Abstract
The Bordetella master virulence regulatory system, BvgAS, controls a spectrum of gene expression states, including the virulent Bvg+ phase, the avirulent Bvg− phase, and at least one Bvg-intermediate (Bvgi) phase. We set out to define the species- and strain-specific features of this regulon based on global gene expression profiling. Rather than functioning as a switch, Bvg controls a remarkable continuum of gene expression states, with hundreds of genes maximally expressed in intermediate phases between the Bvg+ and Bvg− poles. Comparative analysis of Bvg regulation in B. pertussis and B. bronchiseptica revealed a relatively conserved Bvg+ phase transcriptional program and identified previously uncharacterized candidate virulence factors. In contrast, control of Bvg−- and Bvgi-phase genes diverged substantially between species; regulation of metabolic, transporter, and motility loci indicated an increased capacity in B. bronchiseptica, compared to B. pertussis, for ex vivo adaptation. Strain comparisons also demonstrated variation in gene expression patterns within species. Among the genes with the greatest variability in patterns of expression, predicted promoter sequences were nearly identical. Our data suggest that the complement of transcriptional regulators is largely responsible for transcriptional diversity. In support of this hypothesis, many putative transcriptional regulators that were Bvg regulated in B. bronchiseptica were deleted, inactivated, or unregulated by BvgAS in B. pertussis. We propose the concept of a “flexible regulon.” This flexible regulon may prove to be important for pathogen evolution and the diversification of host range specificity.
Bordetellae are gram-negative bacteria that colonize ciliated respiratory epithelial surfaces of a variety of hosts (9). Bordetella bronchiseptica causes infections that are typically chronic and often asymptomatic in a broad range of animals (21). Numerous phylogenetic and genomic analyses (see, e.g., references 13, 43, and 59) have indicated this species to be the progenitor of human-restricted agents of acute respiratory disease in humans: B. pertussis, which causes whooping cough in children and coughing illness in adolescents and adults (63), and B. parapertussis, which confers similar but milder clinical symptoms (22). Despite vaccination, B. pertussis remains a major cause of childhood mortality worldwide and is reemerging in some highly vaccinated populations (7, 42, 63). Even though Bordetella species are closely related at the nucleotide sequence level, they differ in several respects, such as severity of disease and host range specificity. B. pertussis likely evolved from a B. bronchiseptica-like ancestor primarily through genome decay (13, 43). Differential gene expression and coding polymorphisms within expressed loci appear to be the primary determinants of species-specific phenotypes (9, 20, 43).
Expression of nearly all known Bordetella virulence and colonization factors is controlled by the BvgAS signal transduction system (reviewed in reference 8). In response to a variety of environmental signals, the BvgS transmembrane sensor kinase initiates a multistep phosphorylation cascade that ultimately leads to phosphorylation of the DNA-binding response regulator BvgA. Rather than functioning as an on/off switch, BvgAS controls a spectrum of at least three distinct gene expression states along a regulatory continuum (11). When BvgAS is fully active (Bvg+ phase), B. bronchiseptica and B. pertussis express nearly identical sets of toxins and adhesins, including filamentous hemagglutinin, pertactin, fimbriae, and adenylate cyclase toxin. The transcription of a type III secretion system (TTSS) locus is also Bvg activated in both species, although the synthesis of TTSS proteins appears to be posttranscriptionally blocked specifically in B. pertussis (36). A key difference in Bvg-activated gene expression is the pertussis toxin locus (ptx/ptl), the coding sequences of which are intact in both species but expressed only in the Bvg+ phase of B. pertussis (2). Motility loci in B. bronchiseptica (1) and virulence-repressed genes (vrg genes) in B. pertussis (31) are expressed when Bvg is inactive (Bvg− phase). Some genes, such as bipA, are maximally expressed between the Bvg+ and Bvg− poles (Bvg-intermediate phase [Bvgi]) (15, 53).
While the Bvg+ phases of B. bronchiseptica and B. pertussis appear to be adapted to respiratory tract colonization, the Bvg− phase of B. bronchiseptica promotes survival under nutrient-limiting conditions (1, 10, 34, 44). The Bvg− phase of B. pertussis is still relatively uncharacterized, and because B. pertussis is apparently unable to persist outside the host, this phase has been proposed to be an evolutionary remnant (9, 40). The Bvgi phase is thought to be similar in B. bronchiseptica and B. pertussis (17) and may be involved in biofilm formation (25) and aerosol transmission (11, 60).
Although the signals that influence BvgAS in vivo remain unknown, Bvg-regulated gene expression can be precisely regulated in vitro: BvgS is active during growth at 37°C, but temperatures below 26°C or the presence of millimolar amounts of nicotinic acid (NA) or sulfate anion (modulating signals) render BvgS inactive (37, 41). Furthermore, bvgS “phase-locked” mutant alleles can be used to freeze the signaling system in specific positions along the regulatory continuum (9).
Using the complementary approaches of environmental modulation and phase-locked mutants for global expression analysis, we defined the full BvgAS regulons in B. pertussis and B. bronchiseptica. By comparing the species' expression profiles, we sought to identify novel virulence determinants and discover the genetic basis of species-specific phenotypes. To understand the pathogenic and physiological adaptations required for survival in the host environment, we explored the dynamic Bvg-regulated transcription patterns upon environmental modulation and examined regulation of transporter repertoires and metabolic pathways. By identifying novel Bvg− and Bvgi phase genes, we sought to elucidate the roles for these phases in the Bordetella life cycle. To assess intraspecific gene expression diversity, we examined strain-specific Bvg regulation within B. bronchiseptica and B. pertussis. Finally, by comparing promoter sequences and Bvg-regulated transcription factors, we identified possible mechanisms of differential gene expression.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains used in this study are listed in Table 1. Bvg+ and Bvg− phase-locked derivatives of B. pertussis were constructed by introducing the respective bvgS alleles by allelic exchange as described previously (34). Bordetella strains were grown on BG agar (Becton Dickinson, Franklin Lakes, NJ) supplemented with 7.5% or 15% sheep blood for B. bronchiseptica and B. pertussis, respectively. For RNA isolation, cultures were grown overnight at 37°C in modified Stainer-Scholte medium (SSM) and then reinoculated into prewarmed medium at an optical density at 600 nm of approximately 0.05 to 0.1. Cultures were incubated at 37°C until they reached mid-log phase, at which point they were harvested by centrifugation for 30 seconds. (No samples were taken at an optical density at 600 nm exceeding 1.5, at which point the culture is still growing logarithmically under these conditions.) For steady-state modulation experiments, NA was added to both overnight and log-phase cultures at the concentrations indicated below. For phase shift experiments, cultures were grown to mid-log phase in SSM containing 75 mM magnesium sulfate, pelleted by centrifugation, resuspended in prewarmed magnesium sulfate-free SSM, and incubated at 37°C throughout the course of sampling.
TABLE 1.
Strains used in this study
| Species and strain | Phenotype | Genotype | Parent | Source or reference | Alias | Hosta |
|---|---|---|---|---|---|---|
| B. bronchiseptica | ||||||
| RB50 | WTb | 10 | Rabbit | |||
| RB53 | Bvg+ | bvgS-C3 | RB50 | 10 | ||
| RB53i | Bvgi | bvgS-C3, II | RB50 | 11 | ||
| RB54 | Bvg− | ΔbvgS | RB50 | 10 | ||
| Bbr81 | WT | 59 | 732 | Dog | ||
| Bbr77 | WT | 59 | 675 | Human | ||
| B. pertussis | ||||||
| Tohama I | WT | 30 | Human | |||
| ThSc3 | Bvg+ | bvgS-C3 | Tohama I | This study | ||
| ThdS | Bvg− | ΔbvgS | Tohama I | This study | ||
| GMT-1 | WT | GG | 35 | Human | ||
| GSc3 | Bvg+ | bvgS-C3 | GMT-1 | This study | ||
| BPC1.i | Bvgi | bvgS-C3, II | GMT-1 | 60 | ||
| GMTdS | Bvg− | ΔbvgS | GMT-1 | This study |
Host is given only for wild-type strains.
WT, wild type.
Microarray analysis.
