Abstract
The antagonistic interaction between a potential fish probiont, Pseudomonas fluorescens strain AH2, and its target organism, Vibrio anguillarum, was investigated by studying the genetic response of the target organism when it was exposed to the antagonist. We compared the differential display of arbitrarily PCR-amplified gene transcripts in V. anguillarum serotype O1 when it was exposed to AH2 supernatant with the display of transcripts in nonexposed control cultures. Growth of V. anguillarum was immediately arrested when the organism was exposed to 50% (vol/vol) AH2 supernatant. A total of 10 potentially differentially expressed transcripts were identified. Among these we identified a gene homologous to rpoS that was induced in a dose-dependent manner when V. anguillarum was cultured in media supplemented with sterile filtered supernatant from AH2. rpoS was also induced when growth was arrested with the iron chelator 2,2-dipyridyl. A chromosomal transcript homologous to vibE that participates in vibriobactin synthesis in Vibrio cholerae was also upregulated during AH2 exposure. This transcript could represent a functionally active gene in V. anguillarum involved in biosynthesis of anguibactin or another V. anguillarum siderophore. On the pJM1 plasmid of V. anguillarum serotype O1, a pseudogene designated open reading frame E (ORF E) that contains a frameshift mutation was previously identified. The gene homologous to vibE identified in this study, interestingly, also has significant homology to ORF E on the amino acid level and does not possess the frameshift mutation. Thus, the chromosomally encoded vibE homologue could fulfil the role of the inactive plasmid-encoded ORF E pseudogene. Addition of Fe3+ to the system eliminated the growth arrest, and the genes homologous to rpoS and vibE were not induced. To our knowledge, this is the first study linking rpoS induction to iron starvation. Taken together, the results of this study suggest that a major part of the antagonistic property exhibited by strain AH2 is caused by the ability of siderophores in the supernatant to efficiently chelate iron, which results in instant iron deprivation of the pathogen V. anguillarum and complete growth arrest.
Pseudomonads are widespread in aquatic and terrestrial environments and are renowned for their ability to inhibit other microorganisms (6, 13, 48). In particular, it has been suggested that the fluorescent pseudomonads could be used as biocontrol agents against plant diseases (14, 37) or as probiotic organisms to prevent fish diseases (23, 50, 52). The boom in the aquaculture industry, which has been growing almost 10% per year for the last decade (19), has made disease control a major issue. In particular, non-antibiotic-based disease control measures, such as probiotics, have become an important research field due to the concerns raised about the use of antibiotics and the development of antibiotic resistance (24).
Vibrio anguillarum is a marine gram-negative bacterium that causes disease in approximately 50 different fish species (3). Vibriosis is primarily caused by serotypes O1 and O2 and is a typical hemorrhagic septicemia (3). Addition of fluorescent pseudomonads to fish tank water may reduce or delay fish infections with V. anguillarum (23, 52). Little is, however, known about the mechanisms underlying this in vivo effect. Several traits are believed to be involved in the antimicrobial effect of plant biocontrol pseudomonads. These traits include siderophore-based iron chelation (31, 33) and production of antibiotics, such as 2,4-diacetyl-phloroglucinol (46) and phenazines (40).
Iron chelation is an important property both for environmental, nonharmful microorganisms and for pathogenic microorganisms. Due to redox properties, Fe is mostly bound in insoluble complexes, and the concentration of free, accessible iron in most environments is low. Thus, iron limitation is a major constraint for microbial growth in ocean environments (10), and many aquatic microorganisms produce efficient iron chelators (42). Serotype O1 strains of V. anguillarum produce anguibactin encoded by the virulence plasmid (pJM1) (12) or a siderophore(s) encoded by chromosomal genes (30). The plasmid-encoded iron chelation system is essential for virulence of some V. anguillarum strains (12). Despite the ability of V. anguillarum to chelate iron, it has been suggested that part of the probiotic effect of fluorescent pseudomonads could be explained by their more efficient competition for iron (23). Thus, sterile filtered culture supernatants from the fish probiotic Pseudomonas fluorescens strain AH2 were inhibitory to V. anguillarum, whereas supernatants from strain AH2 cultured with iron were not inhibitory (23). It has previously been demonstrated that P. fluorescens AH2 produces several types of siderophores (2).
Knowledge of the mechanisms involved in the interactions between a probiotic biocontrol strain and a pathogen is important in order to evaluate the stability and risk of the treatment and, potentially, to enhance the reaction or characteristics required to obtain an in vivo effect (46). Such mechanisms have traditionally been studied by random or targeted creation of mutants deficient in the antimicrobial property (7). We hypothesized, however, that studies of mechanisms of an interaction also can be carried out by focusing on the target organism and its gene expression in response to the antagonistic strain.
A number of molecular techniques, including serial analysis of gene expression (53), differential display (32), cDNA-amplified fragment length polymorphism (4), RNA-arbitrarily primed PCR (RAP-PCR) (55), and cDNA microarrays (45), can be employed to study the global gene expression profiles and differential regulation of genes in response to, for example, antagonistic effects. Obviously, genomic DNA chips (DNA microarrays) would allow the full genetic response to be evaluated; however, in the case of V. anguillarum such DNA chips are not available, and the genome remains to be fully sequenced. Hence, for this study another technique was selected based on the differential display of arbitrarily PCR-amplified gene transcripts by using the RAP-PCR protocol originally developed by Welsh et al. (55). This technique is readily adaptable to prokaryotes, unlike some of the other methods that rely on the presence of polyadenylated mRNAs. By identifying genes in cultures of V. anguillarum exposed to strain AH2 supernatant that were differentially expressed (i.e., up- or downregulated) compared to the genes in unexposed exponentially growing V. anguillarum, we were able to reveal novel genes and regulation in V. anguillarum that strongly indicated that iron starvation is indeed an important part of the antagonistic effect of the fish probiotic pseudomonad strain AH2 against V. anguillarum.
MATERIALS AND METHODS
Strains, media, and growth conditions.
A virulent strain of the fish pathogen V. anguillarum (serotype O1; strain 90-11-287) (49) was grown in M9 salts (44) supplemented with 0.4% glucose and 0.3% Casamino Acids (M9GC) at 15°C with agitation (200 rpm). This strain causes a positive chrome-Azurol S (CAS) reaction, indicating production of an iron-binding compound(s). P. fluorescens AH2 (22) was likewise grown in M9GC at 15°C with agitation. Prior to inoculation into M9GC, both strains were routinely streaked onto Luria-Bertani agar.
Siderophore assay.
The presence of iron-chelating substances in sterile filtered supernatant was determined by using the spectrophotometric CAS assay (47). Twofold serial dilutions of supernatants were prepared, and each dilution was mixed with an equal volume of the CAS reagent. Absorbance was measured at 630 nm. Each supernatant was tested twice in separate experiments.
AH2 supernatant challenge.
P. fluorescens strain AH2 was grown in M9GC at 15°C to the stationary phase (2 to 3 days) in up to 200 ml of medium. Cells were harvested by centrifugation at 3,000 × g for 5 min, and the supernatant was decanted, vacuum sterile filtered through a 0.22-μm-pore-size polyethersulfone filter system (500 ml; Corning Incorporated, Corning, N.Y.), and kept at 4°C until it was used. AH2 supernatant challenge experiments were initially conducted by using exponentially growing V. anguillarum cultures (optical density at 600 nm [OD600], 0.6 to 0.8) and transferring 20 ml of a culture to 80 ml of medium containing 50 ml of AH2 supernatant and 30 ml of fresh M9GC. In the control 20 ml of an exponentially growing V. anguillarum culture was added to 80 ml of fresh M9GC. The first experiment in this analysis was conducted exclusively to confirm that scaling up the AH2 supernatant challenge conditions from the microtiter plate format to 200-ml cultures did not change the previously observed growth-inhibiting effect and was done only once. The second AH2 supernatant challenge experiment was a time course experiment performed to evaluate the kinetics of the genetic response, in which a single RAP-PCR profile analysis was performed at different times after challenge (5 and 20 min), and it was also done only once. All subsequent AH2 supernatant challenge experiments conducted to confirm identification of differentially expressed genes and to determine the effect of supplementing the medium with 100 μM FeCl3 and the effect of reducing the volume of the AH2 supernatant to 1/100 of the total volume were done at least twice. A final control experiment, in which we examined the effect of controlled iron chelation in a M9GC V. anguilllarum culture using the iron chelator 2,2-dipyridyl (Sigma, Dorset, United Kingdom) and compared the results to the AH2 supernatant-induced physiological and genetic responses, was done once. In general, growth was measured by using optical density and samples withdrawn for RNA extraction from two cultures after a 20-min challenge. In all experiments in which total RNA was isolated, the number of cells used for extraction was kept constant; i.e., in the case of proliferation in one culture, the sample volume was reduced according to the increase in OD600 to keep the number of cells constant. For cultures in which there was no growth the sample size was always 30 ml. Samples used for RNA isolation were fixed by instant transfer to a dry ice-ethanol bath for freezing. Samples were kept at −80°C until they were used.
RNA extraction.
