Abstract
Acyl coenzyme A carboxylase (acyl-CoA carboxylase) was purified from Acidianus brierleyi. The purified enzyme showed a unique subunit structure (three subunits with apparent molecular masses of 62, 59, and 20 kDa) and a molecular mass of approximately 540 kDa, indicating an α4β4γ4 subunit structure. The optimum temperature for the enzyme was 60 to 70°C, and the optimum pH was around 6.4 to 6.9. Interestingly, the purified enzyme also had propionyl-CoA carboxylase activity. The apparent Km for acetyl-CoA was 0.17 ± 0.03 mM, with a Vmax of 43.3 ± 2.8 U mg−1, and the Km for propionyl-CoA was 0.10 ± 0.008 mM, with a Vmax of 40.8 ± 1.0 U mg−1. This result showed that A. brierleyi acyl-CoA carboxylase is a bifunctional enzyme in the modified 3-hydroxypropionate cycle. Both enzymatic activities were inhibited by malonyl-CoA, methymalonyl-CoA, succinyl-CoA, or CoA but not by palmitoyl-CoA. The gene encoding acyl-CoA carboxylase was cloned and characterized. Homology searches of the deduced amino acid sequences of the 62-, 59-, and 20-kDa subunits indicated the presence of functional domains for carboxyltransferase, biotin carboxylase, and biotin carboxyl carrier protein, respectively. Amino acid sequence alignment of acetyl-CoA carboxylases revealed that archaeal acyl-CoA carboxylases are closer to those of Bacteria than to those of Eucarya. The substrate-binding motifs of the enzymes are highly conserved among the three domains. The ATP-binding residues were found in the biotin carboxylase subunit, whereas the conserved biotin-binding site was located on the biotin carboxyl carrier protein. The acyl-CoA-binding site and the carboxybiotin-binding site were found in the carboxyltransferase subunit.
Acidianus brierleyi is a facultatively anaerobic, thermoacidophilic, sulfur-metabolizing archaeon that can grow either heterotrophically or autotrophically at an optimum temperature around 70°C and an optimum pH of 1.5 to 2 (18). Under autotrophic growth, this archaeon utilizes the modified 3-hydroxypropionate cycle for CO2 fixation (21). The 3-hydroxypropionate cycle was originally proposed for a filamentous green non-sulfur-photosynthetic bacterium, Chloroflexus auranticus (63), and has recently been found in the archaea A. brierleyi (21), Metallosphaera sedula, Sulfolobus metallicus, and Acidianus infernus (37). The enzymes for the CO2 fixation of this cycle are acetyl coenzyme A carboxylase (acetyl-CoA carboxylase) and propionyl-CoA carboxylase. Acetyl-CoA carboxylase catalyzes the carboxylation of acetyl-CoA to form malonyl-CoA, whereas propionyl-CoA carboxylase catalyzes the carboxylation of propionyl-CoA to form methylmalonyl-CoA. Although some enzymes in the 3-hydroxypropionate cycle, namely, malonyl-CoA reductase (20) and propionyl-CoA synthase (1), were recently purified from C. auranticus and characterized, the key carboxylating enzymes of this cycle have not been purified yet.
Acetyl-CoA carboxylase and propionyl-CoA carboxylase have been known to be involved in fatty acid biosynthesis (41) and the synthesis of secondary metabolites (51), respectively. These enzymes have been purified and characterized from plants (44, 48), animals (36, 38, 58, 64, 67), yeast (39), algae (52, 69), and bacteria (9, 15, 17, 27, 46, 62). To our knowledge, however, no report of the purification of acetyl-CoA carboxylase or propionyl-CoA carboxylase from Archaea has been published. We report here the purification and molecular characterization of the acyl-CoA carboxylase from A. brierleyi, which plays an important role in the modified 3-hydroxypropionate cycle. Particularly, the bifunctional character of the enzyme (acetyl-CoA carboxylase and propionyl-CoA carboxylase) has been reported. Some characteristics of the enzyme have been shown to be unique among the known acetyl-CoA carboxylases.
MATERIALS AND METHODS
Chemicals and enzymes.