Cy5-labeled cDNA was hybridized to spotted DNA microarrays with a Cy3-labeled reference. The reference sample for these experiments was one of two preparations. In some cases a common reference mixture of RNA samples from modulated and unmodulated B. pertussis Tohama I and B. bronchiseptica RB50 as well as their phase-locked derivatives was labeled with Cy3. In other cases, as described by Talaat et al. (54), we used a Cy3-labeled genomic DNA sample comprised of a 1:1:1 mixture of the three genomes that are represented on the array as described previously (13). All samples in a given experiment (i.e., set of phase-locked strains, NA dose curve, or time course) were processed using the same reference, and data obtained from the two methods were not combined during analysis. Details on nucleic acid isolation, labeling, and microarray construction are given in Methods in the supplemental material.
Background-subtracted Cy5/Cy3 intensity ratios were determined for spots meeting quality control criteria and then averaged across replicates and normalized. For comparison of phase-locked strains, a fold change cutoff of 2.1 was chosen to select Bvg-regulated genes. This threshold was consistent with all but the lowest fold changes of transcripts determined to be significant by significance analysis of microarrays (57) but was more inclusive of known Bvg-regulated genes. For modulation experiments, to be considered Bvg regulated, genes were required to have data from all array hybridizations and a fold change between the maximum and minimum values across the data set exceeding a threshold of 2.4 for B. bronchiseptica and 2.0 for B. pertussis. The selection of cutoffs was chosen empirically to maximize inclusion of known Bvg-regulated genes while minimizing apparent noise. For comparison of B. bronchiseptica strains, genes with at least three measurements per strain and a fold change of at least 2.1 were considered regulated. Data were clustered using the self-organizing tree algorithm (23), after which nodes were reordered manually based on the mean expression profile within each node. Further details are in Methods in the supplemental material.
Gene annotation is as described previously (43), with functional categories according to MultiFun (50). Open reading frame (ORFs) assigned to the “virulence factors” category were reassigned to a functional class reflecting their cellular function (see the tables in the supplemental material). MultiFun categories 7.0.0 (some information, but not classifiable) and 0.0.0 (unknown proteins, no known homologues) were combined into the class “hypothetical ORFs.” Metabolic pathway analysis employed Pathway Tools (29) and automatically generated pathway genome databases from BioCyc (SRI International; [http://biocyc.org/BBRO518/organism-summary?object=BBRO518 and http://biocyc.org/BPER520/organism-summary?object=BPER520]). Further annotation of predicted transporters was obtained from TransportDB (47).
Real-time PCR.
SYBR Green real-time reverse transcription-PCR assays were used to determine transcript levels of selected genes. The relative quantitation method (ΔΔCT) (33) was used to evaluate variation between phase-locked strains relative to each gene examined. Further details are in Methods in the supplemental material.
Motif identification.
Genes were first grouped into putative operons (see Tables S1 and S2 in the supplemental material). MEME (3) was used to search the upstream sequences of coregulated operons for conserved motifs of between 6 and 20 nucleotides (nt). Motifs discovered by this method are weighted statistical models. To assess whether they were specific to the regulated operons, multiple alignments of motifs were input to HMMer (version 1.8.4; S. R. Eddy, Washington University [http://hmmer.wustl.edu]) in order to construct hidden Markov models that were used to search both coregulated genes and unregulated genes. The scores from searches against unregulated genes were used to set the 98th percentile score for each motif. Motifs with scores exceeding this threshold in the coregulated genes were considered significant. Sequence logo displays were created using SEQLOGO (12). Details are in Methods in the supplemental material.
Accession numbers.
Microarray data have been deposited in ArrayExpress as E-MEXP-399, E-TABM-27, E-TABM-28, E-TABM-29, E-TABM-30, and E-TABM-31.
RESULTS
Species-specific regulation by BvgAS.
Genes expressed at either end of the Bvg phase spectrum were identified by microarray-based transcriptional profiling of Bvg+ and Bvg− phase-locked derivatives of B. bronchiseptica RB50 and B. pertussis GMT-1 (Table 1). Data and enhanced figures are available in the supplemental material and at http://asiago.stanford.edu/BvgAS.
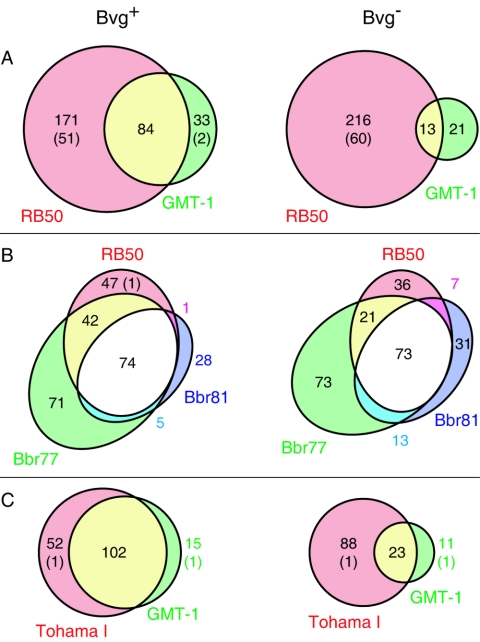
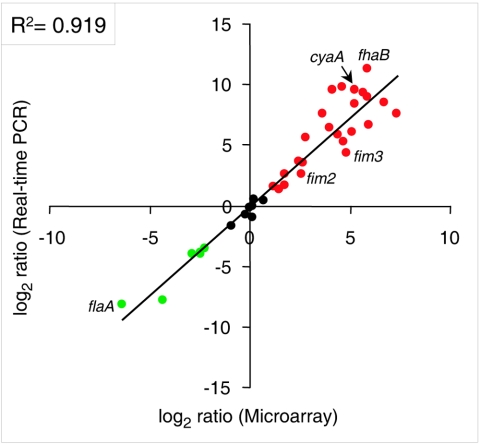
Two hundred eighty-eight genes were classified as Bvg activated and 250 as Bvg repressed (Fig. 1A; see Tables S3 and S4 in the supplemental material). Twenty-nine percent of the Bvg-activated genes were shared by RB50 and GMT-1, but 59% were regulated only in RB50. In contrast, 86% Bvg-repressed genes were so regulated only in RB50. Twenty-nine percent of the genes regulated by BvgAS specifically in RB50 were presumed to be missing from or highly divergent in the GMT-1 genome based upon comparative genome hybridization analysis (Fig. 1A and data not shown). (Such genes were treated as components of the B. bronchiseptica-specific regulon. In the tables in the supplemental material, genes that are absent, by comparative sequencing or comparative genome hybridization analysis, are indicated to distinguish them from those in which transcriptional abundance of an intact gene is being differentially regulated.) Microarray data were validated by quantitative real-time PCR assays of 38 RB50 genes and 29 GMT-1 genes. Microarray and reverse transcription-PCR data were highly concordant for RB50 (r2 = 0.92) (Fig. 2) and GMT-1 (r2 = 0.80) (not shown).
FIG. 1.
Distribution of differentially expressed genes. The distributions of Bvg-activated and Bvg-repressed genes among B. bronchiseptica RB50 and B. pertussis GMT-1 (A), three B. bronchiseptica strains (B), and two B. pertussis strains (C) are shown. Areas within Venn diagrams are drawn approximately to scale, and the number of ORFs in each is indicated. In parentheses are numbers of genes absent from the other genome(s) as determined by genome sequence comparison (RB50 and Tohama I) or comparative genome hybridization (GMT-1).
FIG. 2.
Validation of B. bronchiseptica microarray data by real-time PCR. Ratios of transcript abundance in RB53 (Bvg+) versus RB54 (Bvg−), obtained by microarray analysis (x axis) or real-time PCR (y axis). Red, Bvg activated; green, Bvg repressed. Selected genes are shown for illustrative purposes.
The Bvg-activated genes of B. pertussis and B. bronchiseptica were similarly distributed among functional categories and were enriched for genes predicted to encode proteins involved in folding and ushering and in transport and proteins expressed on the cell surface (see Fig. S1 in the supplemental material). The expected preferential expression of known virulence factors in the Bvg+ phase in both species was corroborated (see Table S5 in the supplemental material). Of the fimbrial major subunit genes, fim2 and fimA were Bvg-activated genes in both species, but fim3, fimX, fimN, and BB3424 were Bvg activated only in RB50.