Culture samples were thawed, and total RNA was extracted from 5 × 108 to 109 cells (corresponding to 30 ml of a culture with an OD600 of approximately 0.15) as previously described (21). The cells were harvested and resuspended in 890 μl of extraction mixture I (80 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 10 mM 2-mercaptoethanol), and this was followed by addition of 1,110 μl of extraction mixture II (0.9% sodium dodecyl sulfate [SDS], 0.41 mg of proteinase K per ml, 18 mM EDTA, 3.6 mM 1,10-phenanthroline, 0.36 mg of heparin per ml) and incubation at 37°C for 20 min and at 65°C for 10 min. One volume of phenol-chloroform-isoamyl alcohol (25:24:1) was added to the extraction mixture, and the resulting aqueous phase was reextracted once with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform. The RNA was precipitated with 0.3 M sodium acetate and 2.5 volumes of 96% ethanol overnight at −20°C, washed once in 70% ethanol, and allowed to dry before resuspension in 50 μl of diethyl pyrocarbonate-treated distilled H2O. Isolated RNA was kept at −80°C until it was used.
RAP-PCR.
RAP-PCR was performed essentially as described by Fleming et al. (18). One microgram of total RNA from exponentially growing V. anguillarum cells or V. anguillarum cells exposed to AH2 supernatant was initially subjected to treatment with 1 U of RNase-free DNase I (Life Technologies, Roskilde, Denmark) for 15 min at 37°C in 10 μl of DNase I buffer (40 mM Tris-HCl [pH 7.9], 10 mM NaCl, 6 mM MgCl2, 10 mM CaCl2) to remove traces of DNA. The DNase was inactivated by adding 1 μl of 25 mM EDTA and heating the sample for 10 min at 65°C before it was placed on ice. Two microliters of the DNase I-treated RNA was used in a 20-μl (total volume) cDNA synthesis reaction mixture comprising 200 μM dATP, 200 μM dCTP, 200 μM dGTP, 200 μM dTTP, 5 mM dithiothreitol, 1× Moloney murine leukemia virus reverse transcriptase (M-MLV-RT) buffer (50 mM Tris-HCl [pH 8.3], 50 mM KCl, 4 mM MgCl2), 0.4 μM reverse primer, and 50 U of M-MLV-RT (Life Technologies). The reactions were performed with a Perkin-Elmer 9600 thermal cycler by using the following procedure: the temperature was ramped from 50 to 30°C over a 15-min period, and this was followed by 1 h of incubation at 37°C and then by 5 min of incubation at 95°C and cooling to 4°C. Three microliters of the reaction mixture was mixed with 27 μl of a mixture to obtain a 30-μl PCR mixture which contained each deoxynucleoside triphosphate at a concentration of 20 μM, 6% dimethyl sulfoxide, 2.5 μCi of [α-33P]dATP (Amersham Biosciences, Hørsholm, Denmark), 0.1% Triton X-100, 2 μM reverse primer, 2 μM forward primer, 1.5 mM MgCl2, 100 mM Tris-HCl (pH 8.3), 50 mM KCl, and 0.3 U of Taq DNA polymerase (Life Technologies). The reactions were performed with a Perkin-Elmer 9600 thermal cycler by using 40 cycles of 94°C for 30 s, 40°C for 1 min, and 72°C for 1 min. The procedure ended with a 10-min hold at 72°C and cooling to 4°C. Parallel negative controls consisted of exactly the same components except for the M-MLV-RT enzyme. Four microliters of each reaction mixture was mixed with 4 μl of loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05 xylene cyanol FF), heated to 94°C for 2 min, and then cooled to 4°C for 5 min. Three microliters of each reaction mixture was loaded on a 6% polyacrylamide-50% urea sequencing gel, and electrophoresis was performed in 1× TBE (90 mM Tris, 90 mM borate, 2 mM EDTA [pH 8.3]) at 55 W for 2.5 h. The gel was dried and exposed to Kodak Biomax MR film overnight.
Primers used in RAP-PCR.
The following primers were selected from a list of short primers optimized for use in prokaryotic differential display techniques by Fislage et al. (17): Ea1 (5′-TTTATCCAGC-3′), Ea2 (5′-ACTTTACGCAG-3′), Ea3 (5′-TTTATCCAGCG-3′), Ea4 (5′-TCAGCGTTTTA-3′), Ea5 (5′-TTTCAGCGCCT-3′), Ea6 (5′-TTTTTTCAGCA-3′), Ea7 (5′-TCTTTTTTACC-3′), Ea8 (5′-ATCATCCAGCA-3′), Ea9 (5′-TTTTACCCAGC-3′), Ea10 (5′-TTCAGCCAGCG-3′), Es3 (5′-GAAGTGCTGG-3′), and Es10 (5′-CTGGAAGAAG-3′). Furthermore, forward (sense) primer SD14 (5′-GGGGAACGACGATG-3′) derived from a comparison of several bacterial mRNA start sites was also included (18, 35). The primers were used in different combinations under each type of conditions to obtain RAP-PCR profiles representing a large fraction of the expressed genes.
Identification of differentially expressed genes.
Differentially expressed genes were detected by side-by-side comparisons of RAP-PCR profiles from V. anguillarum cultures exposed to AH2 supernatant and exponentially growing V. anguillarum cultures. PCR products in a gel that were either present or absent under one of the two conditions were excised from the dried gel with a scalpel. The DNA was eluted by incubation of the filter piece in 50 μl of TE buffer (100 mM Tris-HCl [pH 8], 10 mM EDTA) at 65°C for 2 h. Five microliters of the eluate was used as a template in a reamplification PCR conducted under standard conditions with a mixture containing each deoxynucleoside triphosphate at a concentration of 200 μM, 1.5 mM MgCl2, 2.5 U of Taq DNA polymerase (Life Technologies), and 1× PCR buffer (Life Technologies). Primers were selected depending on the set of primers that generated the profile (e.g., SD14 and Es10) and were each added at a concentration of 1 μM. The reamplification PCR was performed as follows: 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 40°C, and 1 min at 72°C and then 10 min at 72°C and cooling to 4°C. Reamplified cDNA bands were electrophoresed on a 1.5% agarose gel to check for purity. Bands were purified from the gel by using the GFX PCR DNA and a gel band purification kit (Amersham Biosciences). Purified PCR products were cloned in a TA vector-based cloning kit (TOPO TA cloning kit; Invitrogen) used according to the manufacturer's instructions and were sequenced by using a DNA sequencing kit (Amersham Biosciences).
Database searches.
All database searches with identified DNA sequences were done by using WU-BLASTX + BEAUTY against all nonredundant GenBank CDS translations + PDB + SwissProt + PIR + PRF and the Baylor College of Medicine Search Launcer web site (http://searchlauncher.bcm.tmc.edu/seq-search/nucleic_acid-search.html).
PCR-generated probes and radiolabeling.
DNA probes for Northern analyses were generated under standard PCR conditions by using plasmids with the cloned RAP-PCR bands as templates and standard primers [corresponding to M13(-20) (5′-GTAAAACGACGGCCAGT-3′) and M13rev (5′-GGAAACAGCTATGACCATG-3′)] flanking the insertion point of the vector pCR2.1-TOPO. The DNA probe used for Southern blot analysis was generated under standard PCR conditions by using purified genomic DNA from V. anguillarum as the template and primers via1 (5′-TGGGTGAGGAAGCTTACTC-3′) and via2 (5′-CGCAACTGCGTTCACCAAG-3′) designed to amplify a 286-bp region of the A15 RAP-PCR clone. In general, 50 ng of purified PCR product was random primer labeled with Ready-To-Go DNA labeling beads and [α-32P]dCTP (3,000 Ci/mmol; Amersham Biosciences) used according to the manufacturer's instructions. The labeled probes were separated from unincorporated 32P-labeled nucleotides by gel filtration with a NICK column (Amersham Biosciences,), and the efficiency of labeling was subsequently monitored by thin-layer chromatography.
Northern analysis.
Separation of RNA fragments in 1.2% formaldehyde agarose gels and subsequent blotting onto Hybond-N+ nylon membranes (Amersham Biosciences) were done as previously described (20). Prehybridization and hybridization were performed in 0.5 M sodium phosphate (pH 7.2) with 7% SDS at 65°C with PCR-generated probes. Prehybridization was performed for 3 h, and then the prehybridization mixture was replaced by fresh prewarmed (65°C) hybridization mixture with radiolabeled probes added. Hybridization was performed overnight. Washes (three washes, 15 min each) were done by using 20 mM sodium phosphate (pH 7.2) containing 1% SDS at 65°C. The washed membrane filters were subsequently autoradiographed.
Verification of vibE homologue localization in V. anguillarum by Southern analysis.
Purified genomic DNA from V. anguillarum was digested with four different restriction enzymes (EcoRI, NlaIII, SspI, and SacI [New England Biolabs]) and separated by gel electrophoresis in a 2% agarose gel. Digested DNA was transferred to nylon membranes as described by Southern (51). Part of the DNA fragment homologous to vibE (A15) originally identified in the RAP-PCR profile was used as a probe after PCR amplification with primers via1 and via2. The probe was radiolabeled as described above, and prehybridization and hybridization were done in 0.5 M sodium phosphate (pH 7.2) containing 7% SDS at 65°C overnight. The stringency washes consisted of two washes (5 min each) with 2× SSC-0.1% SDS and two washes (15 min each) with 0.1× SSC-0.1% SDS at 65°C (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The membrane was dried, sealed, and autoradiographed.