Sodium [14C]bicarbonate was purchased from Amersham Pharmacia (Little Chalfont, Buckinghamshire, United Kingdom). Scintillation cocktail (Aquasol-2) was purchased from Packard BioScience. Potassium tetrathionate (K2S4O6) was purchased from Merck (Darmstadt, Germany). The solution of 3-hydroxypropionic acid (ca. 30%) was purchased from TCI (Tokyo, Japan). All other reagents were of the highest grade and were purchased from Sigma (St. Louis, Mo.). Nucleotide primers were synthesized by the Sawady company (Tokyo, Japan). Restriction enzymes and the DNA ligation kit were purchased from Toyobo (Osaka, Japan) or TaKaRa (Kyoto, Japan).
Microorganism and preparation of cell extracts.
A. brierleyi (DSM 1651) was grown autotrophically at 70°C in a 3-liter glass jar fermentor with BTM medium as described previously (21). CO2 in the gas mixture (10% CO2 and 90% air; total gas flow rate, 3.5 liters min−1) was utilized as the sole carbon source, and K2S4O6 was used as an energy source. The K2S4O6 concentration was maintained at around 10 to 18 mM throughout the cultivations by analysis with cyanolysis and the spectrophotometric method (24). When the optical density of the culture at 440 nm reached 0.6 to 0.8, the cells were harvested by centrifugation at 7,600 × g for 30 min and washed twice with 100 mM Tricine-KOH buffer (pH 6.0). Cell pellets were resuspended with 3 volumes of the same buffer and stored at −80°C until they were used. For preparation of cell extracts for enzyme purification, a cell mass of approximately 10 g (wet weight) was thawed, and dithiothreitol (DTT) was then added to give a final concentration of 2 mM. The cell suspension was placed on ice-water and disintegrated by sonication (Sonifier 250; Branson). Cell debris was removed by centrifugation at 9,600 × g for 1 h. Then the supernatant was filtered through a 0.20-μm-pore-size surfactant-free cellulose acetate membrane (Corning, Wiesbaden, Germany). The filtrate was designated cell extract.
Gel electrophoresis and Western blot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with 15% polyacrylamide slab gels by the procedure of Laemmli (29). For nondenaturing PAGE, 7.5% acrylamide gels were used with 25 mM Tris-glycine buffer (pH 7.5) as the electrode buffer. Protein bands were detected by staining the gels with Coomassie brilliant blue R-250. In order to detect biotin-containing protein(s) in cell extracts of A. brierleyi, Western blot analysis and in-gel staining were carried out. For the Western blotting, cell extracts (10 μl) were electrophoresed on SDS-15% PAGE gels with the biotinylated protein standard as the molecular weight marker (low-range MW; Bio-Rad, Richmond, Calif.) and were then electroblotted onto a polyvinylidene difluoride membrane by using Trans-blot Cell (both from Bio-Rad). The biotin-containing proteins on the membrane were detected by the procedure described by Menendez et al. (37). For the activity staining, the gel from native PAGE was directly incubated with peroxidase-labeled avidin (Sigma), and biotin-containing proteins were detected by the method described for Western blot analysis.
Enzyme purification.
All of the purification steps were performed at room temperature (25°C) under aerobic conditions. After filtration (see “Microorganism and preparation of cell extracts” above), a cell extract of A. brierleyi was applied to a 1-ml Hi-Trap streptavidin column (Amersham Pharmacia, Uppsala, Sweden) which had been equilibrated with 20 ml of 100 mM Tricine-KOH buffer (pH 7.5) containing 4 mM MgCl2 · 6H2O and 2 mM DTT (buffer A) by using a peristaltic pump with a flow rate of 0.16 ml/min−1. The column was washed extensively with the same buffer, and the biotin-containing proteins were eluted from the column with buffer A containing 1 mM d-biotin. Fractions of approximately 2 ml were collected, and aliquots of each sample were analyzed for the activity of acetyl-CoA carboxylase or propionyl-CoA carboxylase. Active fractions were pooled and concentrated by using the Ultrafree-15 (30-kDa cutoff) centrifugal filter device (Millipore, Bedford, Mass.). The following steps were performed using the fast protein liquid chromatography system with a flow rate of 0.6 ml min−1. The concentrated sample was subjected to a Superose 6 HR 10/30 column (Amersham Pharmacia) which had been equilibrated with 3 column volumes of buffer A. Fractions of approximately 1 ml were collected by the fraction collector, and aliquots of each fraction were analyzed for enzymatic activity. The purity of the enzyme was checked by SDS-PAGE (15% polyacrylamide).