Genes coregulated with bona fide virulence factors may be candidate virulence determinants, particularly if sequence motifs suggest a direct role in pathogenesis. Eight autotransporter genes, including five encoding known virulence factors, were Bvg activated in both species, and two more were Bvg activated in B. bronchiseptica. Several iron acquisition genes were Bvg activated, though the cultures were not iron-depleted, suggesting that Bvg+ phase Bordetella is primed for iron acquisition prior to actual iron restriction. A gene for an aerolysin/pertussis toxin domain-containing protein (BB3242/BP1251), a potential novel toxin, was Bvg activated in both species.
Three putative adhesin genes, i.e., fhaS, fhaL, and BB0110, were Bvg activated in RB50 but not GMT-1. The lipid A palmitoyl transferase gene, pagP, was Bvg activated in RB50, consistent with prior results (45), but was not regulated in GMT-1. Other RB50-specific Bvg-activated genes included genes encoding protein-folding catalysts, catalase, and a DegP family serine protease (MucD), suggesting that B. bronchiseptica may be more resistant to environmental stress in the Bvg+ phase. Of note, two Bvg-activated loci in RB50 were Bvg repressed in GMT-1: osmB, encoding a lipoprotein, and a cluster of three hypothetical ORFs.
Consistent with previous results, the ptx/ptl locus was strongly upregulated in the Bvg+ phase of GMT-1 but not in RB50, in which the promoter is inactive regardless of Bvg phase (2). Other B. pertussis-specific Bvg-activated genes included an outer membrane porin gene (BP0267) and a TonB-dependent iron transporter gene (bfrE).
Bvg-repressed genes were found almost exclusively in B. bronchiseptica. Only 13 genes were Bvg repressed in both species (Fig. 1A), including several genes in the wlb locus, which is required for synthesis of the lipopolysaccharide (LPS) trisaccharide. Phase-specific expression of this locus may contribute to the preferential production of O-antigen-containing LPS in the Bvg− phase (45, 58).
Consistent with previous studies (1), the chemotaxis and flagellar machinery was strongly induced in the B. bronchiseptica Bvg− phase (see Fig. S1 in the supplemental material). Other B. bronchiseptica-specific Bvg-repressed genes of interest are predicted to encode two autotransporters, two iron acquisition proteins, and a 340-kDa protein (BB1186) with multiple hemolysin-type calcium-binding repeats and an associated secretion system. Five genes from the putative capsule biosynthesis locus were upregulated in the Bvg− phase of B. pertussis only, implying phase-specific capsule production.
Patterns of BvgAS-regulated gene expression upon environmental modulation.
The initial experiments assayed the extreme ends of the Bvg phase spectrum and failed to account fully for genes such as bipA that are maximally expressed during other, intermediate phases of Bvg activity. To identify such intermediate-phase genes, expression profiles of Bvgi phase-locked strain derivatives of RB50 and GMT-1 were determined (see Table S6 in the supplemental material). This approach yielded only a few genes with transcript profiles similar to that of bipA, including a gene predicted to encode a probable thiolase (BB4250/BP0422) and, in RB50 only, a predicted phenylacetic acid (paa) degradation operon. Because the use of strains locked at only one intermediate state of Bvg activation may not fully reveal all intermediate-phase genes, we pursued an independent and complementary approach.
Cultures can be experimentally held at any point along the BvgAS activation continuum by chemically modulating BvgS activity (e.g., with NA). The pattern of Bvg-regulated gene expression observed in a series of cultures with decreasing concentrations of modulator is recapitulated in the time-dependent pattern of gene expression following a shift from modulating to nonmodulating conditions (reviewed in reference 9). For example, early Bvg-activated genes (e.g., fhaB) are expressed immediately after a shift and at intermediate concentrations of modulator, while late Bvg-activated genes (e.g., ptx) are expressed at later time points (2 hours or more) after the shift and only at very small amounts of modulator. Here, we use the terms “early” and “late” to describe patterns gleaned from both steady-state modulation and time-dependent shift experiments.
Transcription abundance profiles of RB50 and GMT-1 grown in increasing amounts of NA were determined. Isogenic strain derivatives with a constitutive bvgS allele (Bvgc) were also analyzed to identify genes that respond to NA independently of BvgAS.
(i) B. bronchiseptica.
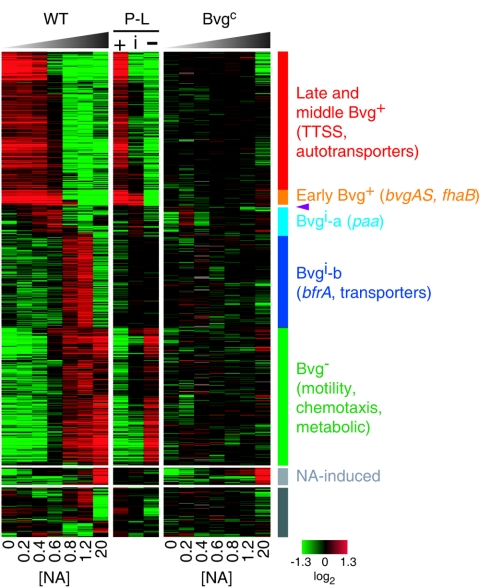
In vitro modulation of RB50 revealed remarkably diverse transcript patterns falling into five dominant gene expression classes (Fig. 3). Bvg-repressed, two classes of Bvg-intermediate, and early and late Bvg-activated gene classes were discerned, each with subtle but clear differences in expression patterns. Most Bvg-repressed genes, including motility and chemotaxis loci, were expressed in the presence of 0.8 mM or more NA, but expression of a few genes, including an ABC transporter locus, was activated only at the highest concentration of NA (20 mM).
FIG. 3.
Patterns of Bvg-regulated gene expression in B. bronchiseptica RB50. Within each experiment, data are centered and shown from Bvg+ (left) to Bvg− (right). Rows correspond to array probes, and columns correspond to experiments. +, phase-locked Bvg+; i, phase-locked Bvgi; −, phase-locked Bvg−. Red, increased transcript abundance; green, decreased transcript abundance; black, no significant change in transcript abundance; gray, no data. NA concentrations (millimolar) are indicated along the bottom. The five major gene expression classes, with some representative loci, and the Bvg-independent NA-induced class are indicated on the right. Purple arrowhead, bipA. The set of genes represented by the gray bar on the right passed filtering criteria but showed no discernible pattern.
The earliest Bvg-activated genes, expressed at NA concentrations of up to 0.8 mM, included bvgAS, fhaB and fimA (members of the same operon), and a putative adhesin gene (BB0110). The cya operon, prn, and other fim genes were expressed at up to 0.6 mM NA, while most remaining Bvg-activated genes, including the TTSS locus, were expressed only at NA concentrations of less than or equal to 0.4 mM. These data suggest a trend in which adhesins are expressed early, whereas toxins and the TTSS are expressed later.
The expression pattern of the prototypical intermediate-phase gene bipA, peaking at 0.6 and 0.8 mM NA, was virtually unique, with only a few loci showing a similar profile. However, 69 genes, including the paa locus and part of the trp locus, were maximally expressed at 0.4 to 0.6 mM NA (Fig. 3, Bvgi-a), and another class of 193 genes displayed maximum expression at 0.8 and 1.2 mM NA (Fig. 3, Bvgi-b). Though conceptually similar, the expression peaks for each of these two distinct intermediate regulatory classes occurred at nonoverlapping concentrations of NA. We refer to these Bvg-regulatory phases and their corresponding gene classes as “Bvgi-a” and “Bvgi-b” to distinguish them from the specific point on the Bvg spectrum represented by the Bvgi phase-locked mutation.
(ii) B. pertussis.