Nucleotide sequence accession numbers.
The nucleotide sequence data for the 10 identified RAP-PCR clones representing expressed mRNAs from V. anguillarum have been deposited in the EMBL database under accession numbers AJ458368 to AJ458377.
RESULTS
Scaling up the AH2 challenge experiment.
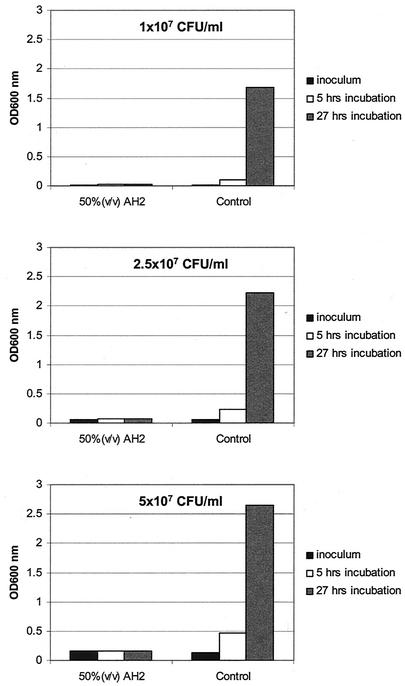
Previous studies of the antagonistic effect of the AH2 supernatant on V. anguillarum were performed in 200-μl formats with microtiter plates by mixing equal volumes of supernatant and fresh substrate (23). Under these conditions the antagonistic effect was unequivocally determined by using an inoculum containing 104 CFU of exponentially growing V. anguillarum per ml. However, larger volumes were needed to conduct RAP-PCR profiling with isolated total RNA, and we therefore determined the effect of scaling up the AH2 challenge experiment. Experiments were conducted with 200-ml mixtures by using three different inocula (107, 2.5 × 107, and 5 × 107 CFU/ml), and the proliferation of the cultures was determined by measuring the optical densities in parallel cultures with and without 50% (vol/vol) AH2 supernatant. Scaling up the challenge experiment clearly did not change the inhibitory effect (Fig. 1) which had been observed in microtiter plates. Growth of V. anguillarum ceased promptly when strain AH2 supernatant was added. The inhibitory effect was observed with all inocula, and subsequent experiments were conducted with an inoculum containing approximately 5 × 107 CFU/ml.
FIG. 1.
Effect of using three different inocula (107, 2.5 × 107, and 5 × 107 CFU/ml) of exponentially growing V. anguillarum with 50% (vol/vol) AH2 supernatant compared to the effect of using fresh M9GC (control). OD600 for both types of cultures and each inoculum were determined at time zero (inoculum) and after 5 and 27 h of incubation of the cultures at 15°C with agitation.
Analysis of the genetic response of V. anguillarum after exposure to AH2 supernatant.
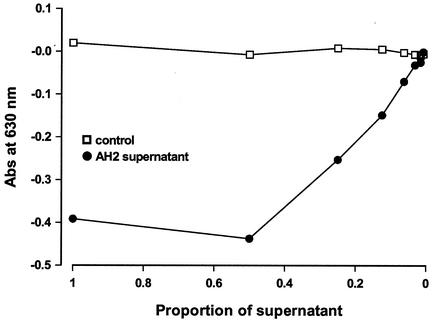
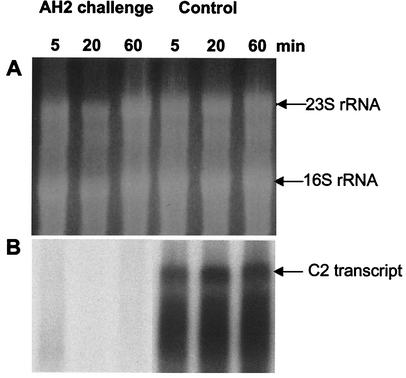
Strain AH2 supernatants were positive for iron chelation, as measured by the CAS assay, and a positive CAS response was also seen in samples diluted up to 30-fold (Fig. 2). V. anguillarum was not killed by the AH2 supernatant but remained viable throughout the experiment (unpublished data). Taking the abruptness of the physiological response to AH2 supernatant into consideration (i.e., the instant cessation of growth), we assumed that the genetic response was equally rapid. Preliminary experiments were used to determine appropriate sampling points by comparing the responses after 5 or 20 min of exposure to fresh substrate and to AH2 supernatant. RAP-PCR profiles were obtained by using one specific primer combination (primers SD14 and Ea5). From this analysis it was evident that differentially expressed genes, as represented by changes in band appearance or disappearance under the two conditions, were most prominently detected with the 20-min samples (data not shown). To further confirm this conclusion, we isolated and sequenced one band that was present only in control cultures (band C2) and used this fragment as a probe in a Northern analysis with total RNA isolated from both cultures exposed to AH2 supernatant and control cultures at 5, 20, and 60 min after exposure (Fig. 3). From this analysis it was evident that the transcript represented by band C2 was expressed during the continued exponential growth in the control culture, while transcription rapidly ceased in the culture exposed to AH2 supernatant; there was only a very faint level of expression after 5 min of exposure to the AH2 supernatant.
FIG. 2.
Absorption in the spectrophotometric CAS assay by twofold dilutions of sterile filtered P. fluorescens strain AH2 supernatant. Abs, absorption.
FIG. 3.
Kinetics of the genetic response of an AH2 supernatant-challenged V. anguillarum culture visualized by using a RAP-PCR clone (C2) as the probe in a Northern analysis. C2 was selected because of its presence only in exponentially growing cultures in preliminary RAP-PCR profiles. Total RNA was isolated at three different times (5, 20, and 60 min) after inoculation into a culture exposed to AH2 supernatant and into a control culture. (A) Formaldehyde-1.2% agarose gel electrophoresis of total RNA isolated from a culture challenged with 50% (vol/vol) AH2 supernatant and from a control culture after 5, 20, and 60 min. (B) Northern analysis in which RAP-PCR clone C2 was used as the probe.
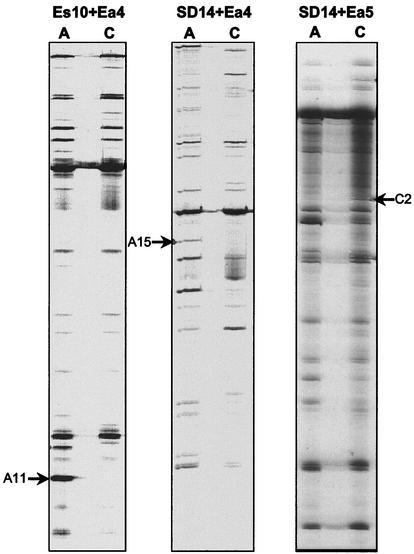
Based on these results, 30 different combinations of primers were used for RAP-PCR profiling of total RNA isolated from control samples and samples exposed to AH2 supernatant taken after 20 min. Table 1 shows the total numbers of bands that were present in the profiles when the different primer combinations were used. The number of bands in a profile was highly dependent on the primers used; e.g., primers Es3 and Ea7 gave 87 bands, and primers Es3 and Ea9 gave only 3 bands. Independent of the number of bands, very few changes were observed when control and challenge cultures were compared, but the changes were distinct (Fig. 4). Altogether, 15 RAP-PCR bands representing potentially differentially expressed genes were identified and subsequently reamplified, cloned, and sequenced. Five of the sequences were derived from the same gene, and two other sequences represented another gene but at different positions, which reduced the total number of different genes represented by RAP-PCR bands to 10.
TABLE 1.
Numbers of RAP-PCR bands in RAP-PCR profiles from V. anguillarum strain 90-11-287 generated by using different forward and reverse primer combinations
| Forward RAP-PCR primer | No. of RAP-PCR bands when the following reverse primers were used:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ea1 | Ea2 | Ea3 | Ea4 | Ea5 | Ea6 | Ea7 | Ea8 | Ea9 | Ea10 | |
| SD14 | 28 | 55 | 36 | 57 | 30 | 60 | 55 | 20 | 46 | 38 |
| Es3 | 17 | 66 | 45 | 24 | 31 | 52 | 87 | 9 | 3 | 60 |
| Es10 | 36 | 50 | 58 | 43 | 15 | 25 | 23 | 58 | 42 | 45 |
FIG. 4.
Examples of side-by-side RAP-PCR profiles generated from V. anguillarum cultures challenged with AH2 supernatant (lanes A) and from control V. anguillarum cultures (lanes C) by using three different sets of primers (Es10 plus Ea4, SD14 plus Ea4, and SD14 plus Ea5). One band that potentially represents a differentially expressed gene in each set of profiles is indicated by an arrow, and the designation of the corresponding RAP-PCR clone is indicated (see Table 2 for detailed information). The panels show only parts of the entire profile lanes.
Sequence analysis and database searches.
