the Renaissance Chemiluminescence Reagent (NEN Life Science Products) and Amersham autoradiography Hyperfilm-MP.
Analysis of nucleotide pools.
Nucleotides were isolated and separated by thin-layer chromatography as described previously (46, 63). 32P-labeled ppGpp, GTP, and ATP were visualized with a STORM phosphorimaging scanner and quantified by Image Quant software (Molecular Dynamics).
QPCR and RNA slot blot hybridization analysis.
Total cellular RNA was isolated from quick-frozen cells by the hot phenol method (60) and used to generate cDNA as described by Lancero et al. (42). One-microliter aliquots of the cDNA synthesis reaction mixtures were used for the subsequent PCR amplifications. PCR mixtures contained gene-specific forward and reverse primers (250 nM, final concentration) and the DyNamo HS SYBR Green qPCR Master Mix (Finnzymes). The primers used for QPCR are listed in Table 1. QPCR was performed with the Opticon 2 system from MJ Research. The rate of accumulation of PCR-generated DNA was measured by continuous monitoring of SYBR Green I (Molecular Probes) fluorescence. To confirm that RNA samples were not contaminated with residual genomic DNA, control cDNA synthesis reaction mixtures that lacked reverse transcriptase were prepared and then analyzed by QPCR as described above for the test samples. Gene expression was quantified by the absolute method of quantification (user bulletin 2, Applied Biosystems), and the expression levels were normalized to levels in wild-type cells at time zero (vegetative growth). Standard curves for each QPCR primer pair were made at 101, 102, 103, 105, 108, 1010, and 1011 plasmid copies/μl. Standard curves for relA primers were made with pMS302R. Slot blot hybridizations were performed on total cellular RNA as described by Kaplan et al. (30). PCR-generated fragments of the sdeK, relA, and nla18 genes were used as probes for slot blot hybridization experiments. The specificity of these probes was confirmed by using yeast mRNA, which yielded no detectable signal.
DNA microarrays.
PCR generated DNA microarrays containing probes to the 7,235 M. xanthus open reading frames identified on the M1genome (26; R. D. Welch, personal communication) were spotted onto poly-l-lysine-coated glass slides by the Stanford Functional Genomics Facility (Stanford, CA). Production of M. xanthus DNA microarrays was done in conjunction with The Myxococcus Microarray Consortium, and construction was based on a pilot array previously described (26). Processing of the DNA arrays, cDNA synthesis, microarray hybridization, and posthybridization processing were performed as described by Jakobsen et al. (26), with the following modifications. Five independent biological replica pairs of wild-type and nla18 mutant strains were used for this analysis, and each independent wild-type-nla18 mutant pair was handled and processed identically. Briefly, each pair of wild-type and nla18 mutant strains was grown at 28°C to a density of 5 × 108 cells/ml, the cells were pelleted by centrifugation, the supernatants were removed, and the cell pellets were quick-frozen in liquid nitrogen. Total cellular RNA was isolated from quick-frozen cells by the hot phenol method (60). Thirty micrograms of total RNA from matched cultures was used to synthesize cDNA with 10 μg of pdN6 primers (Amersham Pharmacia) in the presence of 40 μg/μl RNase inhibitor (Promega). Reverse transcriptase reaction times were modified as follows: 10 min at 37°C, then 42°C for 100 min, followed by a 10-min incubation at 50°C. RNA was hydrolyzed and neutralized as described by Jakobsen et al. (26) and purified with Micron 30 filters (Amicon), and cDNA was eluted and dried with a SpeedVac concentrator (Savant). The dried cDNA was resuspended in 9 μl of 0.1 M sodium bicarbonate (pH 9.0) and incubated for 5 min at 37°C. The cDNA was labeled with Cy3 (DK1622) or Cy5 (AG318) (Amersham Pharmacia) by addition of 2 μl of dye dissolved in 10 μl of dimethyl sulfoxide and incubated for 1 h in the dark. The labeled cDNA was purified with a QIA-quick PCR kit (QIAGEN) as described by the manufacturer and concentrated on a Micron 30 spin filter (Amicon). Labeled cDNA was then dried with a SpeedVac concentrator (Savant) and resuspended in 45 μl of hybridization buffer. Hybridization and posthybridization processing of the slides were performed as described previously (26).
Posthybridized DNA microarrays were scanned with a GenePix 4000A microarray scanner and read by GenePix Pro 3.0 (Axon Inc.). The GenePix array list (gal) file MyxoGALv2.gal, corresponding to the M. xanthus DNA microarrays, was constructed by GalFileMakerv1.2 (DeRisi Lab website; http://derisilab.ucsf.edu). Spots were flagged and removed from analyses based on stringent criteria for shape, signal intensity, and background by GenePix Pro 3.0 (Axon Inc.). Analyses were performed on all unflagged spots. All array analyses, including hierarchical clustering and statistical analysis, were performed by Cluster (Eisen Software; http://rana.lbl.gov/EisenSoftware.htm), Java TreeView software (Alok Saldanha, 2001; http://sourceforge.net/projects/jtreeview), and Significance Analysis of Microarrays (V. G. Tusher, R. Tibshirani and G. Chu, http://www-stat.stanford.edu/∼tibs/SAM).
Isolation of membrane fractions.
Wild-type cells were grown in CTTYE, and nla18 mutant cells were grown in CTTYE containing 40 μg of kanamycin sulfate/ml to a density of 5 × 108 cells/ml. Following centrifugation, the pelleted cells were resuspended in CTTYE and the cell suspension was quick-frozen in liquid nitrogen. The crude cell envelope fraction used for sucrose density gradient centrifugation was prepared by an adaptation of the protocols previously developed by Nikaido (51) and by Orndorff and Dworkin (55). Briefly, after thawing of the frozen samples in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF), the cells were pelleted and resuspended in 8 ml of 10 mM HEPES (pH 7.4)/1 mM PMSF. The cell suspension was then lysed by passage through a French press (SLM Aminco) three times at 14,000 lb/in2. Intact cells were removed from the cell lysates by collecting the supernatant fractions after several rounds of centrifugation at 5,000 × g for 10 min. To pellet the bacterial envelope, the supernatants were centrifuged at 180,000 × g for 1 h at 4°C and the resulting pellet was solubilized overnight with 1 ml of resuspension buffer (10 mM HEPES, 1 mM EDTA, 1 mM PMSF). The membrane fraction was then collected by sucrose gradient centrifugation as described by Osborn et al. (56) with the following steps: 46%, 49%, 52%, 55%, and 58% (wt/wt). Following centrifugation for 18 h in an SW41 rotor (Beckman), a single band was observed at the junction between the 46% and 49% steps. This band was removed, diluted with resuspension buffer, and centrifuged at 180,000 × g for 1 h. Pellets were solubilized at 4°C overnight in 50 μl of resuspension buffer.