Enzyme assay.
The activity of acetyl-CoA carboxylase or propionyl-CoA carboxylase was assayed at 70°C by measuring incorporation of the radioactivity of NaH14CO3 into an acid-stable product according to the procedure of Roessler (52) with slight modifications. An appropriate amount of diluted enzyme was added to a reaction mixture containing 100 mM Tricine-KOH buffer (pH 7.5), 4 mM MgCl2 · 6H2O, 2 mM ATP, 10 mM KCl, 5 mM DTT, and 10 mM NaHCO3 (final concentrations). The mixtures were preincubated in a water bath at 70°C with 10 mM NaH14CO3 for 5 min. Then the reactions were started by addition of a substrate: 0.4 mM acetyl-CoA (for the acetyl-CoA carboxylase assay) or 0.4 mM propionyl-CoA (for the propionyl-CoA carboxylase assay). In a blank, the substrate was replaced with distilled water. The reaction solution (300 μl) was further incubated at 70°C with gentle shaking for 2 min. The reaction was stopped by addition of 200 μl of 2 N HCl and was allowed to stand at room temperature for 15 min. A 200-μl volume of the solution was taken into a scintillation vial and dried at 70°C for 2 h. The residual acid-stable products were dissolved with 0.5 ml of 0.2 N HCl. Then the scintillation cocktail was added, and the total solution was mixed well. The amount of radioactivity counted for the samples minus one for the blank was used to calculate the amount of fixed CO2. One unit of activity was defined as the amount of enzyme required to catalyze the incorporation of 1 μmol of NaH14CO3 per min. This assay was used to measure acetyl-CoA carboxylase or propionyl-CoA carboxylase activities during the purification procedures and to determine the thermostability, optimum pH, and optimum temperature for acetyl-CoA carboxylase activity as well as the effects of various components on the carboxylases.
Characterization of the purified enzyme.
The activity of biotin-dependent carboxylase was measured either in the presence or in the absence of avidin. One unit of avidin (1 nmol) was added to an assay mixture containing the diluted enzyme (0.0024 nmol) before the substrate was added. The activity of acetyl-CoA carboxylase or propionyl-CoA carboxylase was measured as described above. To protect enzyme inhibition, an excessive amount of free biotin (100 nmol) was added to avidin prior to its addition to the enzyme solution.
The molecular mass of the native enzyme was determined by gel filtration on a Sephacryl S-300 column (Amersham Pharmacia) at a flow rate of 1.0 ml min−1 by using 100 mM KH2PO4 containing 200 mM NaCl (pH 7.5) as the elution buffer. The molecular mass of the purified enzyme was calculated from the average relative mobility compared to the molecular weight standards (Bio-Rad). The molecular weight of the subunit was determined by SDS-15% PAGE by using low-range molecular weight markers as standard proteins. The subunit structure of the holoenzyme was estimated from the molar ratio of the amounts of protein in the individual bands on SDS-PAGE.
Optimum pH or optimum temperature was measured by using 2 μl of the purified enzyme (1.3 μg). pH dependence was determined by measuring the activity at 70°C in buffers at various pHs. The buffers used were 100 mM acetate (pH 4.0 to 5.5), 100 mM morpholinoethanesulfonic acid (MES; pH 5.5 to 6.5), 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.0 to 7.5), 100 mM HEPES (pH 6.5 to 8.5), 100 mM N-(2-hydroxyethyl)piperazine-N′-(3-propanesulfonic acid) (EPPS; pH 7.5 to 8.5), 100 mM Tricine (pH 7.5 to 8.5), and 2-(N-cyclohexylamido)ethanesulfonic acid (CHES; pH 8.5 to 10.0). The pH was adjusted at 25°C. The pH shift of the buffers at a high temperature was also measured at 70°C. Temperature dependence at temperatures ranging from 20 to 95°C was determined as described above.
The thermostability of the purified enzyme (65 μg in 0.2 ml; pH 7.5) was determined in sealed vials. Incubation was performed at 50, 60, 70, 80, 90, or 100°C for as long as 120 min. After the heat treatment, the solution was cooled immediately in an ice bath, and the remaining enzymatic activity was measured at 70°C.