The presence of one or several open reading frames (ORFs) in a sequence is required to allow a RAP-PCR band to be identified as an expressed gene (mRNA). Also important is the presence of the primer sequences used for amplification of the bands. We were able to identify the two primer sequences used to amplify the bands in all cases except bands A13 and A15. With bands A13 and A15 only the SD14 primer sequence was identified in both ends. It is, however, possible that this occurred during the amplification step in cases in which the forward primer had a higher affinity for a sequence in the cDNA generated in the initial step than the reverse primer originally used had. In seven of the bands uninterrupted ORFs were identified. In two of the bands (bands C2 and C3) two ORFs that potentially represent parts of polycistronic mRNA were identified. In one band (band A11), 48 nucleotides beyond the stop codon in the 3′ untranslated region of the mRNA were included. Within this sequence stretch we were not able to identify potential stem-loop structures or rho-dependent transcription termination sites, which justified the presence of this region in the mRNA.
Searches of the database revealed a number of similar genes (Table 2), primarily from the close relative Vibrio cholerae, which recently was completely sequenced (26). In three cases (bands A7, C12, and A13) it was not possible to obtain significant hits in the BLASTX analysis. Interestingly, we identified a homologue (band A11) of the V. cholerae sigma factor (rpoS) (57) in V. anguillarum, and furthermore, we identified a homologue (band A15) of the vibE gene of V. cholerae. VibE is involved in the siderophore vibriobactin biosynthesis pathway of V. cholerae (56). Bands C2 and C3, originating from the control cultures, represented two polycistronic mRNAs with significant homology to the rpsD-rpoA and rpsB-tsf genes, respectively, of V. cholerae (26). RAP-PCR band A2 showed significant homology to a peptide ABC transporter, periplasmic peptide-binding protein (DppA) from V. cholerae (26), and band C5 was homologous to the ribosomal small-chain pseudouridine synthase A (RsuA) of V. cholerae (26). In one case (band A1) we found significant homology to a hypothetical protein from V. cholerae with unknown function. In subsequent experiments, we focused on genes with homology to genes with assigned functions in V. cholerae (i.e., A2, C2, C3, C5, A11, and A15), as this allowed us to interpret the genetic response more directly. Obviously, other nonassigned potential genes may also play important roles in the genetic response of V. anguillarum to AH2 supernatant exposure.
TABLE 2.
BLASTX searches with sequences identified in RAP-PCR profiles of V. anguillarum strain 90-11-287 grown in M9GC with or without exposure to P. fluorescens strain AH2 supernatant
| Clone | Accession no. | Origin | Fragment size (nucleotides) | ORF size (nucleotides) | BLASTX results
|
|||
|---|---|---|---|---|---|---|---|---|
| Score | Accession no. | Gene | Organism | |||||
| A1 | AJ458368 | AH2 challenge | 303 | 303 | 425 | AE004426 | Hypothetical protein VCA0994 | V. cholerae |
| A2 | AJ458369 | AH2 challenge | 621 | 621 | 898 | AE004107 | Peptide ABC transporter, periplasmic peptide-binding protein VCO171 (dppA) | V. cholerae |
| C2 | AJ458377 | Control | 402 | 191 | 299 | AE004325 | DNA-directed RNA polymerase, alpha-chain VC2571 (rpoA) | V. cholerae |
| 186 | 291 | AE004325 | Ribosomal protein S4 VC2572 (rpsD) | V. cholerae | ||||
| C3 | AJ458376 | Control | 657 | 114 | 138 | AE004298 | Ribosomal protein S2 VC2260 (rpsB) | V. cholerae |
| 413 | 541 | AE004297 | Translation elongation factor EF-Ts VC2259 (tsf) | V. cholerae | ||||
| C5 | AJ458370 | Control | 327 | 327 | 508 | AE004241 | Ribosomal small-chain pseudouridine synthase A VC1635 (rsuA) | V. cholerae |
| A7 | AJ458371 | AH2 challenge | 249 | 249 | No significant hits (>100) | |||
| A11 | AJ458372 | AH2 challenge | 176 | 128 | 206 | AE004139 | RNA polymerase sigma-38 factor (rpoS) | V. cholerae |
| C12 | AJ458373 | Control | 453 | 453 | No significant hits (>100) | |||
| A13 | AJ458374 | AH2 challenge | 334 | 334 | No significant hits (>100) | |||
| A15 | AJ458375 | AH2 challenge | 316 | 316 | 417 | AE004162 | Vibriobactin-specific 2,3-dihydroxybenzoate-AMP ligase (vibE) | V. cholerae |
Verification of differentially expressed genes.
To verify that the genes of V. anguillarum identified were indeed differentially expressed, we performed a series of Northern analyses in which we compared total RNA fractions isolated 20 min after inoculation from control cultures and from cultures exposed to AH2 supernatant. Differential expression was confirmed for five of the six genes (Table 3); however, the A2 probe gave positive signals for both samples, indicating that this gene was expressed regardless of AH2 supernatant exposure. All the genes expressed only in the control cultures (C2, C3, and C5) are characteristic of actively growing cells. The genes induced or expressed only after exposure to AH2 supernatant (A11 and A15) could indicate why the growth of V. anguillarum is halted by the antagonistic organism P. fluorescens AH2.
TABLE 3.
Verification of differentially expressed genes in V. anguillarum strain 90-11-287 identified in the RAP-PCR analysis by using Northern analyses to compare control cultures and cultures exposed to P. fluorescens AH2 supernatant
| Probe (gene homologue) | Presence in control culture | Presence in culture exposed to AH2 supernatant |
|---|---|---|
| A2 (dppA) | + | + |
| C2 (rpsD-rpoA) | + | − |
| C3 (rpsB-tsf) | + | − |
| C5 (rsuA) | + | − |
| A11 (rpoS) | − | + |
| A15 (vibE) | − | + |
Iron inhibits the antagonistic effect of the AH2 supernatant and alters the AH2 supernatant-induced genetic response.
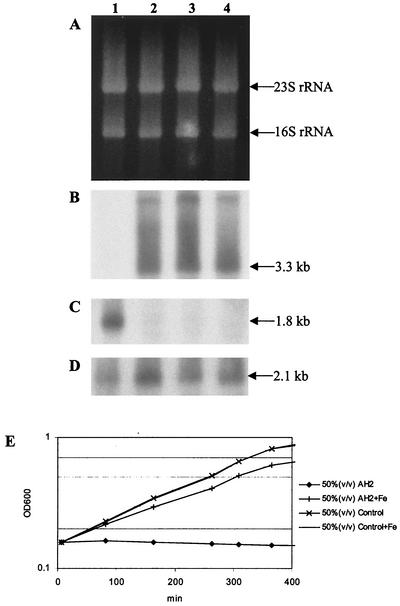
Supplementing the AH2 supernatant-containing medium with excess Fe3+ ions eliminated the growth-inhibiting antagonistic effect of the AH2 supernatant (Fig. 5E). We determined how Fe3+ addition affected expression of RAP-PCR clones A2, C2, and A11, each of which represents a different regulatory pattern in response to AH2 supernatant exposure. Four parallel cultures were inoculated, as described above, with exponentially growing V. anguillarum. For one pair of cultures, one culture was inoculated with 50% (vol/vol) AH2 supernatant and one culture was inoculated with fresh medium; for a second similar pair of cultures, each culture was supplemented with 0.1 mM FeCl3. After 20 min of incubation, samples were taken from each of the four cultures and used for total RNA extraction and subsequent Northern analyses. The optical densities of all four cultures were monitored for several hours after inoculation. The growth of the culture that was exposed to AH2 supernatant and supplemented with 0.1 mM FeCl3 was almost unaffected, like the growth of the two control cultures. The slight reduction in growth rate observed in this culture could be explained by the difference between the volume of fresh M9GC added to the culture and the volume added to the control cultures (Table 4). Growth instantly ceased in the iron-deficient culture exposed to AH2 supernatant (Fig. 5E). Apparent reversal of the physiological response imposed by the AH2 supernatant by addition of excess amounts of Fe3+ ions was also observed in the responses of the genes represented by RAP-PCR clones A2, C2, and A11. C2 potentially represents part of an operon that includes homologues of the rpsD and rpoA genes, and these genes resumed expression in response to iron supplementation and continued growth (Fig. 5B). The expression of dppA homologue A2 was essentially unchanged (Fig. 5D) except for a twofold induction in the iron-supplemented AH2 culture (lane 2). The rpoS homologue A11 was not expressed in the iron-supplemented AH2 supernatant-containing culture (Fig. 4C). Thus, taken together, the data for these three genes indicated that the genetic responses of V. anguillarum cultures exposed to AH2 supernatant after resumption of growth upon addition of 0.1 mM FeCl3 reverted to the expression pattern seen in normally growing unchallenged V. anguillarum cultures. The significance of the small increase in expression of the dppA homologue A2 in iron-supplemented AH2 cultures is difficult to evaluate, since we did not see a similar tendency in the iron-supplemented control culture. This increase could imply that the growth-arresting effect of the AH2 supernatant includes elements other than simple iron starvation.
FIG. 5.