For quick whole-membrane isolations, the total membrane fractions were prepared by the small-scale cell envelope preparation procedure of Morona and Reeves (50). With M. xanthus, addition of lysozyme is not necessary for lysing cells but is important for separation of the membrane from the cell wall, thus avoiding smearing problems on SDS-polyacrylamide gels.
The protein composition of the membrane fractions was analyzed on 7.5% SDS-polyacrylamide gels with a 37.5:1 acrylamide/bisacrylamide ratio. Precision Plus Protein All Blue standards (Bio-Rad) or prestained SDS-polyacrylamide gel electrophoresis (PAGE) broad-range standards (Bio-Rad) were used, and the same total amount of protein was loaded into each lane. Gels were stained with Coomassie brilliant blue R-250 (Kodak) to visualize proteins. Peptide mass mapping by matrix-assisted laser desorption ionization-time of flight mass spectrometry was performed by the Molecular Structure Facility at the University of California, Davis, as described by Shevchenko et al. (61) and analyzed with an ABI 4700 Proteomics Analyzer mass spectrophotometer (Applied Biosystems). Measured monoisotopic masses of tryptic peptides were used as inputs to search the M. xanthus protein database (The Institute for Genomic Research [TIGR]-Monsanto; G. Suen and R. D. Welch, personal communication) with the Mascot search engine and a probability-based scoring algorithm (http://www.matrixscience.com).
Nucleotide sequence accession numbers.
All of the DNA microarray results in this study have been submitted to Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/projects/geo/). Accession numbers are provided in Fig. 5.
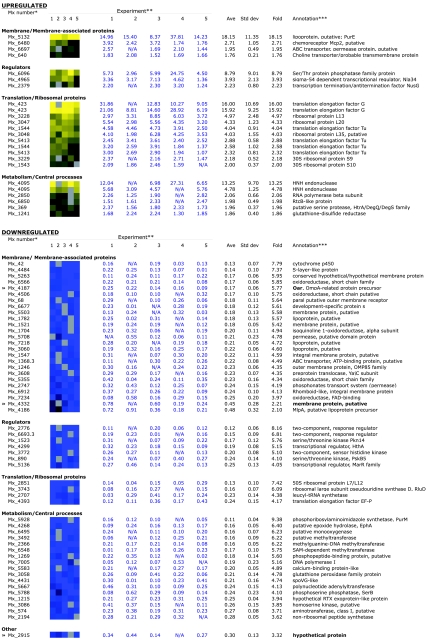
FIG. 5.
Genes showing significant changes in expression. *, Annotation based on M1genome construction of microarrays (26; R. D. Welch, personal communication). Repeated Mx numbers represent duplicate features on the array; not all features are duplicated on each array. **, Five independent biological replicates were prepared as described in Materials and Methods. Averages, standard deviations, and n-fold values are reported. Data are presented graphically as heat maps (yellow is upregulated, blue is downregulated) and as corresponding non-log-transformed ratios of the nla18 mutant to the wild type. A gray box in the head map corresponds to N/A in the table. N/A represents flagged spots on the array (see Materials and Methods). ***, M. xanthus sequence completed by TIGR/Monsanto (personal communication) and submitted to GEO (platform, GPL2848; series, GSE3323; samples, GSM78450, GSM78452, GSM78453, GSM78455, and GSM78456).
RESULTS
Domain structure of Nla18.
The putative domain structure of Nla18 (MXAN3692) based on the completed M. xanthus genome sequence (TIGR website; http://www.tigr.org) was recently described by Jelsbak et al. (27). The C-terminal domain of Nla18 contains a helix-turn-helix motif, which is characteristic of many DNA binding proteins. The central region of the protein contains the highly conserved σ54 activator domain. This domain is required for ATP binding and hydrolysis, which helps σ54-bound RNA polymerase become transcriptionally active. Since these domains are hallmarks of NtrC-like activators, Nla18 is likely to be a bona fide NtrC-like activator protein. Based on a partial sequence of the M. xanthus genome, Caberoy et al. (2) reported that the N-terminal signal recognition domain of Nla18 is about 30 to 40 amino acids (GenBank accession number AY337505). However, the completed M. xanthus genome sequence suggests that the N-terminal signal input domain of the protein is about 210 to 220 amino acids. This region of the Nla18 protein appears to contain a forkhead-associated domain (27), which is a phosphothreonine-specific recognition domain. This finding suggests that the N-terminal signal input domain of Nla18 may interact with a serine/threonine protein kinase, signal transduction proteins that are abundant in M. xanthus.
Developmental gene expression.
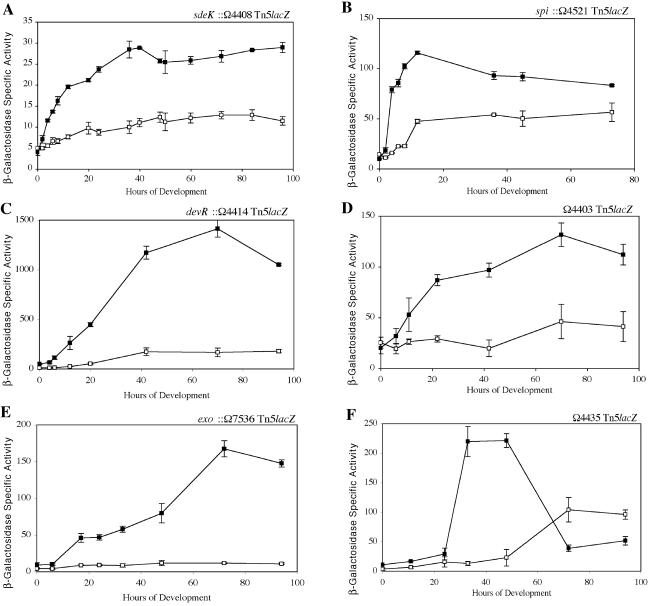
Caberoy et al. (2) found that the nla18 mutant is defective for two important landmark events in fruiting body development, aggregation and sporulation. To determine whether the nla18 mutant is defective for the changes in gene expression that accompany these morphological events, a panel of developmentally regulated lacZ reporter fusions were introduced into nla18 mutant cells. The expression profiles of these lacZ fusions in wild-type and nla18 mutant cells developing on TPM starvation agar are shown in Fig. 1. In wild-type cells, expression of spi::Tn5lacZ and of sdeK::Tn5lacZ was induced prior to the onset of aggregation (Fig. 1A and B), expression of dev::Tn5lacZ and of Ω4403 Tn5lacZ was induced during aggregation (Fig. 1C and D), and expression of exo::Tn5lacZ and of Ω4435 Tn5lacZ was induced as sporulation commences (Fig. 1E and F). In nla18 mutant cells, however, peak expression of the two early reporters spi::Tn5lacZ and sdeK::Tn5lacZ was only 42% to 44% of the peak expression in wild-type cells. The peak expression of the remaining four reporters in nla18 mutant cells ranged from about 10% to 35% of the peak expression observed in wild-type cells. These findings indicate that inactivation of nla18 affects gene expression throughout M. xanthus fruiting body development. They also suggest that Nla18 is required in the early stages of fruiting body development, prior to the start of aggregation.