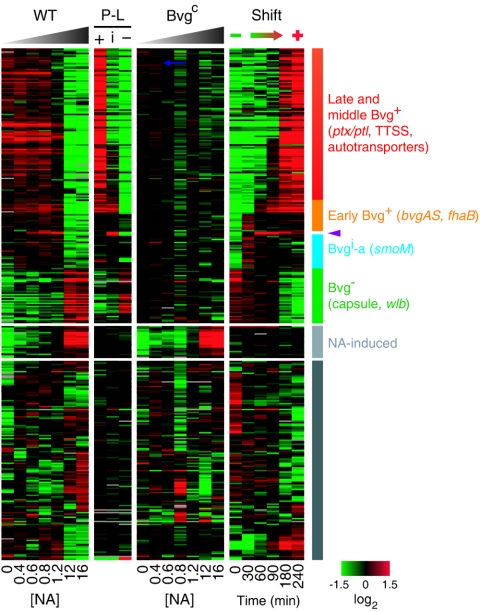
In B. pertussis GMT-1, the Bvg regulatory continuum was assessed both by steady-state NA modulation and by shifting from the Bvg− to the Bvg+ phase following withdrawal of magnesium sulfate from the culture medium. Both treatments resulted in a broad spectrum of expression states that were separable into four dominant gene classes (Fig. 4).
FIG. 4.
Patterns of Bvg-regulated gene expression in B. pertussis GMT-1. Data are centered within each experiment. Results obtained with NA modulation and phase-locked strain analysis are shown from Bvg+ (left) to Bvg− (right), whereas the phase shift experiment is depicted from Bvg− (left) to Bvg+ (right). Rows correspond to array probes, and columns correspond to experiments. +, phase-locked Bvg+; i, phase-locked Bvgi; −, phase-locked Bvg−. Red, increased transcript abundance; green, decreased transcript abundance; black, no significant change in transcript abundance; gray, no data. NA concentrations (millimolar) and time elapsed after shift (minutes) are indicated along the bottom. The four major gene expression classes, with some representative loci, and the Bvg-independent NA-induced class are indicated on the right. Purple arrowhead, bipA. The set of genes represented by the gray bar on the right passed filtering criteria but showed no discernible pattern.
The Bvg+ phase gene expression patterns largely paralleled those of RB50, with a few notable differences. As in B. bronchiseptica, regulatory (bvgAS and bvgR) and adhesin (fhaB and prn) genes were expressed earlier than the TTSS, autotransporter, and toxin (cya and ptx) genes. Modulation of GMT-1 yielded only a small number of Bvg-repressed genes, expression of which was turned off 90 min after shift or in the presence of 1.2 mM or less NA. These included wlbD and the capsule polysaccharide locus.
A few genes were maximally expressed between the Bvg+ and Bvg− phases, but only a putative thiolase gene (BP0422) showed a bipA-like expression pattern. Expression of 33 genes peaked at 30 min postshift and increased slightly in the range of 0.4 to 1.2 mM NA (Fig. 4, Bvgi-a). The Bvg intermediate gene set in GMT-1 was much less extensive than that in RB50 and, except for bipA and BP0422, showed little overlap with the set in RB50.
Regulation of metabolic pathways by BvgAS.
In B. bronchiseptica, genes related to carbon metabolism and transport functions are dynamically regulated across the Bvg continuum. Bvg− phase transcript profiles suggested increased activity of the tricarboxylic acid (TCA) cycle. Upregulation of C4-dicarboxylate, tricarboxylate, and branched-chain amino acid transporters might indicate increased concentrations of TCA intermediates and amino acids that can be degraded to acetyl coenzyme A. The rate of conversion of glycine to C1 units, used to build coenzyme A, may be increased by expression of the glycine cleavage system (gcvTHP). Increased expression of arginase (BB0805) may accelerate the urea cycle, yielding fumarate that can be converted to malate by another Bvg-repressed gene, fumarate hydratase (BB4081). A concordant increase in urea production may explain the observed increase in urease gene expression.
Bvg+ phase transcript profiles indicated a shift in carbon source preference: increased abundance of glutamine transporter (glnHPQ) and glutamate decarboxylase gene transcripts suggests uptake of glutamine and conversion to glutamate, which may be metabolized to TCA cycle reactants. Lactate and fatty acids appear to be preferred alternative carbon sources, as both l-lactate dehydrogenase and enoyl coenzyme A hydratase genes were upregulated. A shift in nitrogen source preference was also apparent. Expression of glnHPQ, together with the upregulation of an ammonia transporter gene (amtB) suggest that the organism is optimized for uptake of glutamine and ammonia in the Bvg+ phase. Ammonia is a readily available source of nitrogen at the airway epithelial surface (16). In addition to a role in nutrition, glnHPQ upregulation might also affect virulence, as glnQ mutant group B streptococci are deficient in both epithelial cell binding and infection of an animal model (55).
Regulation by BvgAS of genes encoding respiratory electron transport chain components was evident. In the Bvg+ phase, gene transcripts for NADH dehydrogenase, l-lactate dehydrogenase, and a putative cytochrome c protein were more abundant, while those for components of the cytochrome bc1 complex, two putative cytochromes, and two ferredoxins were preferentially expressed in the Bvg− phase. In the Bvgi-a phase, genes that encode components of cytochrome bd and cytochrome cbb3 were maximally expressed. Both of these terminal oxidases have very high affinity for oxygen and are maximally transcribed under microaerophilic conditions in other bacteria (see e.g., reference 56). Other electron carriers and cytochrome biosynthetic proteins, as well as a cytochrome-containing periplasmic nitrate reductase (napDABC) implicated in adaptation to anaerobic environments (51), were also preferentially expressed in the Bvgi-a phase.
Regulation of metabolic and transport functions by BvgAS was much less apparent in B. pertussis. The only such components of the B. pertussis Bvg+ phase transcript pattern shared with B. bronchiseptica were the amtB ammonium transporter gene and the heme biosynthesis locus. Nutrient transporter operons and cytochrome complex genes were not Bvg regulated in GMT-1. These results suggested that the physiological state of B. pertussis is not appreciably regulated by BvgAS and may help to explain the inability of B. pertussis to survive in nutrient-limited environments.
Intraspecies variation in BvgAS regulation.
Differences in Bvg-regulated gene expression between Bordetella species prompted the examination of differential regulation among isolates of the same species. Different regulatory patterns within a species might reflect ongoing microevolution and could cast light upon mechanisms of host adaptation. Diversification of gene expression profiles may also influence pathogenicity, as has been recently suggested for Mycobacterium tuberculosis (18).
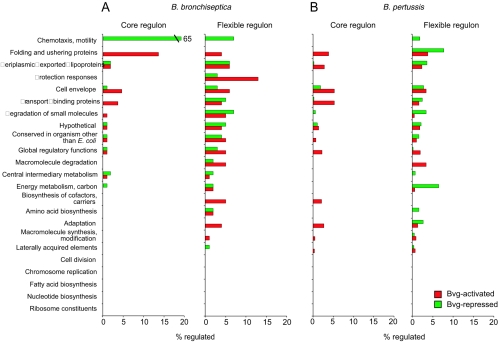
Bvg-regulated gene expression in RB50 (isolated from a rabbit) was compared to that in phylogenetically distant strains isolated from a human (Bbr77) and from a dog (Bbr81), which were grown in the presence or absence of NA to induce the Bvg− and Bvg+ phases, respectively. Comparison of Bvg-regulated genes revealed a surprising amount of diversity among the three isolates (Fig. 1B; see Fig. S2 in the supplemental material). Of 268 genes that were Bvg activated in at least one strain, only 74 were similarly regulated in all three strains. Several genes that are predicted to encode factors involved in protein folding and ushering (dsbA, dsbG, and BB3803) and bfrD, encoding a putative TonB-dependent iron receptor, were among these genes (Fig. 5A). These also included most known and suspected virulence factors, including seven autotransporters and the TTSS, as well as BB0110, encoding a putative adhesin, and BB3242, encoding a putative toxin. Notable exceptions were fhaS, fimX, and BB0450, which were activated to a lesser extent in Bbr81, and dnt, which is missing from the Bbr77 genome and does not appear to be regulated in Bbr81. The strain-specific Bvg-activated genes were rather evenly distributed among most nonessential functional classes (Fig. 5A) and included risA (only in RB50) and bipA and ompQ (only in RB50 and Bbr77).