Effect of supplementing V. anguillarum challenged with AH2 supernatant with 0.1 mM FeCl3 on the expression of three different genes identified by RAP-PCR. (A) Formaldehyde-1.2% agarose gel electrophoresis of total RNA isolated from cultures challenged with 50% (vol/vol) AH2 supernatant without (lane 1) or with (lane 2) 0.1 mM FeCl3 and from control cultures with fresh medium without (lane 3) or with (lane 4) 0.1 mM FeCl3. (B to D) Northern analyses in which the C2 (B), A11 (C), or A2 (D) RAP-PCR clone was used as the probe. Predicted sizes of the transcripts are indicated on the right in panels B to D. (E) Growth curves of the four cultures analyzed.
TABLE 4.
Compositions of different cultures of V. anguillarum used in this study
| Culture | H2O (ml) | M9GC (ml) | Inoculum (ml) | AH2 supernatant (ml) | 0.01 M FeCl3 (ml) |
|---|---|---|---|---|---|
| 50% (vol/vol) AH2 | 0 | 30 | 20 | 50 | 0 |
| 10% (vol/vol) AH2 | 40 | 30 | 20 | 10 | 0 |
| 1% (vol/vol) AH2 | 49 | 30 | 20 | 1 | 0 |
| 50% (vol/vol) control | 0 | 80 | 20 | 0 | 0 |
| 10% (vol/vol) control | 40 | 40 | 20 | 0 | 0 |
| 1% (vol/vol) control | 49 | 31 | 20 | 0 | 0 |
| 50% (vol/vol) AH2 + Fe | 0 | 30 | 20 | 50 | 1 |
| 50% (vol/vol) control + Fe | 0 | 80 | 20 | 0 | 1 |
Investigating the response of V. anguillarum to different doses of the AH2 supernatant.
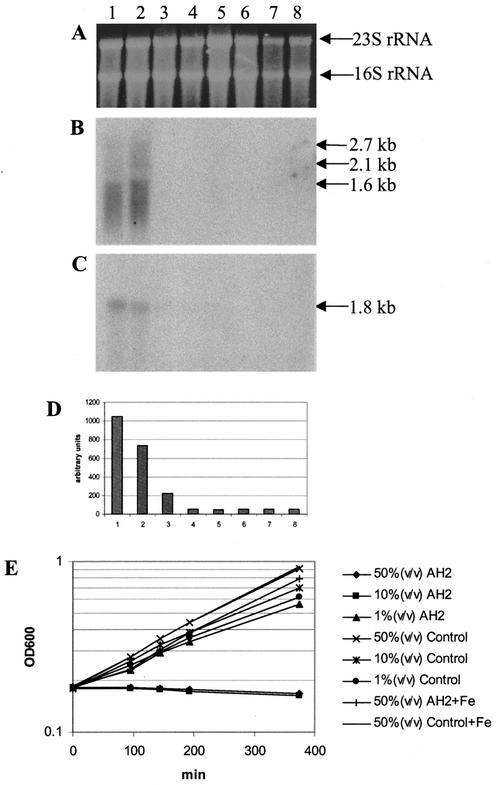
Up to 30- to 50-fold dilution of the AH2 supernatant still resulted in chemical detection of iron-chelating ability by the CAS assay (Fig. 2). We therefore investigated the expression or induction of the genes homologous to rpoS (band A11) and vibE (band A15) in exponentially growing V. anguillarum when it was exposed to different concentrations of the AH2 supernatant. Eight V. anguillarum cultures, including cultures exposed to AH2 supernatant, control cultures, and cultures supplemented with 0.1 mM FeCl3, were set up (Table 4). Total RNA was isolated from each of the cultures 20 min after inoculation (Fig. 6A) and prepared for Northern analysis by using A15 (Fig. 6B) and A11 (Fig. 6C) as the probes. Growth of the eight cultures was subsequently monitored for several hours (Fig. 6E). Focusing on the three cultures exposed to AH2 supernatant without iron supplementation, we observed that at a 100-fold dilution (i.e., 1% [vol/vol] AH2 supernatant), the antagonistic effect was eliminated. Both the 10-fold dilution and the 2-fold dilution, on the other hand, still had antagonistic effects. Minor differences in the growth rate appeared to correlate with differences in the amount of fresh M9GC supplied to the individual cultures.
FIG. 6.
Northern analysis of RAP-PCR clones A11 and A15 after exposure of exponentially growing V. anguillarum to three different doses of AH2 supernatant (see Table 4 for information on the compositions of the different cultures). Lane 1 and bar 1, 50% (vol/vol) AH2 supernatant; lane 2 and bar 2, 10% (vol/vol) AH2 supernatant; lane 3 and bar 3, 1% (vol/vol) AH2 supernatant; lane 4 and bar 4, 50% (vol/vol) control; lane 5 and bar 5, 10% (vol/vol) control; lane 6 and bar 6, 1% (vol/vol) control; lane 7 and bar 7, 50% (vol/vol) AH2 supernatant plus Fe; lane 8 and bar 8, 50% (vol/vol) control plus Fe. (A) Formaldehyde-1.2% agarose gel electrophoresis of total RNA isolated from each of the eight cultures after 20 min of incubation. (B and C) Northern analyses in which the A15 (B) or A11 (C) RAP-PCR clone was used as the probe. (D) Quantitative measurements of the A11-associated hybridization signals in the Northern blot shown in panel C. (E) Growth curves of the eight cultures analyzed.
The Northern analysis in which A15 was used as the probe provided evidence that expression of the gene homologous to vibE was induced only when growth had ceased. In the presence of 1% (vol/vol) AH2 supernatant, no expression was observed, as was the case in the culture exposed to AH2 supernatant and supplemented with excess iron ions. The size of the largest transcript (2.7 kb) in the A15 hybridization (Fig. 6B) corresponded nicely with the predicted size of an mRNA in V. cholerae containing the cotranscribed genes vibC and vibE as proposed by Wyckoff et al. (56), suggesting that the genes in V. anguillarum are organized in a similar manner. We also observed smaller transcripts and a smear below 1.6 kb, indicating that the mRNA was degraded. This finding does not, however, rule out the possibility that this mRNA encodes functional proteins.
Only in cultures in which the AH2 supernatant resulted in growth arrest could we identify significant expression of the rpoS homologue, A11; however, unlike the level of expression of the vibE homologue, it appeared that the level of expression of the rpoS homologue was slightly reduced in the culture containing 10% (vol/vol) AH2 supernatant compared to the level in the culture containing 50% (vol/vol) AH2 supernatant. When analyzing a Northern blot quantitatively with a phosphorimaging device, we observed a faint signal from the culture containing 1% (vol/vol) AH2 supernatant, whereas all other control cultures, as well as the iron-supplemented cultures, gave only background signals (Fig. 6D). These results suggest that expression of the rpoS homologue is positively correlated with the amount of AH2 supernatant added to a culture, and even at very low levels (i.e., a 100-fold dilution), the supernatant still affected the culture, although the ability of the cells to grow was not affected.
2,2-Dipyridyl-induced iron chelation in M9GC V. anguillarum control cultures stimulates the stationary-phase response through rpoS.
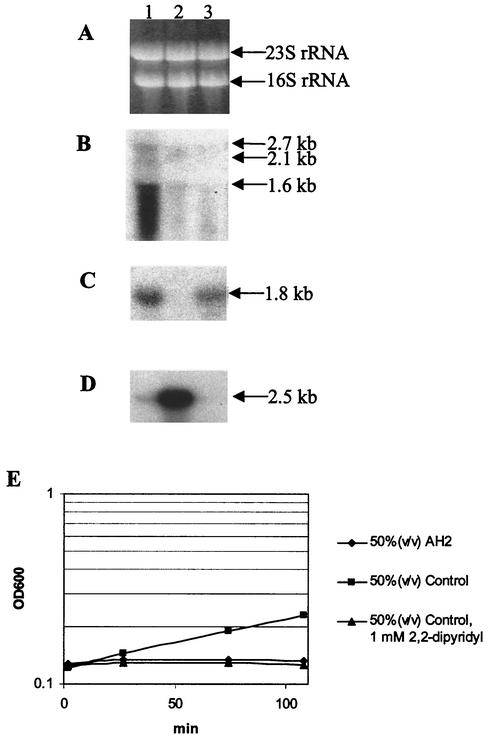
The results described above indicated that iron deprivation is essential for the mechanism of the AH2 supernatant effect on V. anguillarum. We investigated this phenomenon by comparing the effect of the commonly used iron chelator 2,2-dipyridyl to the effect of AH2 supernatant by performing Northern analyses with A11, A15, and C3 as the probes. Prior to the experiment we performed a titration of 2,2-dipyridyl to determine the concentration needed to inhibit growth of V. anguillarum and maintain a viable culture. Concentrations between 1 and 0.25 mM inhibited growth, whereas 0.125 mM had only a marginal effect. Addition of 1 mM 2,2-dipyridyl to a V. anguillarum cell suspension had no effect on the viability of cells, and this concentration was chosen for subsequent experiments (Table 5). Growth was monitored for approximately 2 h after inoculation (Fig. 7E), and total RNA was extracted from samples from the three cultures taken after 20 min (Fig. 7A). The Northern analysis in which C3 was used as the probe (Fig. 7D) revealed an expected pattern of signals that included a very bright signal from a 2.5-kb transcript in the 50% (vol/vol) control culture, indicating growth, and only very faint signals in the growth-inhibited cultures (50% [vol/vol] AH2 supernatant and 50% [vol/vol] control supplemented with 1 mM 2,2-dipyridyl). When we used probe A11 homologous to rpoS (Fig. 7C), we observed clear AH2 supernatant-mediated induction of rpoS, and interestingly, we observed similar although slightly reduced induction when the culture was exposed to 1 mM 2,2-dipyridyl.