FIG. 1.
Developmental expression in wild-type cells and nla18 mutant cells. Expression of Tn5lacZ reporter gene fusions was monitored at various times of development on TPM agar. Mean β-galactosidase specific activities were determined from three independent experiments. Error bars are the standard deviations of the means. Black squares show β-galactosidase specific activities for strains carrying the wild-type nla18+ alleles, whereas empty squares show β-galactosidase specific activities for strains carrying the nla18 mutant alleles. The specific fusions monitored were sdeK::Ω4408 Tn5lacZ (A), spi::Ω4521 Tn5lacZ (B), devR::Ω4414 Tn5lacZ (C), Ω4403 Tn5lacZ (D), exo::Ω7536 Tn5lacZ (E), and Ω4435 Tn5lacZ (F).
FruA is a response regulator that plays a critical role in the C-signal transduction pathway, the cell-cell signaling system that regulates changes in gene expression during aggregation and sporulation (10, 54, 64). Since gene expression during aggregation and sporulation appears to be severely impaired in nla18 mutant cells, we examined whether they express FruA. Isogenic wild-type and nla18 mutant cells were harvested after 18 h (when FruA levels peak) and 24 h of development on TPM agar, the cells were lysed, and whole-cell extracts were probed with anti-FruA antibody. The FruA expression profiles shown in Fig. 2 revealed that nla18 mutant cells were producing little or no FruA after 18 and 24 h of development. Similar results were observed when nla18 mutant cells were given additional time to develop (data not shown), suggesting that FruA production in the nla18 mutant is abolished.
FIG. 2.
FruA protein levels in wild-type cells and nla18 mutant cells developing on TPM agar. Whole-cell lysates were prepared from strains DK1622 (nla18+ fruA+), DK11063 (nla18+ fruA), and AG318 (nla18 fruA+) at the indicated times of development. Protein samples were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with anti-FruA antibody as described in Materials and Methods. The same total amount of protein was loaded into each lane. The experiments were repeated three times, and a representative experiment is shown.
A-factor production.
Genes on the early developmental pathway such as spi and fruA are activated in response to A-factor (or A-signal) production. The inability of the nla18 mutant to express normal levels of these genes suggested that this mutant is defective for production of A-factor. To test this hypothesis, A-factor assays were performed as previously described (23, 57) with conditioned medium from wild-type DK101 (nla18+ asgA+), AG339 (nla18 asgA+), or DK476 (nla18+ asgA) cells as the source of A-factor. The ability of conditioned medium to rescue β-galactosidase production in an asg mutant (DK4323) carrying the A-factor-dependent spi::Ω4521 Tn5lacZ transcriptional fusion was used to determine A-factor activity; 1 U of β-galactosidase specific activity is equal to 1 U of A-factor activity. The results of the A-factor assays are shown in Table 2. The levels of A-factor produced by nla18 mutant cells were about 37% of those in wild-type cells. However, nla18 mutant cells generated 2.7-fold more A-factor than asgA cells, which are known to be defective for production of A-factor. Thus, it seems that A-factor production in nla18 mutant cells is impaired, which is consistent with the defects in early developmental gene expression (Fig. 1 and 2).
TABLE 2.
A-factor activity in wild-type cells and mutant cells
| Strain | A-factor activity (U/ml)a |
|---|---|
| DK101 (nla18+asgA+) | 32.2 ± 4.2 |
| AG339 (nla18 asgA+) | 12.0 ± 0.6 |
| DK476 (nla18+asgA) | 4.4 ± 0.2 |
The values shown are means derived from three independent experiments. Standard deviations are shown next to the means.
ppGpp accumulation.
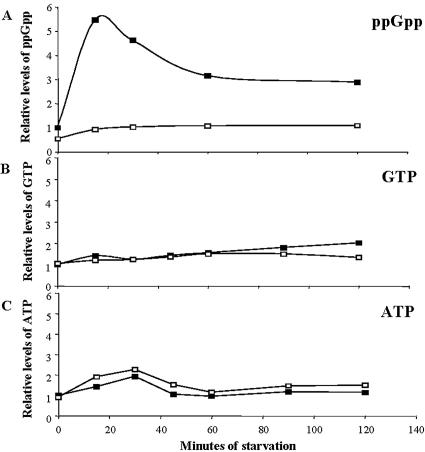
Inactivation of nla18 affects early developmental gene expression and A-signal production, events that are dependent on accumulation of the intracellular starvation signal (p)ppGpp (23, 63). Therefore, we hypothesized that the nla18 mutant may also be defective for (p)ppGpp accumulation. To test this hypothesis, we assayed the relative levels of ppGpp in nla18 mutant cells during vegetative growth and starvation. In the experiment shown in Fig. 3, wild-type DK101 cells and nla18 mutant cells were grown in nutrient broth and subjected to a nutrient downshift, and the relative amounts of ppGpp were measured (Fig. 3A). In wild-type cells, the levels of ppGpp increased about sixfold after 15 min of starvation, followed by a decrease to new steady-state levels that were about threefold higher than the vegetative growth levels. The levels of ppGpp in nla18 mutant cells were about 50% of the wild-type levels during vegetative growth. Furthermore, the peak poststarvation levels of ppGpp in nla18 mutant cells were about 18% of the wild-type peak, indicating that nla18 mutant cells failed to fully initiate a starvation response. No additional rise in ppGpp levels was observed in nla18 mutant cells when we extended the period of starvation (data not shown). Presumably, the relatively low levels of ppGpp in nla18 mutant cells affect downstream developmental gene expression and cell-cell signal production.