FIG. 5.
Functional categorization of ORFs in core and flexible BvgAS regulons. Bvg-regulated genes (red, Bvg activated; green, Bvg repressed) were grouped by functional categories (43, 50). Data are expressed as the percentage that is regulated by BvgAS among all annotated ORFs in each class. (A) B. bronchiseptica. ORFs in the core regulon were Bvg regulated by strains RB50, Bbr77, and Bbr81. Those in the flexible regulon were Bvg regulated in either one or two of the three strains. (B) B. pertussis. ORFs in the core regulon were Bvg regulated by strains GMT-1 and Tohama I. Those in the flexible regulon were Bvg regulated in only one of the two strains.
Of 254 genes that were Bvg repressed in at least one strain, only 73 were similarly regulated in all three strains. These included chemotaxis, motility, LPS biosynthesis, sulfate utilization, and carbon metabolism loci (Fig. 5A). Strain-specific Bvg-repressed genes were distributed over many functional classes, with substantial diversity in expression of genes related to small-molecule degradation and transport (Fig. 5A). Finally, seven genes were Bvg activated in one strain and Bvg repressed in another. In most cases, the gene was not Bvg regulated in the third strain.
In order to investigate B. pertussis strain-specific BvgAS regulation, GMT-1, a recent clinical strain, was compared to Tohama I, a strain isolated before widespread pertussis vaccination and subsequently maintained as a laboratory strain. Bvg+ and Bvg− phase-locked derivatives of Tohama I were analyzed as described for GMT-1. Many differences between strains were observed, particularly in the Bvg− phase, in which only 23 of the 113 genes repressed in Tohama I were also repressed in GMT-1 (Fig. 1C; see Fig. S4 in the supplemental material). The conserved Bvg-repressed genes mainly belonged to cell envelope, small-molecule degradation, and hypothetical categories (Fig. 5B). Among the strain-specific Bvg-repressed genes were the previously described but functionally uncharacterized vrg6, vrg18, vrg24, and vrg73 (31), which were Bvg repressed only in Tohama I. Another notable difference between strains was a more extensive and complete Bvg repression of the capsule locus in Tohama I.
Sixty percent of the total B. pertussis Bvg-activated genes were similarly regulated in the two B. pertussis strains (Fig. 1C). Like in B. bronchiseptica, the predominant functional classes among this group were folding and ushering, cell envelope, and transport (Fig. 5B). Most known virulence factors and autotransporters that are present in the B. pertussis genome, including the TTSS, were Bvg activated by both strains, as was BB3242, encoding a putative toxin. Interestingly, fimX, fhaS, and dnt were Bvg activated only in Tohama I. fim3 was Bvg activated in GMT-1 but Bvg repressed in Tohama I, as previously described (24).
Mechanisms of differential transcriptional regulation. (i) Comparative analysis of putative transcriptional regulatory sequences.
Species- or strain-specific differences in Bvg regulation of a given locus have at least three plausible explanations, with the trivial one being the absence of the gene from one of the genomes. Differential expression of genes present in both genomes can be attributed to either sequence divergence in cis-regulatory regions or variation in the levels, activity, or encoding of transcriptional regulatory proteins.
Among 61 loci that were differentially regulated between RB50 and Tohama I, 41 were present in both genomes. Five of the loci that were Bvg activated specifically in RB50 had either an IS481 element or a prophage near the 5′ end of the ORF in Tohama I, suggesting that the cis-regulatory domains have been disrupted or rearranged in Tohama I (see Table S7 in the supplemental material). At two loci, fim3 and BB2270/BP1793, large gaps distinguished the B. bronchiseptica and B. pertussis upstream regions, implicating cis-regulatory variation as a likely determinant of differential regulation. Twenty-five of the upstream intergenic regions differed only by single-nucleotide gaps or mismatches, with nucleotide identities ranging from 93.7% to 99.5%. The remaining eight putative promoter regions were identical in the two genomes. For identical upstream regions and, most likely, those with a small number of mismatches, differential regulation can probably be attributed to variation in expression or activity of a transcriptional regulator.
(ii) Bvg-regulated transcription factors.
Not all Bvg-regulated genes are directly regulated by the binding of BvgA to their promoters (9). Bvg-regulated expression of downstream regulators may lead to fine-tuning of the virulence program, for example, by influencing timing of gene expression or integrating environmental signals.
Genes for the known downstream regulators, BvgR (39) and BtrS (36), and 10 other putative transcriptional regulators were found to be Bvg activated in both B. bronchiseptica RB50 and B. pertussis Tohama I (see Table S8 in the supplemental material). Genes for 3 putative regulators were Bvg activated specifically in Tohama I, while those for 15 regulators, 8 of which are absent or interrupted in Tohama I, were Bvg activated only in RB50. Eighteen RB50 genes for transcriptional regulators, 11 of which are absent or interrupted in Tohama I, were Bvg repressed, while only 1 was Bvg repressed in Tohama I. As many as 11 transcriptional regulator genes, 7 of which are deleted or disrupted in the B. pertussis genome, were maximally expressed in B. bronchiseptica in the presence of intermediate NA concentrations, of which only btr was regulated in B. pertussis. Comparison of the sequences upstream of the differentially expressed regulatory genes that are intact in both species revealed perfect conservation at four loci and a minimum of 97% identity for the others (see Table S7 in the supplemental material), suggesting that divergence in cis-regulatory motifs is not likely to be solely responsible for the observed differences in expression. We propose that the species-specific differences in Bvg-regulated global transcriptional profiles are largely due to the deletion and inactivation in Tohama I of more than two dozen transcriptional regulators that are Bvg regulated in RB50.
Differential expression of transcriptional regulators between strains of the same species was also observed. Genes for seven putative regulatory proteins that were Bvg activated in either Tohama I or GMT-1 were differentially regulated between the two isolates (see Table S8 in the supplemental material). Among B. bronchiseptica isolates, 22 likely transcriptional regulatory protein genes that were Bvg activated, and 13 that were Bvg repressed, were differentially regulated. Thus, transcriptional regulation of downstream regulators by BvgAS can vary dramatically from strain to strain, which may profoundly affect global transcriptional profiles and the phenotypes conferred.
(iii) Identification of putative regulatory motifs.
Although the binding of BvgA to a few virulence factor gene promoters has been well characterized, little is known about regulatory control of most Bvg-regulated genes. To identify putative cis-regulatory sequences, the upstream regions of Bvg-regulated transcription units were searched for common motifs by using the pattern detection algorithm MEME (3). Motifs that were significantly enriched among an expression class were identified by comparing the distributions of motif scores in the upstream regions of Bvg-regulated versus Bvg-unregulated genes.
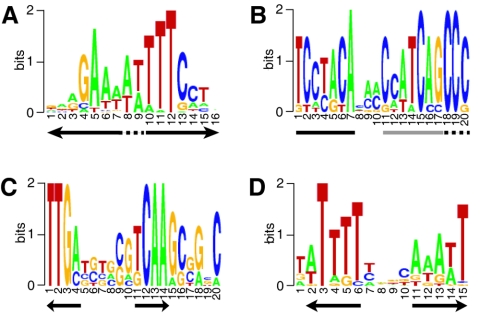
Some regions upstream of Bvg-activated and Bvg-repressed genes were enriched for motifs reminiscent of the traditional BvgA-binding consensus site (TTT[C/G]NTA) and suggestive of an inverted repeat. A weighted statistical model constructed from high-scoring putative BvgA half-sites identified the experimentally confirmed high-affinity BvgA-binding sites in the promoters of bvgA, fhaB, cyaA, and bipA (4, 15, 27, 28) (see Table S9 in the supplemental material). These sites consisted of a high-scoring heptad adjacent to a lower scoring inverted heptad (Fig. 6A). A putative binding motif was found upstream of prn and 11 other Bvg-activated RB50 genes and of 6 other Bvg-activated Tohama I genes at an average distance of 168 nt from the predicted start codon. The majority of the putative BvgA-binding motifs identified by this approach did not contain substitutions in their high-scoring heptads that have been shown to abolish in vivo activity of the fhaB promoter (6). However, several of the motifs did harbor such substitutions (e.g., A, T, or G instead of C in position 4), suggesting that these motifs may not actually represent functional BvgA-binding sites (see Table S9 in the supplemental material).