TABLE 5.
Compositions of cultures of V. anguillarum used in control experiment with specific iron chelation in which 2,2-dipyridyl was used
| Culture | M9GC (ml) | M9GC with 1.25 mM 2,2-dipyridyl (ml) | Inoculum (ml) | AH2 supernatant (ml) |
|---|---|---|---|---|
| 50% (vol/vol) AH2 | 30 | 0 | 20 | 50 |
| 50% (vol/vol) control | 80 | 0 | 20 | 0 |
| 50% (vol/vol) control with 2,2-dipyridyl | 0 | 80 | 20 | 0 |
FIG. 7.
Northern analysis of RAP-PCR clones A15, A11, and C3 after exposure of exponentially growing V. anguillarum to three different media (see Table 5 for information on the compositions of the different cultures). Lane 1, 50% (vol/vol) AH2 supernatant; lane 2, 50% (vol/vol) control; lane 3, 50% (vol/vol) control plus 1 mM 2,2-dipyridyl. (A) Formaldehyde-1.2% agarose gel electrophoresis of total RNA isolated from each of the three cultures after 20 min of incubation. (B to D) Northern analysis in which the A15 (B), A11 (C), or C3 (D) RAP-PCR clone was used as the probe. (E) Growth curves of the three cultures analyzed.
The results of the Northern analysis when probe A15 homologous to vibE was used confirmed that there was clear induction of a gene homologous to vibE in V. anguillarum when it was exposed to AH2 supernatant (Fig. 7B), and again we observed a tendency toward degradation of the vibE transcripts. However, unlike the experiment shown in Fig. 6B, we observed a basal level of expression of this gene in the control culture, but we believe that this was due to differences in the sensitivities of the two different Northern analyses. From a quantitative analysis of the Northern blot in Fig. 7B we concluded that the gene was induced at least fivefold when it was exposed to AH2 supernatant. No induction was seen in the 2,2-dipyridyl-containing control culture. Either this reflected differences in the efficiency with which the AH2 supernatant affected the genetic response compared to the effect of 1 mM 2,2-dipyridyl, or another component, aside from the lack of iron, in the AH2 supernatant stimulated expression of the vibE homologue of V. anguillarum.
The vibE homologue A15 is not identical to ORF E on virulence plasmid pJM1 of V. anguillarum.
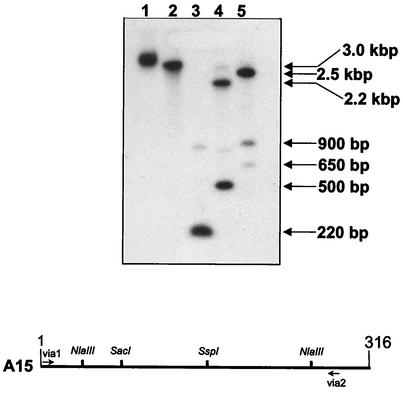
Recently, a region on virulence plasmid pJM1 of V. anguillarum was sequenced and characterized (54). One ORF (ORF E) was identified in this region that had significant homology to the genes vibE of V. cholerae and entE of Escherichia coli, which are involved in siderophore synthesis of vibriobactin and enterobactin, respectively. ORF E contains a frameshift mutation and is therefore not believed to encode any functional protein, suggesting that this gene is not essential (54). The A15 RAP-PCR clone identified in this work gave a significant hit to VibE (Table 2 and Fig. 8A), and the similarity of A15 to ORF E was investigated. Alignment of the 104-amino-acid overlap of the A15 and ORF E products gave a clear indication that these two regions are not identical but are homologous, and interestingly, we demonstrated that the A15 sequence does not contain the same frameshift mutation as that observed in ORF E (Fig. 8B). ORF E and A15 have 70% identity and 75% similarity, while VibE and the A15 product have 74% identity and 87% similarity (Fig. 8). A Southern analysis was performed with a 286-bp region of A15 as the probe to confirm that A15 represented a chromosomally encoded gene (Fig. 9). Purified genomic DNA of V. anguillarum was digested with four different restriction enzymes (EcoRI, NlaIII, SspI, and SacI). EcoRI was not expected to have any recognition sites within A15, while NlaIII was expected to have two sites and SspI and SacI were each expected to have one site. The Southern analysis confirmed that the A15 region is indeed part of the V. anguillarum genome (Fig. 9) and that it potentially represents part of a functionally active gene (e.g., angE) encoding a protein involved in the synthesis of anguibactin or another siderophore of V. anguillarum.
FIG. 8.
Pairwise amino acid sequence alignments of the 104-amino-acid region of the product of the uninterrupted ORF of RAP-PCR clone A15 with VibE from V. cholerae (A) and with ORF E from virulence plasmid pJM1 of V. anguillarum (B). Identical residues are indicated by boldface type, and conservative amino acid substitutions are indicated by plus signs. In panel B, an arrow indicates the position of the frameshift of ORF E, as reported by Welch et al. (54).
FIG. 9.
Southern analysis performed with a 286-bp region of RAP-PCR clone A15 as the probe against undigested genomic DNA from V. anguillarum (lane 1) and genomic DNA from V. anguillarum digested with EcoRI (lane 2), NlaIII (lane 3), SspI (lane 4), and SacI (lane 5). The sizes of bands are indicated on the right, and the positions of known NlaIII, SspI, and SacI sites within the 316-nucleotide A15 clone are indicated at the bottom. The positions of primers via1 and via2 used to generate the probe are also indicated at the bottom.
DISCUSSION
With this study we demonstrated the potential of using molecular techniques to unravel putative mechanisms of the antagonistic behavior of a probiotic strain against a pathogenic strain, not by focusing on the antagonist but instead by characterizing the genetic response of the pathogen. By selecting this strategy we obviously excluded the possibility of obtaining more direct evidence of the antagonistic mechanism exhibited by the AH2 strain, which would have been possible if we had constructed, selected for, and characterized noninhibitory mutants of the AH2 strain. However, for the random mutagenesis approach to be successful there is an absolute requirement of access to reliable and efficient genetic tools, and this is not always possible, particularly not with natural strains, such as P. fluorescens AH2. Genetic manipulation of strain AH2 (e.g., by Tn5 mutagenesis) has proven to be difficult (5).
By using the RAP-PCR-based differential display technique we were able to identify a small subset of genes that were either absent (i.e., not expressed or expressed at levels below the detection limit) or present (i.e., expressed or induced) in one of the two conditions of V. anguillarum that we tested. The changes observed reflected rearrangements of the transcriptome that V. anguillarum exhibited in going from an actively exponentially growing phase to an instantly growth-inhibited stage due to the AH2 supernatant. The physiological response (i.e., the prompt arrest of growth) was also displayed in the genetic response by the lack of transcription of RAP-PCR clones C2 and C3, which encode homologues of the rpoA-rpsD genes (corresponding to part of the α operon of E. coli) and the rpsB-tsf genes (corresponding to the S2 operon of E. coli), respectively. In E. coli expression of these genes is known to be coordinated with the synthesis of ribosomal proteins as a function of the growth rate (28, 34), and it is therefore not surprising that transcription of these genes in V. anguillarum stops when growth stops. This is a trivial observation, but nevertheless a connection between a physiological response and a genetic response was made.
Other less trivial observations concerning the genetic response were made based on the RAP-PCR profiles. First, an rpoS homologue was induced in the pathogen after strain AH2 supernatant exposure, and second, a vibE homologue was induced in V. anguillarum exposed to AH2 supernatant.
We demonstrate here that during stress response the fish pathogen V. anguillarum expresses a gene homologous to rpoS. The results of the 2,2-dipyridyl-mediated growth arrest analysis demonstrate to our knowledge for the first time that the stationary-phase response through stimulation of rpoS can be induced specifically by iron starvation. Also, the data indicate that the rpoS induction seen when V. anguillarum is exposed to AH2 supernatant could be caused by the iron-chelating properties of the supernatant. The rpoS gene, which has been intensely studied in E. coli and Salmonella, encodes RpoS, which is also called the alternative sigma factor S (σS). RpoS is required for the global response of these bacteria in the stationary phase and for responses to several stress factors, such as oxidative damage, salinity, and starvation (15, 38, 43). RpoS mutants typically show reduced survival during stress exposure, and also virulence factors may be modulated by RpoS. Thus, Iriarte et al. (27) demonstrated that an RpoS mutant of Yersinia enterocolitica did not produce enterotoxin. The rpoS gene has been sequenced in environmental bacteria, such as Pseudomonas putida (41), and in nonenteric pathogens, such as V. cholerae (57), in which it plays a role in starvation and stress survival like the role that it plays in enteric pathogens. Our data indicate that iron starvation is the major mechanism governing the growth arrest of V. anguillarum when it is exposed to strain AH2. Most starvation studies have focused on rpoS as induced by carbon, nitrogen, or phosphorus limitation, and it has not previously been shown that rpoS induction specifically is linked to iron starvation as the cause of growth arrest. The rpoS induction in V. anguillarum was a function of the amount of AH2 supernatant (i.e., there was a dose-response relationship) and was eliminated when iron was added to the culture. V. anguillarum is almost constantly exposed to conditions of nutrient or mineral limitation. In the planktonic state in the ocean, many nutrients are limited, and the organism is exposed to UV light, fluctuating temperatures, etc. In its infectious stage it must survive host defense mechanisms, including iron depletion conditions. Thus, V. anguillarum must be equipped with a broad, well-functioning stress response system. It would be interesting to determine if rpoS in V. anguillarum determines survival and has any influence on virulence. Unpublished rpoS sequences have been submitted for another fish pathogen, Vibrio harveyi (accession number AF321124), but the role of RpoS in this organism and the factors that induce its expression are unknown. Nelson et al. (36) concluded that the starvation stress response in V. anguillarum was different from that of other bacterial species since starvation caused only a temporal increase in resistance to other stress factors, such as ethanol exposure. Our finding that rpoS is present indicates that the response of V. anguillarum may not be very different, and further studies with genetic tools should elucidate the importance of RpoS in this organism.