FIG. 3.
Relative levels of nucleotides in wild-type cells and nla18 mutant cells. Nucleotides were isolated from DK101 (nla18+ relA+) and AG339 (nla18 relA+) cells at different times following starvation and analyzed as described in Materials and Methods. Signal intensities were normalized to that of vegetative wild-type cells at 0 min (vegetative growth). These assays were repeated four times, and a representative sample set is shown. Black squares show the relative levels of ppGpp, GTP, and ATP in DK101 cells, and empty squares show the relative levels of ppGpp, GTP, and ATP in AG339 cells. The levels of ppGpp were similar when the nla18 insertion was placed in a DK101 or DK1622 strain background (data not shown). Strain MS1000 served as the negative control for the ppGpp assays (data not shown).
RelA converts GTP into (p)ppGpp with ATP as the pyrophosphate donor (3, 63). Hence, GTP and ATP are essential precursors for the synthesis of the (p)ppGpp starvation signal. To determine whether nla18 mutant cells are defective in the production of these ppGpp precursors, we quantified the relative levels of GTP and ATP in nla18 mutant and wild-type cells. As shown in Fig. 3B and C, GTP and ATP levels in nla18 mutant cells were similar to those found in wild-type cells, indicating that nla18 mutant cells produce the nucleotides required for RelA-dependent synthesis of ppGpp.
Expression of genes implicated in ppGpp accumulation.
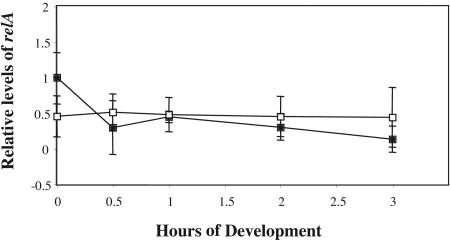
One testable model of Nla18 function is that Nla18 modulates ppGpp levels by regulating expression of the relA gene. To test this hypothesis, we monitored relA mRNA levels in wild-type cells and nla18 mutant cells during vegetative growth and development by QPCR (Fig. 4). The QPCR studies revealed that wild-type cells and nla18 mutant cells expressed similar levels of relA mRNA. The results were confirmed by RNA slot blot hybridization studies (data not shown). Based on the results of these expression studies, we conclude that relA is not under transcriptional control of Nla18, implying that Nla18 modulates ppGpp levels by an alternative mechanism (see Discussion).
FIG. 4.
Expression of relA in wild-type cells and nla18 mutant cells. QPCR was used to examine developmental expression of relA in the wild-type and nla18 mutant backgrounds as described in Materials and Methods. Expression of the relA gene was determined by the absolute method of quantification, and the expression levels were normalized to levels in wild-type cells at time zero (vegetative growth). Expression of relA in wild-type strain DK1622 (black squares) and nla18 mutant strain AG318 (empty squares) was determined with primers relAfwd and relArev (Table 1). The values shown are means derived from four replicates. Error bars are standard deviations of the means.
In M. xanthus, several other genes have been implicated in ppGpp regulation, including socE (5) and mx_1594. The product of the mx_1594 gene has strong sequence similarity to the N-terminal hydrolase domain of E. coli SpoT (K. A. O'Connor and D. R. Zusman, personal communication; M. E. Diodati and M. Singer, unpublished data), a protein that modulates (p)ppGpp levels in response to starvation stimuli (14). To determine whether inactivation of nla18 affects expression of socE or mx_1594, we monitored the levels of socE and mx_1594 mRNAs in wild-type cells and nla18 mutant cells during vegetative growth and development by QPCR. Similar levels of both socE and mx_1594 mRNAs were detected in wild-type cells and nla18 mutant cells (data not shown), indicating that nla18 inactivation does not affect socE or mx_1594 expression.
Finally, for completeness, we performed the reciprocal of the experiment above, assaying for whether nla18 expression is under the control of relA. By both QPCR and RNA slot blot hybridization analysis, we determined that nla18 mRNA levels are not affected by a relA deletion (data not shown). These data suggest that nla18 is not downstream of relA on the M. xanthus developmental pathway.
Vegetative growth.
We found that the phenotypes caused by nla18 inactivation are not specific to the M. xanthus developmental process; an nla18 mutation alters colony color, cell cohesion, and the vegetative growth rate of M. xanthus cells. The growth defect is dependent upon growth media and temperature. When they are grown in CTT broth at 32°C, which are standard laboratory conditions, nla18 mutant cells have a generation time of approximately 14 to 16 h, whereas wild-type cells have a generation time of 5 h. This defect is less severe when nla18 mutant cells are grown at 28°C and the CTT broth is supplemented with yeast extract (CTTYE broth). Under these conditions, nla18 mutant cells have a generation time of 10.5 to 12 h, whereas wild-type cells have a generation time of 5 to 6 h. In addition, nla18 mutant cells display a 60- to 72-h lag phase prior to exponential growth, while wild-type cells begin exponential growth after 4 to 5 h.
Gene expression during vegetative growth.
Our preliminary phenotypic analysis of the nla18 mutant revealed a vegetative growth rate reduction, implying that Nla18 plays an important role in regulating gene expression in vegetative cells. We used a global DNA microarray approach to examine vegetative gene expression patterns in nla18 mutant cells, in an attempt to identify genes under Nla18 control. As described in Materials and Methods, wild-type and nla18 mutant cells were grown to a density of 5 × 108 cells/ml (mid-exponential phase), total cellular RNA was harvested from these cells, and the RNA was used for DNA microarray studies. More than 700 genes showed altered patterns of expression in nla18 mutant cells compared to wild-type cells, some of which are presented in Fig. 5. A complete listing of all of the significant down-regulated and up-regulated genes is provided in Tables S1 and S2, respectively, in the supplemental material, and a complete list of all of the genes on the array with data in at least four of the five experiments is presented in Table S3 in the supplemental material. Inactivation of nla18 affected the expression of several genes whose products are likely to be required for protein synthesis. Perhaps misregulation of these genes in nla18 mutant cells perturbs the M. xanthus translation machinery, affecting the ribosome-associated RelA protein's ability to monitor the translation state of cells and its ability to synthesize (p)ppGpp. Inactivation of nla18 also affected the expression of genes that are likely to encode membrane and membrane-associated proteins, the largest class of nla18-dependent genes with known or predicted functions. Among these genes, expression of oar, which encodes an OmpA-related protein (48), is one of the most severely impacted. In addition, expression of mlpA, a putative lipoprotein gene in the oar operon (21), was down about 2.1-fold in the nla18 mutant (Fig. 5). Other categories of genes whose expression was altered in nla18 mutant cells include a number of putative regulatory genes and genes that are likely to encode metabolic enzymes.