FIG. 6.
Putative cis-regulatory motifs. Multiple alignments are represented as sequence logos. x axis, nucleotide position within motif; y axis, bit score. Arrows indicate putative half-sites. (A) BvgA-binding motif from regions upstream of Bvg-activated genes. Dashed line, variable-length (0- to 2-nt) spacer between half-sites. (B) Fim box. Solid lines, box 1 and box 2; dashed line, beginning of poly(C) tract. (C and D) FNR box (C) and palindromic motif (D) from regions upstream of Bvgi-b genes.
In the upstream regions of genes that were Bvg activated at early time points, the half-sites of the predicted BvgA-binding site were adjacent, while upstream of late Bvg-activated genes, the half-sites tended to be separated by one or two bases, suggesting that extra bases between the half-sites may reduce affinity for BvgA dimers and lead to delayed transcriptional activation. The BvgA motif model also identified the high-affinity binding site in the bipA promoter and putative sites upstream of nine other RB50 Bvgi-a and Bvgi-b phase genes.
Putative BvgA-binding sites were also found upstream of Bvg-repressed genes (see Table S9 in the supplemental material). However, sites from Bvg+ and Bvg− phase genes were indistinguishable (data not shown) and distances to the predicted start codons were similar, suggesting that the putative BvgA-binding site position relative to the core promoter may not determine activation versus repression. We speculate that BvgA may act as a transcriptional repressor, even when bound far upstream of the core promoter, implying a different mechanism than the RNA polymerase interference proposed for repression of bipA (15, 62).
Surprisingly, only 11% of the Bvg+ and Bvg− phase gene upstream regions possessed discernible high-affinity BvgA-binding sites as defined by the motif model, suggesting that the actual BvgA-binding motif may be highly degenerate, an interpretation supported by the failure of this model to identify a BvgA-binding site in the bvgR and ptx promoters, both of which bind BvgA (5, 38). Alternatively, expression of many Bvg-regulated genes may be governed by downstream transcriptional regulators.
A motif corresponding to the Fim box (48) (Fig. 6B) was found upstream of all fimbrial major subunit genes except fimA, as well as four other RB50 Bvg-activated genes (fhaS, tcfA, the glutamate decarboxylase gene, and an autotransporter gene) and two Tohama I Bvg-activated genes (vag8 and bapC), suggesting that these confirmed and likely virulence factors may be coregulated with fimbriae. Although fim3 is Bvg repressed in Tohama I, the promoter has an intact Fim box. This motif was also found upstream of three RB50 Bvg-repressed genes as well as the Tohama I Bvg-repressed capsular polysaccharide locus promoter, suggesting that its cognate binding factor may promote transcriptional repression as well as activation.
Among the 25 RB50 Bvgi-a phase operons, a single motif was present in seven upstream regions, including those of several respiratory complex loci (Fig. 6C). This highly conserved, palindromic motif corresponds to the FNR box, the binding site for Fnr, a highly conserved transcriptional regulator that controls respiration pathways under low-oxygen conditions in Escherichia coli and a number of other bacterial species (52). The Bordetella homologue of Fnr, Btr, was coexpressed with its putative target genes, suggesting that Btr may activate these genes.
Among the promoters of RB50 loci maximally expressed at 0.8 to 1.2 mM NA, three palindromic motifs were identified, together accounting for 15% of the genes in this class. One of these motifs (Fig. 6D) has the length and spacing typically observed for LysR family transcription factor-binding sites, suggesting that it might be the recognition site for one of the several LysR family proteins maximally expressed in this phase.
Overall, putative regulatory motifs that were significantly overrepresented among the Bvg-regulated gene class were identified upstream of only one-third of the Bvg-regulated genes. This relatively low percentage of Bvg-regulated loci may reflect degeneracy of recognition sites, cooperativity between multiple poorly conserved sites, or the existence of many small subregulons, each with a unique regulator. Identifying the cis-regulatory sequences of the remaining loci will require further exploration, and validation of the sites described here will require direct experimentation.
DISCUSSION
Differentiation of Bvg-regulated gene expression.
Comparative global expression analysis revealed unexpected complexity of the Bordetella BvgAS regulon and insights into possible evolutionary paths for virulence regulation among strains and species of an important group of bacterial pathogens. In particular, B. bronchiseptica, a pathogen and persistent commensal in a broad set of animal species, displayed a wide variety of gene expression profiles in vitro. These profiles may reflect spatially and/or temporally defined patterns during the infectious cycle. The spatial model proposes that colonization of different airway niches and the process of transmission require overlapping yet distinct sets of gene products, the expression of which is regulated by BvgAS in response to environmental conditions characteristic of those anatomic locations. Alternatively, or in addition, BvgAS may regulate a gene expression program in a temporally defined manner, facilitating the establishment of infection (49). The steady-state modulation and phase shift experiments described in this study can be considered in vitro counterparts of the spatial and temporal models, respectively. Although these models are not necessarily mutually exclusive, expression of metabolic genes, especially respiratory chain components, suggests that the phases are tuned to different environmental niches, favoring spatially defined regulation.
Patterns of Bvg-activated gene expression in B. bronchiseptica and B. pertussis were similar, emphasizing that the Bvg+ phase serves similar functions in their respective life cycles. The differentially expressed Bvg-activated genes identified here are candidate determinants of Bordetella host specificity. For example, B. bronchiseptica-specific expression of the putative adhesin BB0110 and several fim genes could contribute to its wider host range compared to B. pertussis.
In contrast, the Bvg− phases of the two species were remarkably different. In B. bronchiseptica, the multitude of Bvg-repressed metabolic, transport, motility, and chemotaxis loci suggests that this phase is optimized for nutrient scavenging and survival ex vivo. However, these gene classes have been shown to be important for in vivo survival in other systems (see, e.g., references 14 and 26), so a role for the Bvg− phase within the host cannot be ruled out. The number and diversity of B. pertussis Bvg− phase genes were limited, reflecting either a different function for the Bvg− phase or the loss of expression, subsequent to host restriction, of loci that facilitate survival ex vivo. The near-complete discordance of Bvg− phase expression patterns of B. pertussis strains GMT-1 and Tohama I supports the latter scenario, as lack of selective pressure is likely to result in random loss and heterogeneity of Bvg− phase factors. These data support the suggestion that the B. pertussis Bvg− phase is an evolutionary remnant (9, 40). Of note, B. bronchiseptica strains also differed in Bvg− phase expression patterns, but to a lesser extent than B. pertussis, possibly reflecting a preference of these strains for different ex vivo niches or redundancy in the Bvg-repressed gene repertoire or, alternatively, suggesting partial random degradation of the Bvg− phase in this species as well.
This is the most extensive comparative analysis of the Bordetella transitional phases along the Bvg regulatory continuum. Prior to this study, with the exception of a few genes (see, e.g., references 15, 46, and 49), expression patterns between the Bvg+ and Bvg− phases had not been determined. Only a single intermediate-phase gene, bipA, had been identified, and the Bvgi phases of B. pertussis and B. bronchiseptica were thought to be fairly similar. In our study, features of the intermediate Bvg phases were among the more unexpected findings. In B. bronchiseptica RB50 there are at least two distinct gene expression states between the Bvg+ and Bvg− poles, comprising 262 genes. These genes may facilitate survival in different environmental niches through activation of specific metabolic and transport pathways. An even higher-resolution examination of the Bvg regulation spectrum may reveal further distinct phenotypic phases. The ability of BvgAS to transduce environmental signals into a series of discrete gene expression states could also be a feature of other regulatory systems. However, because gene regulation studies have historically focused on the endpoints of the spectrum, this has, as yet, remained unrecognized. We propose that examination of transcriptional profiles of other regulatory systems at intermediate levels of environmental signals may reveal regulatory spectra analogous to that described here for BvgAS.