The effects of various substances on acetyl-CoA carboxylase activity were examined at 70°C after preincubation of the purified enzyme with the following reagents on ice for 5 min: ATP at 0.01 to 10 mM, metal ions (MgCl2, MnCl2, FeCl2, CoCl2, CuCl2, CaCl2, or NiCl2) at 0.5 to 20 mM, salts (NaCl or KCl) at 2.5 to 200 mM, sulfate at 10 to 300 mM, citrate at 2.5 to 40 mM, and DTT at 0.5 to 20 mM.
The steady-state kinetic parameters(Km and Vmax) were measured at optimum conditions for acetyl-CoA carboxylase activity. Reactions were carried out at 1.0 mM ATP and various substrate (acetyl-CoA or propionyl-CoA) concentrations from 0.025 to 5.0 mM. To determine the Km values for ATP, 2.2 mM acetyl-CoA or 1.2 mM propionyl-CoA was used and the ATP concentration was varied from 0.01 to 4.0 mM. The kinetic parameters were calculated by fitting initial rate data to the Michaelis-Menten equation as described by Sakoda and Hiromi (54).
The effects of the metabolites in the 3-hydroxypropionate cycle and related substances (see Results) on the activities of acetyl-CoA and propionyl-CoA carboxylases were also examined. The assay mixture which contained the components in optimum concentrations was treated with the metabolites (at 0.5 mM for CoA derivatives and 1.0 mM for the other compounds) for 5 min on ice prior to measurement of enzymatic activities.
Protein quantitation.
Protein concentrations were determined by the Bio-Rad protein assay method with the bovine serum albumin of Bio-Rad protein assay standard II as a standard protein.
N-terminal amino acid sequencing.
The purified enzyme (6 μg) was separated on an SDS-15% PAGE gel and transferred to a Sequi-Blot polyvinylidene difluoride membrane (Bio-Rad) by using a semiblot dryer (ATTO, Tokyo, Japan). Electroblotting was carried out for 3 h with blotting buffer (Towbin buffer, comprising 25 mM Tris, 0.2 M glycine, and 20% [vol/vol] methanol). Protein bands were visualized by staining with Coomassie brilliant blue R-250. Protein bands corresponding to each subunit were excised, and their N termini were sequenced by automated Edman degradation using a model 491 protein sequencer (Procise; Applied Biosystems).
Preparation of DNA probe for cloning of acyl-CoA carboxylase genes.
Degenerate oligonucleotide primers were designed based on the N-terminal sequences of the acyl-CoA carboxylase. The upstream primer was 5′-GCXYTXGTXGCXAAYMGXGGXGARATHGC-3′, and the downstream primer was 5′-GCCATDATRTAXGTRTCXCCXGTRTCXGCRTA-3′. The 1.6-kb fragments were obtained from PCR amplification using A. brierleyi genomic DNA as a template, and the amplified fragments were then recovered from an agarose gel by using the Geneclean II kit (Bio 101, La Jolla, Calif.). The PCR product was cloned into the pGEM T-Easy vector (Promega, Madison, Wis.) according to the manufacturer's instructions. Several positive clones were picked up, and plasmids were isolated by the alkaline lysis method. For DNA sequencing, plasmids with an insert were prepared by using the Wizard DNA purification kit (Promega). The 1.4-kb EcoRI fragments from the plasmids were labeled with a PCR digoxigenin labeling kit (Roche, Mannheim, Germany) and used as the DNA probe for Southern hybridization. All of the other DNA manipulation techniques were performed according to standard protocols (55).
Cloning of acyl-CoA carboxylase genes from A. brierleyi genomic DNA.
The genomic DNA of A. brierleyi was prepared by the method of Marmur (35). The DNA was digested with various restriction enzymes and probed with the labeled DNA for the Southern hybridization experiment. Detection of the possible acyl-CoA carboxylase gene was performed by developing the color of alkaline phosphatase-conjugated anti-digoxigenin with the substrates 5-bromo-4-chloro-3-indolyl-phosphate (X-phosphate) and nitroblue tetrazolium. Two positive XbaI fragments of 8.0 and 3.5 kb were chosen to construct the genomic DNA library by using pUC19 as a cloning vector and Escherichia coli JM109 as a host strain. Transformants with positive signals were screened by colony hybridization using the labeled DNA fragment as described above. The small fragment (3.5 kb) was subcloned by digestion with EcoRI, and the large fragment (8.0 kb) was subcloned by digestion with PstI. All subcloned fragments were ligated into pUC19 and sequenced.