Membrane protein profiles.
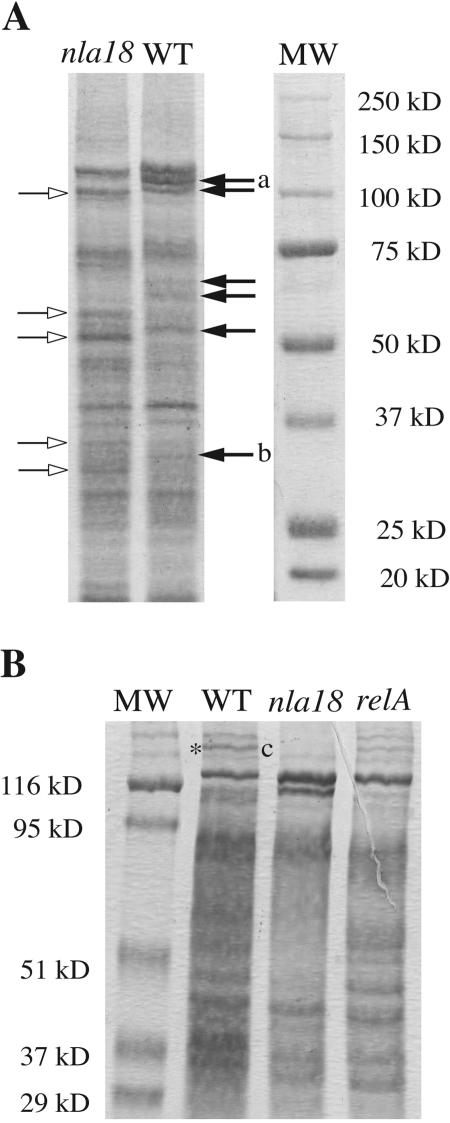
Because the DNA microarrays indicate that expression of many genes encoding putative membrane and membrane-associated proteins is impaired in nla18 mutant cells, we compared the membrane protein profile of nla18 mutant cells to that of wild-type cells by two different membrane protein preparation methods (see Materials and Methods). Previous studies have demonstrated that the presence or absence of membrane proteins can vary based on the method of preparation (51, 55, 56). The data shown in Fig. 6A revealed that the membrane protein profiles of nla18 mutant cells and wild-type cells were different. Many bands appeared to be underrepresented or missing in the nla18 lanes. Several other protein bands are overrepresented in the nla18 lanes, suggesting a complex role for the Nla18 activator in the regulation of these proteins.
FIG. 6.
Membrane protein profiles of wild-type (WT) and mutant cells. (A) Membranes of wild-type (DK1622) and nla18 mutant (AG318) cells were isolated by sucrose density gradient centrifugation and run on a 7.5% SDS-PAGE gel. The same total amount of protein was loaded into each lane. Proteins were visualized with Coomassie brilliant blue R-250 stain, and Precision Plus Protein All Blue standards (Bio-Rad) were used as a reference (MW). The empty and black arrows represent protein bands whose expression appears to increase or decrease in the nla18 mutant, respectively. Bands excised and identified by peptide mass mapping are labeled “a” and “b.” The larger band, band a, is the Oar protein, and band b is Mx_2915, which is a hypothetical protein. The absence of these bands in the nla18 mutant is consistent with our DNA microarray data (Fig. 5). (B) Membranes of isogenic wild-type (DK101), nla18 mutant (AG339), and relA mutant (MS1000) cells were isolated via quick whole-membrane preparation as described in Materials and Methods and run on a 7.5% SDS-PAGE gel. The same total amount of protein was loaded into each lane. Proteins were visualized with Coomassie brilliant blue R-250 stain, and prestained SDS-PAGE broad-range standards (Bio-Rad) were used as a reference (MW). The band excised and identified by peptide mass mapping is flanked by an asterisk and the letter “c.” This band, Mx_4332 (a putative membrane protein), is present in the relA mutant, albeit at a lower intensity than in the wild type. The absence of this band in the nla18 mutant is consistent with our DNA microarray data (Fig. 5).
Previous work with Vibrio cholerae suggests that defects in ppGpp accumulation may lead to changes in membrane protein profiles (22). Therefore, it is possible that the ppGpp accumulation defect in nla18 mutant cells is responsible for the altered membrane protein profile. To explore this idea, we compared the membrane profiles of isogenic wild-type, nla18 mutant, and ΔrelA1 mutant strains (Fig. 6B). Under these conditions, the relative levels of most membrane proteins in ΔrelA1 mutant cells were similar to those in wild-type cells. In contrast, the membrane protein profile of nla18 mutant cells was quite different from that of wild-type cells. Furthermore, band c (Mx_2195), which is differentially expressed in ΔrelA1 mutant cells, is absent in nla18 mutant cells (Fig. 6B). These data imply that the altered membrane protein profile of nla18 mutant cells is not simply due to a decrease in ppGpp levels.
To further examine the differences in membrane protein profiles and to confirm the results of the DNA microarray analysis, we attempted to identify three proteins missing in nla18 mutant cells but present in both wild-type and relA cells. Three wild-type protein bands were excised and digested with trypsin, and the peptide fragments were subjected to matrix-assisted laser desorption ionization-time of flight mass spectrometry. Three proteins were identified: the OmpA-related protein Oar (Mx_4187), a hypothetical protein (Mx_2915), and the putative membrane protein Mx_4332 (Fig. 6A and B, bands a, b, and c, respectively). These results are consistent with the DNA microarray analysis showing that expression of the mx_4187 (Oar), mx_2915, and mx_4332 genes is reduced between 2.2-fold and 5.8-fold (Fig. 5), which supports the hypothesis that nla18 mutant cells have a general defect in the expression of genes that code for membrane and membrane-associated proteins.