Because intermediate-phase gene expression patterns were found to differ significantly between the two species, these phases may provide species-specific functions in addition to the putative roles in aerosol transmission and biofilm formation (25, 60). Preferential expression in the Bvgi-a phase of B. bronchiseptica of genes for respiratory terminal oxidases with high oxygen affinity suggests that this phase may be optimized for survival in microaerophilic conditions, which may be encountered at the airway epithelium due to the accumulation of mucus and debris. B. pertussis intermediate-phase expression patterns did not group into discrete classes as in B. bronchiseptica, nor did they suggest an obvious physiological role, suggesting that this phase may be an evolutionary remnant in B. pertussis.
Exploring the flexible regulon.
This study joins a growing number of reports (see, e.g., references 18, 19, and 61) in demonstrating that global transcript or protein profiles among strains of the same species can be highly variable. Consequently, gene expression data from a single strain may not reflect the prevailing expression profile of that species. For example, the most frequently studied B. pertussis Bvg-repressed genes were identified in the B. pertussis type strain 18-323 (31), yet the present data did not reveal Bvg repression of any of these loci in GMT-1, raising the question of whether Bvg-regulated expression of these genes is a feature of modern clinical isolates.
Among strains of a single microbial species, genomic content can vary substantially (32). Genes that are present in all isolates (i.e., the “core genome”) are assumed to be phylogenetically conserved, while those that are variably present (i.e., the “flexible genome”) are proposed to be horizontally acquired or differentially lost within the species. The components of the flexible genome are thought to confer differences in phenotypes such as virulence and host range. We favor an analogous distinction for the description of gene regulation patterns found in related strains in response to a stimulus and propose that a global gene expression profile comprises a “core regulon” that is shared among all strains and a “flexible regulon” that varies between strains. Regulated expression of each gene in the core regulon is presumed to be required by every isolate of a species. For a pathogen grown under conditions that promote virulence, the core regulon includes factors that are required for efficient infection and transmission. Therefore, components of the core regulon are the best candidate vaccine or therapeutic targets. On the other hand, regulated expression of the genes in the flexible regulon is not required by every strain, and variation in the expression of these genes is tolerated within the population. In a pathogen population, the flexible regulon could include determinants of host specificity, thus facilitating its spread into new hosts or, conversely, its restriction to a single host. More generally, the flexible regulon could reflect adaptation to specific habitats by subpopulations. We believe that differentially expressed loci are an informative class of Bvg-regulated genes and may prove critical in our understanding of evolution and diversification of host range specificity between and within Bordetella species.
One important determinant of the flexible regulon is the collection of downstream transcriptional regulators that are themselves regulated, directly or indirectly, by the core top-level regulators. Because each transcription factor may be capable of modifying the expression patterns of many individual loci, a single mutation that changes the concentration or activity of a regulator can have widespread effects on the global expression pattern. Indeed, we observed differential regulation of many putative transcriptional regulators, both within and between Bordetella species. This variability in regulatory protein repertoires could represent an efficient and powerful way to generate phenotypic diversity during evolution and may be one of the predominant mechanisms of differential gene expression in the bordetellae.
Supplementary Material
Acknowledgments
We thank Mary Brinig and Mari Nakamura for microarray production and critical review of the manuscript, Sin-Yee Liew for technical assistance, Peggy Cotter (University of California, Santa Barbara) for strains, and Peter Karp (SRI International) for providing Bordetella pathway genome databases before publication.
C.A.C. was supported by Research Training Fellowships from the American Lung Association and ALA-California. H.J.B. was funded (in part) by a TALENT fellowship from The Netherlands Organization for Scientific Research (NWO). J.F.M. was supported by NIH/NIAID grants AI38417 and AI061598. D.A.R. was supported by NIH/NIAID grants AI54970 and AI057188.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Akerley, B. J., P. A. Cotter, and J. F. Miller. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80:611-620. [DOI] [PubMed] [Google Scholar]
- 2.Arico, B., and R. Rappuoli. 1987. Bordetella parapertussis and Bordetella bronchiseptica contain transcriptionally silent pertussis toxin genes. J. Bacteriol. 169:2847-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, p. 28-36, In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 4.Boucher, P. E., K. Murakami, A. Ishihama, and S. Stibitz. 1997. Nature of DNA binding and RNA polymerase interaction of the Bordetella pertussis BvgA transcriptional activator at the fha promoter. J. Bacteriol. 179:1755-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boucher, P. E., and S. Stibitz. 1995. Synergistic binding of RNA polymerase and BvgA phosphate to the pertussis toxin promoter of Bordetella pertussis. J. Bacteriol. 177:6486-6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boucher, P. E., M. S. Yang, and S. Stibitz. 2001. Mutational analysis of the high-affinity BvgA binding site in the fha promoter of Bordetella pertussis. Mol. Microbiol. 40:991-999. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2002. Pertussis—United States, 1997-2000. Morb. Mortal. Wkly. Rep. 51:73-76. [PubMed] [Google Scholar]
- 8.Cotter, P. A., and A. M. Jones. 2003. Phosphorelay control of virulence gene expression in Bordetella. Trends Microbiol. 11:367-373. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, P. A., and J. F. Miller. 2001. Bordetella, p. 620-674. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, Calif.
- 10.Cotter, P. A., and J. F. Miller. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62:3381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotter, P. A., and J. F. Miller. 1997. A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol. Microbiol. 24:671-685. [DOI] [PubMed] [Google Scholar]
- 12.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cummings, C. A., M. M. Brinig, P. W. Lepp, S. van de Pas, and D. A. Relman. 2004. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J. Bacteriol. 186:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73:241-268. [DOI] [PubMed] [Google Scholar]
- 15.Deora, R., H. J. Bootsma, J. F. Miller, and P. A. Cotter. 2001. Diversity in the Bordetella virulence regulon: transcriptional control of a Bvg-intermediate phase gene. Mol. Microbiol. 40:669-683. [DOI] [PubMed] [Google Scholar]
- 16.Dwyer, T. M. 2004. Sampling airway surface liquid: non-volatiles in the exhaled breath condensate. Lung 182:241-250. [DOI] [PubMed] [Google Scholar]
- 17.Fuchslocher, B., L. L. Millar, and P. A. Cotter. 2003. Comparison of bipA alleles within and across Bordetella species. Infect. Immun. 71:3043-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, Q., K. E. Kripke, A. J. Saldanha, W. Yan, S. Holmes, and P. M. Small. 2005. Gene expression diversity among Mycobacterium tuberculosis clinical isolates. Microbiology 151:5-14. [DOI] [PubMed] [Google Scholar]
- 19.Gaynor, E. C., S. Cawthraw, G. Manning, J. K. MacKichan, S. Falkow, and D. G. Newell. 2004. The genome-sequenced variant of Campylobacter jejuni NCTC 11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J. Bacteriol. 186:503-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerlach, G., F. von Wintzingerode, B. Middendorf, and R. Gross. 2001. Evolutionary trends in the genus Bordetella. Microbes Infect. 3:61-72. [DOI] [PubMed] [Google Scholar]
- 21.Goodnow, R. A. 1980. Biology of Bordetella bronchiseptica. Microbiol. Rev. 44:722-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heininger, U., K. Stehr, S. Schmitt-Grohe, C. Lorenz, R. Rost, P. D. Christenson, M. Uberall, and J. D. Cherry. 1994. Clinical characteristics of illness caused by Bordetella parapertussis compared with illness caused by Bordetella pertussis. Pediatr. Infect. Dis. J. 13:306-309. [DOI] [PubMed] [Google Scholar]