DNA sequencing and sequence analysis.
DNA sequencing was carried out by the primer walking method and was done from both strands with a DNA sequencing kit (Big Dye terminator sequencing ready reaction kit; Applied Biosystems) using specific primers. The nucleotide sequences of the acyl-CoA carboxylase genes and flanking regions were determined by a model 377 DNA sequencer (ABI PRISM; Applied Biosystems). A homology search was carried out by using the BLAST program at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/BLAST/). Identification of open reading frames and multiple sequence alignment were performed using the GENETYX-MAC program (Software Development, Tokyo, Japan).
Organisms used for amino acid sequence alignment and accession numbers.
The following organisms (with GenBank accession numbers in parentheses) were used for amino acid sequence alignment: Arabidopsis thaliana (AF360279), Brassica napus (AY034410), Glycine max (soybean; AF163150), Oryza sativa (rice; AC092548), Gallus yallus (chicken; J03541), Rattus norvegicus (rat; J03808), Cyclotella cryptica (diatom; L20784), Saccharomyces cerevisiae (yeast; NC001146), Aspergillus niger (mold; Y15996), Anabaena sp. (L14862), Nostoc sp. (NC003272), Synechocystis sp. (NC000911), Aquifex aeolicus (NC000918), Bacillus subtilis (NC000964), Corynebacterium glutamicum (U35023), E. coli K-12 (NC000913), Lactobacillus plantarum (AB025973), Mycobacterium tuberculosis (NC000962), Myxococcus xanthus (AB039884), Neisseria meningitidis (NC003112), Pseudomonas aeruginosa (NC002516), Salmonella enterica serovar Typhimurium (NC003197), Vibrio cholerae (NC002505), A. brierleyi (this study), Archaeoglobus fulgidus (NC000917), Halobacterium sp. (AE005066), Methanobacterium thermoautotrophicum (NC000916), S. metallicus (AF042099), Sulfolobus solfataricus (AE006641), and Sulfolobus tokodaii (NC003106).
Nucleotide sequence accession number.
The sequences of A. brierleyi acyl-CoA carboxylase genes and flanking regions were submitted to the DNA Data Bank of Japan (DDBJ) under accession no. AB088419.
RESULTS
Detection of biotin-containing proteins.
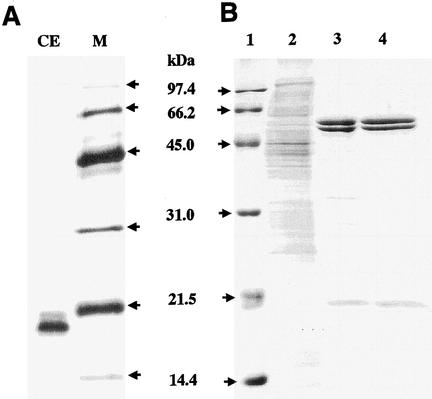
A single biotinylated protein (Fig. 1A) was detected in the cell extract of A. brierleyi by Western blot analysis using a streptavidin-peroxidase conjugate. The biotinylated protein on an SDS-15% PAGE gel was shown to have a molecular mass of approximately 20 kDa. In-gel staining after native PAGE (data not shown) confirmed that A. brierleyi had a single biotinylated protein within the cells. Interestingly, the activities of both acetyl-CoA and propionyl-CoA carboxylases were detected in the cell extract.
FIG. 1.
(A) Detection of biotin-containing proteins in an A. brierleyi cell extract by Western blot analysis. The cell extract was separated on an SDS-15% PAGE gel and then probed with a streptavidin-peroxidase conjugate. A single biotinylated protein was detected by the procedure described in Materials and Methods. (B) SDS-PAGE of the purified acyl-CoA carboxylase from A. brierleyi. Lane 1, molecular mass markers; lane 2, 4 μg of cell extract; lane 3, 4 μg of partially purified enzyme from the streptavidin column; lane 4, 4 μg of the purified enzyme from the Superose 6 column. Proteins were separated on a 15% polyacrylamide gel and stained with Coomassie brilliant blue.