DISCUSSION
M. xanthus genomic analyses have identified 52 NtrC-like activators (27), proteins required for transcription from σ54-dependent promoters (49, 70). Because RpoN (σ54) has previously been shown to be essential in M. xanthus, it has been suggested that one or more of these NtrC-like activators are likely to be essential (31). While mutational analyses have uncovered 15 NtrC-like activators that are required for normal development, these studies failed to identify an M. xanthus activator protein that is absolutely required for vegetative growth (2, 15, 18, 20, 26, 27, 36, 67, 69). Interestingly, mutations in two (nla4 and nla18) of these 15 activator genes cause severe vegetative phenotypes, as well as developmental defects. Based on these phenotypes, we propose that inactivation of both nla4 and nla18 may be lethal like null mutations in rpoN.
In this paper, we establish connections among the key NtrC-like activator Nla18, the starvation response, and vegetative gene expression. When starved for nutrients, nla18 mutant cells accumulate about sixfold less ppGpp than their wild-type counterparts. This result suggests that inactivation of nla18 affects the earliest stages of fruiting body development, when M. xanthus cells are assessing the status of available nutrients.
Our results demonstrate that nla18 and relA strains have several phenotypes in common: they are severely impaired for (p)ppGpp accumulation, and they are defective for vegetative growth and development (23, 46, 47, 63). Like relA mutations (5, 23), mutations in nla18 affect the normal function of M. xanthus cell-cell signaling systems. Cells carrying an inactivated copy of nla18 fail to produce normal levels of A-signal, a cell density signal that is required early in fruiting body development (39, 40, 41, 57). In addition, nla18 mutant cells produce little or no FruA, a response regulator that is essential for the C-signal response pathway (10, 64). Since the C-signaling system guides aggregation and sporulation, the lack of FruA in nla18 mutant cells is likely to have dire consequences for the later stages of development. This idea is consistent with the finding that inactivation of nla18 affects aggregation and sporulation (2).
The fact that the vegetative and developmental phenotypes that we observed for nla18 mutant cells were similar to relA cells suggested to us that Nla18 may be directly or indirectly involved in the ppGpp starvation response. The nla18 mutation could affect ppGpp levels by altering the expression patterns of genes coding for regulators of ppGpp synthesis/turnover such as relA, socE, mx_1594 (putative SpoT hydrolase domain-containing gene), and csgA (5, 23). When we examined the levels of relA, mx_1594, and socE mRNAs in nla18 mutant and wild-type cells, we found no significant differences during vegetative growth or development. These results rule out the simple hypothesis that during vegetative growth, Nla18 is working through any of these known or predicted regulators of ppGpp accumulation. It is a formal possibility that during development, Nla18 is working through CsgA; there is sufficient signal for extracellular complementation (2); however, it is not known what level of CsgA is necessary to maintain a stringent response. All of our data, taken as a whole, suggest that the effect the nla18 mutation has on the stringent response is probably quite complex and that indirect effects play a significant role (see below).
How might the nla18 mutation affect ppGpp accumulation? The results of our DNA microarray analysis of vegetative gene expression patterns indicate that inactivation of nla18 affects the expression of several genes whose products are likely to be required for protein synthesis (e.g., EF-Tu, EF-G, and RluD). In nla18 mutant cells, the expression of some of these genes increases while the expression of others decreases relative to that in the wild type (Fig. 5; Tables S1 and S2 in the supplemental material), suggesting that nla18 mutant cells fail to properly regulate genes that encode important components of the M. xanthus translation machinery. It is possible that the differential expression patterns observed for nla18 mutant and wild-type cells are due to growth rate effects; the generation time of the nla18 mutant is more than twice the generation time of a wild-type strain. However, the fact that nla18 mutant cells overexpress some translation-associated genes and underexpress others argues that the altered expression patterns in nla18 mutant cells are not simply due to growth rate effects. Therefore, we propose that Nla18 either directly or indirectly modulates the expression of the M. xanthus translational machinery itself. In the absence of Nla18, the ribosome-associated RelA protein is unable to properly monitor the translation state of cells and adjust ppGpp levels accordingly. Examining the distribution of the various populations of ribosomes, polysomes, and their subunits in the nla18 mutant cells and wild-type cells will allow us to directly test this hypothesis.
In addition to genes required for protein synthesis, nla18 inactivation strongly affects the vegetative expression of a large set of genes for putative membrane and membrane-associated proteins. These results were corroborated by our analysis of the membrane protein profile of nla18 mutant cells; the membrane protein profile of nla18 mutant cells is quite different from that of wild-type cells. Work on the OmpA-related protein Oar supports our findings (21, 48). Strains carrying deletions in oar and mlpA, genes whose expression is affected by nla18 inactivation, show a developmental delay similar to that of the nla18 mutant (21, 48). Furthermore, the oar mlpA double mutant is defective for sporulation, although its sporulation defect is not as severe as that of the nla18 mutant.
Because the membrane protein profile of relA cells is similar to that of wild-type cells, it seems unlikely that the altered membrane protein profile of nla18 mutant cells is simply due to a decrease in ppGpp levels. However, it has been shown in E. coli that relA mutations can affect phospholipid and fatty acid production (9, 52, 53, 65). It is therefore possible that the M. xanthus relA mutant has a compromised membrane that was not detectable in our protein analysis. This caveat aside, our data strongly suggest that the nla18 mutant's vegetative and developmental phenotypes are due to at least two defects: (i) decreased levels of ppGpp, a defect that may be caused by the misexpression of components of the translational machinery, and (ii) the altered expression patterns of genes that code for membrane and membrane-associated proteins.
These two defects may be linked; the misexpression of genes such as those encoding transporters could have significant effects on the nla18 mutant cell's ability to acquire nutrients from the environment, causing unbalanced growth and defects in ppGpp synthesis. However, the fact that nla18 mutant cells grow on minimal A1 medium indicates that these defects are not simply due to auxotrophy (2). There is precedence for the regulation of important membrane and/or membrane-associated proteins by the σ54 system. In Borrelia and Pseudomonas, it has been reported that several genes for membrane and membrane-associated proteins are directly controlled by RpoN (25, 71). Our next challenges are to determine which genes uncovered in these studies are directly regulated by Nla18 and to elucidate the signal transduction networks that modulate Nla18 activity.