The A15 RAP-PCR clone has significant homology on the amino acid level to the corresponding vibE and entE genes of V. cholerae and E. coli, respectively. These genes are both constituents of the siderophore-based iron-sequestering system for these two bacterial species using vibriobactin (25) and enterobactin (39), respectively, and it was natural to hypothesize that this novel gene in V. anguillarum has a role in iron uptake. Basically, in many microorganisms regulated systems make sure that under iron-deprived conditions efficient assimilation of iron is maintained by induction of the synthesis of low-molecular-weight iron-chelating siderophores (29). Through the years several studies have specifically focused on the iron uptake systems of V. anguillarum (9, 11, 12, 30, 54). It has been established that V. anguillarum serotype O1 strains possess two different iron uptake systems, one encoded by chromosome genes and one encoded by plasmid genes (11, 30). The chromosome-mediated iron uptake system in V. anguillarum produces siderophores functionally related to enterobactin (30), and the plasmid (pJM1)-encoded system produces the catechol-type siderophore anguibactin (1), which is structurally different from enterobactin and vibriobactin.
Recently, Welch et al. (54) provided evidence of the essential role of two overlapping genes (angB and angG) on virulence plasmid pJM1 in siderophore biosynthesis of anguibactin. In this study a region including these two genes was sequenced on the pJM1 plasmid, and in addition to other genes a third gene, designated ORF E, was also identified. ORF E contained a frameshift mutation and was therefore considered to be nonfunctional. ORF E showed significant homology to vibE, as well as to the A15 clone product on the amino acid level (Fig. 8B). However, A15 does not contain the frameshift mutation, and therefore, it most likely encodes a functional protein and resides on the chromosome of V. anguillarum, exactly like vibE in V. cholerae. VibE is a 2,3-dihydroxybenzoate-AMP ligase that, as indicated by Wyckoff et al. (56), plays a role in the vibriobactin biosynthesis pathway going from 2,3-dihydroxybenzoic acid to vibriobactin. It is interesting to speculate that the function of the vibE homologue in V. anguillarum is similar. This suggests that anguibactin biosynthesis in V. anguillarum could be controlled by genes that reside on both pJM1 and the chromosome. A similar coupling of chromosome-mediated and plasmid-mediated biosynthesis of anguibactin was reported by Chen et al. (9), but it involves the chromosomal gene product AroC of V. anguillarum, which catalyzes the synthesis of a precursor known as chorismate to 2,3-dihydroxybenzoic acid.
Welch et al. (54) discussed the presence of pseudogenes like ORF E on the pJM1 plasmid and their possible origin as a function of transposition events and horizontal transfer of genes potentially originating in V. cholerae or an ancestral organism. It was suggested that these genes could have become nonfunctional during evolution because, as these authors state, they are nonessential. Our study demonstrated that the lack of function of the pseudogenes on the plasmid could be due to the presence of chromosomally encoded functionally identical genes.
We were able to demonstrate by supplementing the AH2 supernatant with excess FeCl3 that growth of V. anguillarum was maintained after exposure and induction of the rpoS and vibE homologues did not occur. Thus, simply providing sufficient iron ions ensured continued growth and apparently eliminated the antagonistic effect of the AH2 supernatant. Furthermore, we observed a dose-dependent response of V. anguillarum when increasingly diluted AH2 supernatants were added. rpoS transcription was gradually diminished with increasing dilution of AH2 supernatant, and even at a 100-fold dilution, at which growth was possible, a small increase in rpoS transcription above the level in unexposed cultures could be identified.
However, when we attempted to mimic the presumed iron-chelating ability of the AH2 supernatant by using the commonly used chemical iron chelator 2,2-dipyridyl, we observed differences in the behavior of the two systems with respect to the effect on induction of the vibE homologue in V. anguillarum. Hence, presumably not just simple iron deficiency induces the vibE homologue, but a combination of a lack of iron and the presence of other components of the AH2 supernatant is required to induce the vibE homologue. From other studies (8, 16) siderophores are known to positively affect the growth of bacteria other than the siderophore-producing organism. The examples include specific cross feeding (16) or stimulation of a non-siderophore-producing bacterium by addition of siderophore-containing supernatants from another producing organism (8). Although growth of V. anguillarum is inhibited after exposure to the siderophore-containing AH2 supernatant, the presence of siderophores of strain AH2 origin might positively affect the V. anguillarum siderophore synthesis apparatus, leading to production of novel V. anguillarum siderophores (e.g., anguibactin) and hence induction of the vibE homologue. Obviously, more thorough investigations are needed to confirm this.
Based on these results it is tempting to propose a model for the mechanism of the antagonistic effect of the AH2 supernatant against V. anguillarum proliferation. The AH2 supernatant contains iron-complexing compounds (i.e., siderophores) in huge excess, and when introduced into an exponentially growing culture of V. anguillarum, it immediately binds the available free iron ions, which leads to an iron-deprived environment. In response to this, V. anguillarum stops growing, and a stress response through rpoS is induced. In parallel, siderophore biosynthetic genes are induced, as indicated by the presence of mRNA from the gene homologous to vibE. Assuming that V. anguillarum siderophores are synthesized and secreted into the medium, these compounds are not capable of outcompeting the chelation exerted by the strain AH2-derived siderophores, nor is V. anguillarum capable of utilizing the complex-bound iron. When excess Fe3+ ions are added, efficient uptake of iron resumes and growth continues. It follows that trials investigating the in vivo effect on vibriosis of, for example, pseudomonads that overproduce siderophores (or in which the siderophore capacity is diminished) should be conducted to verify our suggestions.
Acknowledgments
The technicians Karen F. Appel, Dorte Lauritsen, Jette Melchiorsen, and Trine Møller are thanked for their skillful technical assistance.
The Danish Ministry of Food, Agriculture and Fisheries financially supported this study.
REFERENCES
- 1.Actis, L. A., W. Fish, J. H. Crosa, K. Kellerman, S. R. Ellenberger, F. M. Hauser, and J. Sanders-Loehr. 1986. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(pJM1). J. Bacteriol. 167:57-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthoni, U., T. Børresen, C. Christophersen, L. Gram, and P. H. Nielsen. 1990. Is trimethylamine oxide a reliable indicator for the marine origin of fish? Comp. Biochem. Physiol. B 97:569-571. [Google Scholar]
- 3.Austin, B., and D. A. Austin. 1999. Bacterial fish pathogens: diseases of farmed and wild fish. Springer Verlag, Berlin, Germany.
- 4.Bachem, C. W. B., R. S. van der Hoeven, S. M. de Bruijn, D. Vreugdenhil, M. Zabeau, and R. Visser. 1996. Visualisation of differential gene expression using a novel method of RNA finger-printing based on AFLP: analysis of gene expression during potato tuber development. Plant J. 9:745-753. [DOI] [PubMed] [Google Scholar]
- 5.Buch, C. 2001. Genetic characterisation of fish probiotics. Optimisation of transposon Tn5 mutagenesis of Pseudomonas fluorescens AH2. M.Sc. thesis. Technical University of Denmark, Kgs. Lyngby, Denmark.