Purification of acyl-CoA carboxylase.
Acyl-CoA carboxylase was purified by affinity chromatography and gel filtration chromatography (Table 1). A typical elution profile from streptavidin column chromatography resulted in a broad peak as determined by absorbance at 280 nm when the enzyme was eluted with 1.0 mM d-biotin. The partially purified enzyme was almost homogeneous as judged by SDS-15% PAGE (Fig. 1B). With respect to acetyl-CoA and propionyl-CoA carboxylation, all active fractions had both acetyl-CoA and propionyl-CoA carboxylase activities. The concentrated fractions from a streptavidin column were further subjected to a Superose 6 column. During purification by this column, the enzyme was eluted as a single sharp protein peak, which still had two carboxylase activities. The activities of the two carboxylases were completely inhibited by addition of 1 nmol of avidin to the assay mixture. However, the enzymatic activities could be restored almost to 100% if avidin was incubated with an excessive amount of free biotin (100 nmol) prior to its addition to the enzyme. This result showed that the purified enzyme is a biotin-dependent carboxylase. When we calculated the amount of acyl-CoA carboxylase (in terms of acetyl-CoA carboxylase) by using the numbers in Table 1, acyl-CoA carboxylase was shown to constitute approximately 4% of the total proteins in the cell extract. It had been indicated that an abundant amount of acyl-CoA carboxylase was induced during autotrophic growth to function as a carboxylating enzyme in the modified 3-hydroxypropionate cycle in A. brierleyi (21). The specific activities of acetyl-CoA and propionyl-CoA carboxylases in the combined fractions were 14.5 ± 1.01 and 11.9 ± 0.92 μmol of fixed CO2 min−1 mg−1, respectively. Also, the ratio between the acetyl-CoA and propionyl-CoA carboxylase activities was almost constant throughout the purifications (Table 1).
TABLE 1.
Purification of the A. brierleyi acyl-CoA carboxylasea
| Step | Amt of protein (mg) | Activity (U)b of:
|
Sp act (U/mg) of:
|
ACC/PCC ratio | ACC yield (%) | ACC purification (fold) | ||
|---|---|---|---|---|---|---|---|---|
| ACC | PCC | ACC | PCC | |||||
| Cell extract | 558 | 340 | 298 | 0.6 | 0.5 | 1.20 | 100 | 1 |
| Streptavidin | 4.14 | 52.5 | 41.2 | 12.7 | 10.0 | 1.27 | 15 | 21.2 |
| Superose 6 | 1.96 | 28.4 | 23.4 | 14.5 | 11.9 | 1.22 | 8.4 | 24.2 |
From ca. 10 g of cells (wet weight).
The activities of acetyl-CoA carboxylase (ACC) and propionyl-CoA carboxylase (PCC) were measured at 70°C. One unit of enzymatic activity is defined as the amount of enzyme which incorporates 1 μmol of NaH14CO3 per min.
Molecular properties of acyl-CoA carboxylase.
Since the purified enzyme was shown to be the biotin-dependent carboxylase and was dependent on ATP, bicarbonate, and acetyl-CoA or propionyl-CoA as the substrates (data not shown), the purified enzyme should be recognized as acyl-CoA carboxylase, although the end product has not been determined. The purified enzyme was found to be composed of three subunits when it was subjected to SDS-15% PAGE. The estimated molecular masses of the subunits were found to be 62, 59, and 20 kDa. The 20-kDa peptide was shown to contain biotin as a prosthetic group based on specific detection with a streptavidin-peroxidase conjugate (data not shown). Comparison of the subunit structures of all biotin enzymes (66) showed that acyl-CoA carboxylase of A. brierleyi exhibits a unique subunit structure which lies between the eukaryotic type of acetyl-CoA carboxylase (with three functional units in one protein [α]) and the bacterial type (with three functional units in four different proteins [αβγ1γ2]). Overall, A. brierleyi acyl-CoA carboxylase seems to be close to the bacterial acetyl-CoA carboxylase (37).
The apparent native molecular mass of acyl-CoA carboxylase was approximately 530 to 540 kDa. The ratio of the amounts of 62-, 59-, and 20-kDa subunits on Coomassie brilliant blue-stained gels was estimated to be approximately 3:3:1. The expected ratio for equimolar amounts of subunits is 1:1:1. This result fits an α4β4γ4 subunit structure.