Supplementary Material
Acknowledgments
We thank L. Søgaard-Andersen for providing FruA antibody and R. Welch, J. Jakobsen, B. Goldman, TIGR, and the Monsanto Company for providing access to the M. xanthus genome sequence prior to publication. We thank J. Bragg for help with microarray analyses, G. Suen for Perl scripts, and G. Siu for technical assistance.
This work was supported by National Institute of General Medical Sciences Public Health Service grants T32GM0737 and GM56765B to M. E. Diodati, National Institute of General Medical Sciences Public Health Service grant GM54592 to M. Singer, and National Science Foundation grant MCB-0444154 to A. Garza.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bretscher, A. P., and D. Kaiser. 1978. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J. Bacteriol. 133:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caberoy, N. B., R. D. Welch, J. S. Jakobsen, and S. C. Slater, and A. G. Garza. 2003. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J. Bacteriol. 185:6083-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1494. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 4.Chatterji, D., and A. K. Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4:160-165. [DOI] [PubMed] [Google Scholar]
- 5.Crawford, E. W., and L. J. Shimkets. 2000. The stringent response in Myxococcus xanthus is regulated by SocE and the CsgA C-signaling protein. Genes Dev. 14:483-492. [PMC free article] [PubMed] [Google Scholar]
- 6.Downard, J., S. V. Ramaswamy, and K-S. Kil. 1993. Identification of esg, a genetic locus involved in cell-cell signaling during Myxococcus xanthus development. J. Bacteriol. 175:7762-7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dworkin, M. 1962. Nutritional requirements for vegetative growth of Myxococcus xanthus. J. Bacteriol. 288:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dworkin, M. 1996. Recent advances in the social and developmental biology of myxobacteria. Microbiol. Rev. 60:70-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichel, J., Y.-Y. Chang, D. Riesenberg, and J. E. Cronan, Jr. 1999. Effect of ppGpp on Escherichia coli cyclopropane fatty acid synthesis is mediated through the RpoS sigma factor. J. Bacteriol. 181:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellehauge, E., M. Nørregaard-Madsen, and L. Søgaard-Andersen. 1998. The FruA signal transduction protein provides a checkpoint for the temporal coordination of intercellular signals in M. xanthus development. Mol. Microbiol. 30:807-813. [DOI] [PubMed] [Google Scholar]
- 11.Garza, A. G., J. S. Pollack, B. Z. Harris, A. Lee, I. M. Keseler, E. F. Licking, and M. Singer. 1998. SdeK is required for early fruiting body development in Myxococcus xanthus. J. Bacteriol. 180:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garza, A. G., B. Z. Harris, and B. M. Greenberg. 2000. Control of asgE expression during growth and development of Myxococcus xanthus. J. Bacteriol. 182:6622-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garza, A. G., B. Z. Harris, J. S. Pollack, and M. Singer. 2000. The asgE locus is required for cell-cell signalling during Myxococcus xanthus development. Mol. Microbiol. 35:812-824. [DOI] [PubMed] [Google Scholar]
- 14.Gentry, D. R., and M. Cashel. 1996. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol. Microbiol. 19:1373-1384. [DOI] [PubMed] [Google Scholar]
- 15.Gorski, L., and D. Kaiser. 1998. Targeted mutagenesis of σ54 activator proteins in Myxococcus xanthus. J. Bacteriol. 180:5896-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gronewold, T. M., and D. Kaiser. 2001. The act operon controls the level and time of C-signal production for Myxococcus xanthus development. Mol. Microbiol. 40:744-756. [DOI] [PubMed] [Google Scholar]
- 17.Gulati, P., D. Xu, and H. B. Kaplan. 1995. Identification of the minimum regulatory region of a Myxococcus xanthus A-signal-dependent developmental gene. J. Bacteriol. 177:4645-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, D., Y. Wu, and H. B. Kaplan. 2000. Identification and characterization of genes required for early Myxococcus xanthus gene expression. J. Bacteriol. 182:4564-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagen, D. C., A. P. Bretscher, and D. Kaiser. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 20.Hager, E., H. Tse, and R. E. Gill. 2001. Identification and characterization of spdR mutations that bypass the BsgA protease-dependent regulation of developmental gene expression in Myxococcus xanthus. Mol. Microbiol. 39:765-780. [DOI] [PubMed] [Google Scholar]
- 21.Hanlon, W. A., M. Martinez-Canamero, M. Inouye, and S. Inouye. 1995. MlpA, a lipoprotein required for normal development of Myxococcus xanthus. J. Bacteriol. 177:7150-7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haralalka, S., S. Nandi, and K. B. Rupak. 2003. Mutation in the relA gene of Vibrio cholerae affects in vitro and in vivo expression of virulence factors. J. Bacteriol. 185:4672-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris, B. Z., D. Kaiser, and M. Singer. 1998. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 12:1022-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodgkin, J., and D. Kaiser. 1977. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. USA 74:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubner, A., X. Yang, D. M. Nolen, T. G. Popova, F. C. Cabello, and M. V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA 98:12724-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakobsen, J. S., L. Jelsbak, L. Jelsbak, R. D. Welch, C. Cummings, B. Goldman, E. Stark, S. Slater, and D. Kaiser. 2004. σ54 enhancer binding proteins and Myxococcus xanthus fruiting body development. J. Bacteriol. 186:4361-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jelsbak, L., M. Givskov, and D. Kaiser. 2005. Enhancer-binding proteins with a forkhead-associated domain and the σ54 regulon in Myxococcus xanthus fruiting body development. Proc. Natl. Acad. Sci. USA 102:3010-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julien, B. J., D. Kaiser, and A. G. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan, H. B., A. Kuspa, and D. Kaiser. 1991. Suppressors that permit A-signal independent developmental gene expression in Myxococcus xanthus. J. Bacteriol. 173:1460-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keseler, I. M., and D. Kaiser. 1995. An early A-signal-dependent gene in Myxococcus xanthus has a σ54-like promoter. J. Bacteriol. 177:4638-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keseler, I. M., and D. Kaiser. 1997. σ54, a vital protein for Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 94:1979-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, S. K., and D. Kaiser. 1990. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell 61:19-26. [DOI] [PubMed] [Google Scholar]