- 23.Herrero, J., A. Valencia, and J. Dopazo. 2001. A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics 17:126-136. [DOI] [PubMed] [Google Scholar]
- 24.Hot, D., R. Antoine, G. Renauld-Mongenie, V. Caro, B. Hennuy, E. Levillain, L. Huot, G. Wittmann, D. Poncet, F. Jacob-Dubuisson, C. Guyard, F. Rimlinger, L. Aujame, E. Godfroid, N. Guiso, M. J. Quentin-Millet, Y. Lemoine, and C. Locht. 2003. Differential modulation of Bordetella pertussis virulence genes as evidenced by DNA microarray analysis. Mol. Genet. Genomics 269:475-486. [DOI] [PubMed] [Google Scholar]
- 25.Irie, Y., S. Mattoo, and M. H. Yuk. 2004. The Bvg virulence control system regulates biofilm formation in Bordetella bronchiseptica. J. Bacteriol. 186:5692-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Josenhans, C., and S. Suerbaum. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605-614. [DOI] [PubMed] [Google Scholar]
- 27.Karimova, G., J. Bellalou, and A. Ullmann. 1996. Phosphorylation-dependent binding of BvgA to the upstream region of the cyaA gene of Bordetella pertussis. Mol. Microbiol. 20:489-496. [DOI] [PubMed] [Google Scholar]
- 28.Karimova, G., and A. Ullmann. 1997. Characterization of DNA binding sites for the BvgA protein of Bordetella pertussis. J. Bacteriol. 179:3790-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karp, P. D., S. Paley, and P. Romero. 2002. The Pathway Tools software. Bioinformatics 18(Suppl. 1):S225-S232. [DOI] [PubMed] [Google Scholar]
- 30.Kasuga, T., Y. Nakase, K. Ukishima, and K. Takatsu. 1954. Studies on Haemophilus pertussis. V. Relation between the phase of bacilli and the progress of the whooping-cough. Kitasato Arch. Exp. Med. 27:57-62. [PubMed] [Google Scholar]
- 31.Knapp, S., and J. J. Mekalanos. 1988. Two trans-acting regulatory genes (vir and mod) control antigenic modulation in Bordetella pertussis. J. Bacteriol. 170:5059-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan, R., and P. R. Reeves. 2000. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 8:396-401. [DOI] [PubMed] [Google Scholar]
- 33.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 34.Martinez de Tejada, G., P. A. Cotter, U. Heininger, A. Camilli, B. J. Akerley, J. J. Mekalanos, and J. F. Miller. 1998. Neither the Bvg− phase nor the vrg6 locus of Bordetella pertussis is required for respiratory infection in mice. Infect. Immun. 66:2762-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez de Tejada, G., J. F. Miller, and P. A. Cotter. 1996. Comparative analysis of the virulence control systems of Bordetella pertussis and Bordetella bronchiseptica. Mol. Microbiol. 22:895-908. [DOI] [PubMed] [Google Scholar]
- 36.Mattoo, S., M. H. Yuk, L. L. Huang, and J. F. Miller. 2004. Regulation of type III secretion in Bordetella. Mol. Microbiol. 52:1201-1214. [DOI] [PubMed] [Google Scholar]
- 37.Melton, A. R., and A. A. Weiss. 1989. Environmental regulation of expression of virulence determinants in Bordetella pertussis. J. Bacteriol. 171:6206-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merkel, T. J., P. E. Boucher, S. Stibitz, and V. K. Grippe. 2003. Analysis of bvgR expression in Bordetella pertussis. J. Bacteriol. 185:6902-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merkel, T. J., and S. Stibitz. 1995. Identification of a locus required for the regulation of bvg-repressed genes in Bordetella pertussis. J. Bacteriol. 177:2727-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merkel, T. J., S. Stibitz, J. M. Keith, M. Leef, and R. Shahin. 1998. Contribution of regulation by the bvg locus to respiratory infection of mice by Bordetella pertussis. Infect. Immun. 66:4367-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, J. F., S. A. Johnson, W. J. Black, D. T. Beattie, J. J. Mekalanos, and S. Falkow. 1992. Constitutive sensory transduction mutations in the Bordetella pertussis bvgS gene. J. Bacteriol. 174:970-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mooi, F. R., I. H. van Loo, and A. J. King. 2001. Adaptation of Bordetella pertussis to vaccination: a cause for its reemergence? Emerg. Infect. Dis. 7:526-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. G. Holden, C. R. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeño-Tárraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, R. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 44.Porter, J. F., R. Parton, and A. C. Wardlaw. 1991. Growth and survival of Bordetella bronchiseptica in natural waters and in buffered saline without added nutrients. Appl. Environ. Microbiol. 57:1202-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Preston, A., E. Maxim, E. Toland, E. J. Pishko, E. T. Harvill, M. Caroff, and D. J. Maskell. 2003. Bordetella bronchiseptica PagP is a Bvg-regulated lipid A palmitoyl transferase that is required for persistent colonization of the mouse respiratory tract. Mol. Microbiol. 48:725-736. [DOI] [PubMed] [Google Scholar]
- 46.Prugnola, A., B. Arico, R. Manetti, R. Rappuoli, and V. Scarlato. 1995. Response of the bvg regulon of Bordetella pertussis to different temperatures and short-term temperature shifts. Microbiology 141:2529-2534. [DOI] [PubMed] [Google Scholar]
- 47.Ren, Q., K. H. Kang, and I. T. Paulsen. 2004. TransportDB: a relational database of cellular membrane transport systems. Nucleic Acids Res. 32:D284-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riboli, B., P. Pedroni, A. Cuzzoni, G. Grandi, and F. de Ferra. 1991. Expression of Bordetella pertussis fimbrial (fim) genes in Bordetella bronchiseptica: fimX is expressed at a low level and vir-regulated. Microb. Pathog. 10:393-403. [DOI] [PubMed] [Google Scholar]
- 49.Scarlato, V., B. Arico, A. Prugnola, and R. Rappuoli. 1991. Sequential activation and environmental regulation of virulence genes in Bordetella pertussis. EMBO J. 10:3971-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serres, M. H., and M. Riley. 2000. MultiFun, a multifunctional classification scheme for Escherichia coli K-12 gene products. Microb. Comp. Genomics 5:205-222. [DOI] [PubMed] [Google Scholar]
- 51.Siddiqui, R. A., U. Warnecke-Eberz, A. Hengsberger, B. Schneider, S. Kostka, and B. Friedrich. 1993. Structure and function of a periplasmic nitrate reductase in Alcaligenes eutrophus H16. J. Bacteriol. 175:5867-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spiro, S. 1994. The FNR family of transcriptional regulators. Antonie Leeuwenhoek 66:23-36. [DOI] [PubMed] [Google Scholar]
- 53.Stockbauer, K. E., B. Fuchslocher, J. F. Miller, and P. A. Cotter. 2001. Identification and characterization of BipA, a Bordetella Bvg-intermediate phase protein. Mol. Microbiol. 39:65-78. [DOI] [PubMed] [Google Scholar]
- 54.Talaat, A. M., S. T. Howard, W. ten Hale, R. Lyons, H. Garner, and S. A. Johnston. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamura, G. S., A. Nittayajarn, and D. L. Schoentag. 2002. A glutamine transport gene, glnQ, is required for fibronectin adherence and virulence of group B streptococci. Infect. Immun. 70:2877-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tseng, C. P., J. Albrecht, and R. P. Gunsalus. 1996. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J. Bacteriol. 178:1094-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van den Akker, W. M. 1998. Lipopolysaccharide expression within the genus Bordetella: influence of temperature and phase variation. Microbiology 144:1527-1535. [DOI] [PubMed] [Google Scholar]
- 59.van der Zee, A., F. Mooi, J. Van Embden, and J. Musser. 1997. Molecular evolution and host adaptation of Bordetella spp.: phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J. Bacteriol. 179:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vergara-Irigaray, N., A. Chavarri-Martinez, J. Rodriguez-Cuesta, J. F. Miller, P. A. Cotter, and G. Martinez de Tejada. 2005. Evaluation of the role of the Bvg intermediate phase in Bordetella pertussis during experimental respiratory infection. Infect. Immun. 73:748-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wehmhoner, D., S. Haussler, B. Tummler, L. Jansch, F. Bredenbruch, J. Wehland, and I. Steinmetz. 2003. Inter- and intraclonal diversity of the Pseudomonas aeruginosa proteome manifests within the secretome. J. Bacteriol. 185:5807-5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams, C. L., P. E. Boucher, S. Stibitz, and P. A. Cotter. 2005. BvgA functions as both an activator and a repressor to control Bvgi phase expression of bipA in Bordetella pertussis. Mol. Microbiol. 56:175-188. [DOI] [PubMed] [Google Scholar]
- 63.Yeh, S. H. 2003. Pertussis: persistent pathogen, imperfect vaccines. Expert Rev. Vaccines 2:113-127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.