The N-terminal amino acid sequence from each subunit was determined and compared to those in the database. The N-terminal amino acid of the 62-kDa subunit was blocked, whereas the N-terminal amino acid sequence of the 59-kDa subunit was PPFSRVLVANRGEIAT (75% identical to the biotin carboxylase [BC] of S. metallicus; see Materials and Methods) and that of the 20-kDa subunit was MKLYRAYADTGDTYIMAIDS (55% identical to the biotin carboxyl carrier protein [BCCP] of S. metallicus).
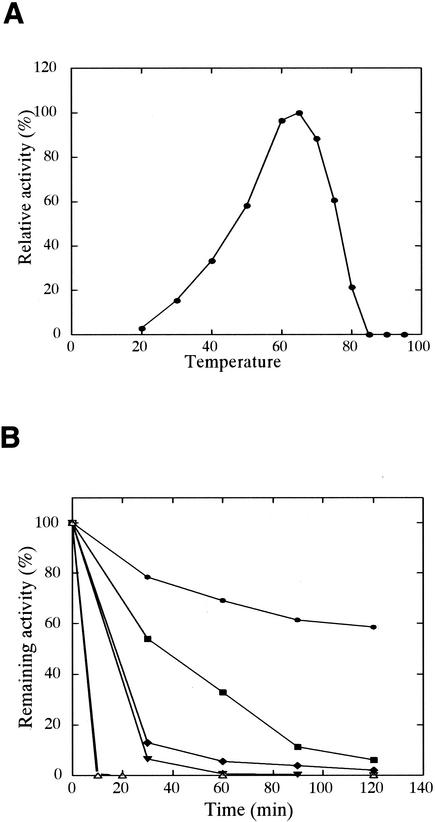
Optimum temperature and stability.
When acyl-CoA carboxylase from A. brierleyi was assayed at temperatures ranging from 20 to 100°C, it exhibited the highest activity at 65°C (Fig. 2A). The activity of acetyl-CoA carboxylase drastically decreased when it was assayed at temperatures higher than its growth temperature (70°C).
FIG. 2.
Determination of the optimum temperature (°C) (A) and thermostability (B) of acyl-CoA carboxylase. To determine temperature dependence, the enzyme was preincubated and its activity was measured at the tested temperature. To determine thermostability, the enzyme was treated at various temperatures: 50°C (•), 60°C (▪), 70°C (♦), 80°C (▾), 90°C (○), and 100°C (▵). The remaining enzymatic activity was measured at 70°C as described in Materials and Methods.
The thermostability of the purified enzyme was determined at 50 to 100°C (Fig. 2B) in 100 mM Tricine-KOH (pH 7.5) containing 10 mM KCl. The activity of acetyl-CoA carboxylase gradually decreased at 50°C, with a residual activity of around 60% after 2 h of incubation. At 60°C, the residual activity of the enzyme was 50% after incubation for 35 to 40 min, whereas the residual activities at 70 and 80°C were less than 15%. The enzyme completely lost its activity within 10 min when it was incubated at 90°C or above.
Catalytic properties.
The optimum pH of the purified enzyme was slightly acidic (pH 6.4 to 6.9 at 70°C). At the optimum pH, the acetyl-CoA carboxylase activities in HEPES, Tricine, and EPPS were not significantly different. Therefore, Tricine buffer was chosen throughout the characterization of the purified enzyme as performed in the previous study (21).
The purified enzyme was absolutely dependent on acetyl-CoA or propionyl-CoA, ATP, Mg2+, and HCO3− for activity. The enzyme exhibited the maximum activity when the ATP concentration was 1.0 to 1.5 mM. An excess of ATP (>2.0 mM) had an inhibitory effect on the enzymatic activities. Therefore, we chose the optimum concentration of ATP at 1.0 mM for further study of enzyme kinetics. The enzyme was quite specific for ATP. In fact, neither ADP, AMP, GTP, CTP, UTP, nor dTTP (0.2 mM) could be substituted for ATP.