- 34.Kim, S. K., and D. Kaiser. 1990. Cell alignment is required in differentiation of Myxococcus xanthus. Science 249:926-928. [DOI] [PubMed] [Google Scholar]
- 35.Kim, S. K., and D. Kaiser. 1991. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J. Bacteriol. 173:1722-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirby, J. R., and D. R. Zusman. 2003. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 100:2008-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 38.Kroos, L., and D. Kaiser. 1987. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1:840-854. [DOI] [PubMed] [Google Scholar]
- 39.Kuspa, A., L. Kroos, and D. Kaiser 1986. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev. Biol. 117:267-276. [DOI] [PubMed] [Google Scholar]
- 40.Kuspa, A., L. Plamann, and D. Kaiser 1992. Identification of heat-stable A-factor from Myxococcus xanthus. J. Bacteriol. 174:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuspa, A., L. Plamann, and D. Kaiser. 1992. A-signalling and the cell density requirement for Myxococcus xanthus development. J. Bacteriol. 174:7360-7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lancero, H., N. B. Caberoy, S. Castañeda, Y. Li, A. Lu, D. Dutton, X. Y. Duan, H. B. Kaplan, W. Shi, and A. G. Garza. 2004. Characterization of a Myxococcus xanthus mutant that is defective for adventurous and social motilities. Microbiology 150:4085-4093. [DOI] [PubMed] [Google Scholar]
- 43.LaRossa, R., J. Kuner, D. Hagen, C. Manoil, and D. Kaiser. 1983. Developmental cell interactions of Myxococcus xanthus: analysis of mutants. J. Bacteriol. 153:1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, S., B. U. Lee, and L. J. Shimkets. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 45.Licking, E., L. Gorski, and D. Kaiser. 2000. A common step for changing cell shape in fruiting body and starvation-independent sporulation of Myxococcus xanthus. J. Bacteriol. 182:3553-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manoil, C., and D. Kaiser. 1980. Accumulation of guanosine tetraphosphate and guanosine pentaphosphate in Myxococcus xanthus during starvation and myxospore formation. J. Bacteriol. 141:297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manoil, C., and D. Kaiser. 1980. Guanosine pentaphosphate and guanosine tetraphosphate accumulation and induction of Myxococcus xanthus fruiting body development. J. Bacteriol. 141:305-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Canamero, M., J. Muñoz-Dorado, E. Farez-Vidal, M. Inouye, and S. Inouye. 1993. Oar, a 115-kilodalton membrane protein required for development in Myxococcus xanthus. J. Bacteriol. 175:4756-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morett, E., and L. Segovia. 1993. The σ54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 175:6067-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morona, R., and P. Reeves. 1982. The tolC locus of Escherichia coli affects the expression of three major outer membrane proteins. J. Bacteriol. 150:1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nikaido, H. 1994. Isolation of outer membranes. Methods Enzymol. 235:225-234. [DOI] [PubMed] [Google Scholar]
- 52.Nunn, W. D., and J. E. Cronan, Jr. 1976. Evidence for a direct effect on fatty acid synthesis in relA gene control of membrane phospholipid synthesis. J. Mol. Biol. 102:167-172. [DOI] [PubMed] [Google Scholar]
- 53.Nunn, W. D., and J. E. Cronan, Jr. 1976. Regulation of membrane phospholipid synthesis by the relA gene: dependence on ppGpp levels. Biochemistry 15:2546-2550. [DOI] [PubMed] [Google Scholar]
- 54.Ogawa, M., S. Fujitani, X. Mao, S. Inouye, and T. Komano. 1996. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol. Microbiol. 22:757-767. [DOI] [PubMed] [Google Scholar]
- 55.Orndorff, P. E., and M. Dworkin. 1980. Separation and properties of the cytoplasmic and outer membranes of vegetative cells of Myxococcus xanthus. J. Bacteriol. 141:914-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Osborn, M. J., J. E. Gander, E. Parisi, and J. Carson. 1972. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem. 247:3962-3972. [PubMed] [Google Scholar]
- 57.Plamann, L., A. Kuspa, and D. Kaiser. 1992. Proteins that rescue A-signal defective mutants of Myxococcus xanthus. J. Bacteriol. 174:3311-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plamann, L., J. M. Davis, B. Cantwell, and J. Mayor. 1994. Evidence that asgB encodes a DNA-binding protein essential for growth and development of Myxococcus xanthus. J. Bacteriol. 176:2013-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Romeo, J. M., and D. R. Zusman. 1991. Transcription of the myxobacterial hemagglutinin gene is mediated by a σ54-like promoter and a cis-acting upstream regulatory region of DNA. J. Bacteriol. 193:2969-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 61.Shevchenko, A., M. Wilm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 62.Shimkets, L. J. 1990. Social and developmental biology of the myxobacteria. Microbiol. Rev. 54:473-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singer, M., and D. Kaiser. 1995. Ectopic production of guanosine penta- and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 9:1633-1644. [DOI] [PubMed] [Google Scholar]
- 64.Søgaard-Andersen, L., F. J. Slack, H. Kimsey, and D. Kaiser. 1996. Intercellular C-signaling in Myxococcus xanthus involves a branched signal transduction pathway. Genes Dev. 10:740-754. [DOI] [PubMed] [Google Scholar]
- 65.Spencer, A., E. Muller, J. E. Cronan, Jr., and T. A. Gross. 1977. relA gene control of the synthesis of lipid A fatty acyl moieties. J. Bacteriol. 130:114-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spratt, B. G., P. J. Hedge, S. T. Heesen, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogs of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41:337-342. [DOI] [PubMed] [Google Scholar]
- 67.Sun, H., and W. Shi. 2001. Analyses of mrp genes during Myxococcus xanthus development. J. Bacteriol. 179:6733-6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ueki, T., S. Inouye, and M. Inouye. 1996. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183:153-157. [DOI] [PubMed] [Google Scholar]
- 69.Wu, S. S., and D. Kaiser. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J. Bacteriol. 179:7748-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu, H., and T. R. Hoover. 2001. Transcriptional regulation at a distance in bacteria. Curr. Opin. Microbiol. 4:138-144. [DOI] [PubMed] [Google Scholar]
- 71.Yamano, Y., T. Nishikawa, and Y. Komatsu. 1998. Involvement of the RpoN protein in the transcription of the oprE gene in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 162:31-37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.