- 6.Budzikiewicz, H. 1993. Secondary metabolites from fluorescent pseudomonads. FEMS Microbiol. Rev. 104:209-228. [DOI] [PubMed] [Google Scholar]
- 7.Buysens, S., K. Heugens, J. Poppe, and M. Höfte. 1996. Involvement of pyochelin and pyoverdin in suppression of Pythium-induced damping-off by tomato by Pseudomonas aeruginosa 7NSK2. Appl. Environ. Microbiol. 62:865-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Champomier-Vergé, M.-C., A. Stintzi, and J.-M. Meyer. 1996. Acquisition of iron by the non-siderophore producing Pseudomonas fragi. Microbiology 142:1191-1199. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Q., L. A. Actis, M. E. Tolmasky, and J. H. Crosa. 1994. Chromosome-mediated 2,3-dihydroxybenzoic acid is a precursor in the biosynthesis of the plasmid-mediated siderophore anguibactin in Vibrio anguillarum. J. Bacteriol. 176:4226-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Church, M. J., D. A. Hutchins, and H. W. Ducklow. 2000. Limitation of bacterial growth by dissolved organic matter and iron in the Southern Ocean. Appl. Environ. Microbiol. 66:455-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conchas, R. F., M. L. Lemos, J. L. Barja, and A. E. Toranzo. 1991. Distribution of plasmid- and chromosome-mediated iron uptake systems in Vibrio anguillarum strains of different origins. Appl. Environ. Microbiol. 57:2956-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crosa, J. H. 1980. A plasmid associated with virulence in the marine fish pathogen Vibrio anguillarum specifies an iron sequestering system. Nature (London) 283:566-568. [DOI] [PubMed] [Google Scholar]
- 13.Ellis, R., T. M. Timms-Wilson, and M. J. Bailey. 2000. Identification of conserved traits in fluorescent pseudomonads with antifungal activity. Environ. Microbiol. 2:274-284. [DOI] [PubMed] [Google Scholar]
- 14.Expert, J. M., and B. Digat. 1995. Biocontrol of Sclerotina wilt of sunflower by Pseudomonas fluorescens and Pseudomonas putida strains. Can. J. Microbiol. 41:685-691. [Google Scholar]
- 15.Ferenci, T. 2001. Hungry bacteria—definition and properties of a nutritional state. Environ. Microbiol. 3:605-611. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez, A. I. G., A. F. Fernandez, M. J. Perez, T. P. Nieto, and A. E. Elis. 1998. Siderophore production by Aeromonas salmonicida subsp. salmonicida. Lack of specificity. Dis. Aquat. Organisms 33:87-92. [DOI] [PubMed] [Google Scholar]
- 17.Fislage, R., M. Berceanu, Y. Humboldt, M. Wendt, and H. Oberender. 1997. Primer design for a prokaryotic differential display RT-PCR. Nucleic Acids Res. 25:1830-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming, J. T., W.-H. Yao, and G. S. Sayler. 1998. Optimization of differential display of prokaryotic mRNA: application to pure culture and soil microcosms. Appl. Environ. Microbiol. 64:3698-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Food and Agriculture Organization. 2000. The state of world fisheries and aquaculture 2000. Food and Agriculture Organization, Rome, Italy.
- 20.Fourney, R. M., J. Miyakoshi, R. S. Day III, and M. S. Paterson. 1988. Northern blotting: efficient RNA staining and transfer. Focus 10:5-7. [Google Scholar]
- 21.Gopalakrishna, Y., D. Langley, and N. Sarkar. 1981. Detection of high levels of polyadenylate-containing RNA in bacteria by the use of single-step RNA isolation procedure. Nucleic Acids Res. 9:3545-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gram, L., C. Wedell-Neergaard, and H. H. Huss. 1990. The bacteriology of fresh and spoiling Lake Victorian Nile perch (Lates niloticus). Int. J. Food Microbiol. 10:303-316. [DOI] [PubMed] [Google Scholar]
- 23.Gram, L., J. Melchiorsen, B. Spanggaard, I. Huber, and T. F. Nielsen. 1999. Inhibition of Vibrio anguillarum by Pseudomonas fluorescens AH2, a possible probiotic treatment of fish. Appl. Environ. Microbiol. 65:969-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gram, L., and E. Ringø. Prospects of fish probiotics. In W. Holzapfel and P. Naughton (ed.), Microbial ecology of the growing animal, in press. Elsevier, Amsterdam, The Netherlands.
- 25.Griffiths, G. L., S. P. Sigel, S. M. Payne, and J. B. Neilands. 1984. Vibriobactin, a siderophore from Vibrio cholerae. J. Biol. Chem. 259:383-385. [PubMed] [Google Scholar]
- 26.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iriarte, M., I. Stainier, and G. R. Cornelis. 1995. The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect. Immun. 63:1840-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jinks-Robertson, S., and M. Nomura. 1982. Ribosomal protein S4 acts in trans as a translational repressor to regulate expression of the alpha operon in Escherichia coli. J. Bacteriol. 151:193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lankford, C. E. 1973. Bacterial assimilation of iron. Crit. Rev. Microbiol. 2:273-331. [Google Scholar]
- 30.Lemos, M. L., P. Salinas, A. E. Toranzo, J. L. Barja, and J. H. Crosa. 1988. Chromosome-mediated iron uptake system in pathogenic strains of Vibrio anguillarum. J. Bacteriol. 170:1920-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leong, J. 1986. Siderophores: their biochemistry and possible role in the biocontrol of plant pathogens. Annu. Rev. Phytopathol. 24:187-209. [Google Scholar]
- 32.Liang, P., and A. B. Pardee. 1992. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257:967-971. [DOI] [PubMed] [Google Scholar]
- 33.Loper, J. E., and L. S. Buyer. 1991. Siderophores in microbial interactions on plant surfaces. Mol. Plant-Microbe Interact. 4:5-13. [Google Scholar]
- 34.Miyajima, A., and Y. Kaziro. 1978. Coordination of levels of elongation factors Tu, Ts and G, and ribosomal protein S1 in Escherichia coli. J. Biochem. 83:453-462. [DOI] [PubMed] [Google Scholar]
- 35.Neidhardt, C. F., J. C. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell, a molecular approach, p. 1-29. Sinauer Associates, Inc., Sunderland, Mass.
- 36.Nelson, D. R., Y. Sadlowski, M. Eguchi, and S. Kjelleberg. 1997. The starvation-stress response of Vibrio (Listonella) anguillarum. Microbiology 143:2305-2312. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen, M. N., J. Sørensen, J. Fels, and H. C. Pedersen. 1998. Secondary metabolite- and endochitinase-dependent antagonism toward plant-pathogenic microfungi of Pseudomonas fluorescens isolates from sugar beet rhizosphere. Appl. Environ. Microbiol. 64:3563-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nyström, T. 1999. Starvation, cessation of growth and bacterial aging. Curr. Opin. Microbiol. 2:214-219. [DOI] [PubMed] [Google Scholar]
- 39.O'Brien, L. G., and F. Gibson. 1970. The structure of enterochelin and related 2,3-dihydroxy-N-benzoylserine conjugates from Escherichia coli. Biochim. Biophys. Acta 215:393-402. [DOI] [PubMed] [Google Scholar]
- 40.Pierson, L. S., III, and L. Thomashow. 1992. Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30-84. Mol. Plant-Microbe Interact. 5:330-339. [DOI] [PubMed] [Google Scholar]
- 41.Ramos-González, M. I., and S. Molin. 1998. Cloning, sequencing and phenotypic characterization of the rpoS gene from Pseudomonas putida KT2440. J. Bacteriol. 180:3421-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reid, R. T., D. H. Live, D. J. Faulkner, and A. Butler. 1993. A siderophore from a marine bacterium with an exceptional ferric ion affinity constant. Nature 366:455-458. [DOI] [PubMed] [Google Scholar]
- 43.Rozen, Y., and S. Belkin. 2001. Survival of enteric bacteria in seawater. FEMS Microbiol. Rev. 25:513-529. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 46.Schnider, U., C. Keel, C. Blumer, J. Troxler, G. Défago, and D. Haas. 1995. Amplification of the housekeeping sigma factor in Pseudomonas fluorescens CHA0 enhances antibiotic production and improves biocontrol abilities. J. Bacteriol. 177:5387-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 48.Shanahan, P., D. J. O'Sullivan, P. Simpson, J. D. Glennon, and F. O'Gara. 1992. Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol. 58:353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skov, M. N., K. Pedersen, and J. L. Larsen. 1995. Comparison of pulsed-field gel electrophoresis, ribotyping, and plasmid profiling for typing of Vibrio anguillarum serovar O1. Appl. Environ. Microbiol. 61:1540-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, P., and S. Davey. 1993. Evidence for the competitive exclusion of Aeromonas salmonicida from fish with stress-inducible furunculosis by a fluorescent pseudomonad. J. Fish Dis. 16:521-524. [Google Scholar]
- 51.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 52.Spanggaard, B., I. Huber, J. Nielsen, E. B. Sieck, C. B. Pipper, T. Martinussen, W. J. Slierendrecht, and L. Gram. 2001. The probiotic potential against vibriosis of the indigenous microflora of rainbow trout. Environ. Microbiol. 3:755-765. [DOI] [PubMed] [Google Scholar]
- 53.Velculescu, V. E., L. Zhang, B. Vogelstein, and K. W. Kinzler. 1995. Serial analysis of gene expression. Science 270:484-487. [DOI] [PubMed] [Google Scholar]
- 54.Welch, T. J., S. Chai, and J. H. Crosa. 2000. The overlapping angB and angG genes are encoded within the trans-acting factor region of the virulence plasmid in Vibrio anguillarum: essential role in siderophore biosynthesis. J. Bacteriol. 182:6762-6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welsh, J., K. Chada, S. S. Dalal, R. Cheng, D. Ralph, and M. McClelland. 1992. Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res. 20:4965-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyckoff, E. E., J. A. Stoebner, K. E. Reed, and S. M. Payne. 1997. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J. Bacteriol. 179:7055-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yildiz, F. H., and G. K. Schoolnik. 1998. Role of rpoS in stress survival and virulence of Vibrio cholerae. J. Bacteriol. 180:773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]