The effects of divalent cations (as the chloride salts) and their concentrations were examined in 100 mM Tricine buffer containing 2.0 mM ATP, 10.0 mM KCl, and 5.0 mM DTT. The maximum acetyl-CoA carboxylase activity was obtained at 4.0 mM Mg2+, whereas 1.0 mM Mn2+ gave 82% of the maximum activity obtained with the Mg2+ ion. Both Mg2+ and Mn2+ were inhibitory when each was present in the assay at a concentration higher than 4.0 or 2.0 mM, respectively. Moreover, 26% of the maximum activity was observed when Co2+ (10 to 15 mM) was used as a divalent cation in an assay mixture. Other cations such as Fe2+, Cu2+, Ca2+, and Ni2+ (0.5 to 20 mM) had no effect on acetyl-CoA carboxylase activity. The effects of the monovalent cations Na+ and K+ (both were used as chloride salts) were determined at concentrations ranging from 0 to 200 mM. Both Na+ and K+ had inhibitory effects on acetyl-CoA carboxylase activity with similar patterns. The enzyme was less than 50% activated when the concentration was higher than 40 mM. Citrate and sulfate (both were used as K+ ions) inhibited acetyl-CoA carboxylase activity by more than 50% at concentrations of 3 and 25 mM, respectively. DTT also inhibited acetyl-CoA carboxylase slightly, by approximately 12%, at a final concentration of 5 to 10 mM.
Kinetic constants were determined with the optimum concentrations of the components in the following assay mixture: 100 mM Tricine-KOH (pH 7.5) containing 1.0 mM ATP, 4.0 mM MgCl2 · 6H2O, and 10.0 mM NaHCO3. The purified enzyme exhibited typical Michaelis-Menten kinetics when it was assayed with increasing concentrations of the substrate (acetyl-CoA or propionyl-CoA) at a constant concentration of ATP (1.0 mM). Propionyl-CoA carboxylase activity was slightly higher than acetyl-CoA carboxylase activity at low substrate concentrations (0.05 to 0.50 mM), whereas the activity of acetyl-CoA carboxylase was higher than that of propionyl-CoA carboxylase when the substrate concentration was above 3.0 mM. The apparent Km for acetyl-CoA was 0.17 ± 0.03 mM, with a Vmax of 43.3 ± 2.8 U mg−1, and the Km for propionyl-CoA was 0.10 ± 0.008 mM, with a Vmax of 40.8 ± 1.0 U mg−1. These results indicate that propionyl-CoA is preferable to acetyl-CoA as a substrate for the enzyme. The Km values for ATP of acetyl-CoA carboxylase (0.04 ± 0.002 mM) and propionyl-CoA carboxylase (0.05 ± 0.005 mM) were quite similar. The enzyme also carboxylated a small amount of n-butyryl-CoA (0.5 mM) as a substrate at a low rate (4.6%) relative to that for acetyl-CoA or propionyl-CoA. Neither succinyl-CoA, palmitoyl-CoA, acetate (plus 0.5 mM CoASH), propionate (plus 0.5 mM CoASH), nor pyruvate was carboxylated by the enzyme.
Effects of metabolites on the purified enzyme.
Malonyl-CoA (0.5 mM), methylmalonyl-CoA (0.5 mM), succinyl-CoA (0.5 mM), and CoASH (1 mM) were shown to be negative effectors for both acetyl-CoA and propionyl-CoA carboxylases when 0.4 mM acetyl-CoA or 0.4 mM propionyl-CoA was used as a substrate, respectively. The activity of acetyl-CoA carboxylase was strongly inhibited by succinyl-CoA (84%) and methylmalonyl-CoA (79%), whereas inhibition of propionyl-CoA carboxylase activity was 57% for succinyl-CoA and 60% for methylmalonyl-CoA. Interestingly, malonyl-CoA and CoASH inhibited acetyl-CoA carboxylase activity (50 to 57%), but only 11 to 13% inhibition of propionyl-CoA carboxylase was observed. We also found that 0.5 mM palmitoyl-CoA, a specific inhibitor of most acetyl-CoA carboxylases, had a slight stimulative effect on acetyl-CoA carboxylase (116%) and propionyl-CoA carboxylase (135%) from A. brierleyi. Other effectors such as NADP+, NADPH, 3-hydroxypropionate, succinate, fumarate, malate, and glyoxylate had no significant effect on acetyl-CoA carboxylase or propionyl-CoA carboxylase activity.
Cloning and nucleotide sequencing of acyl-CoA carboxylase genes.
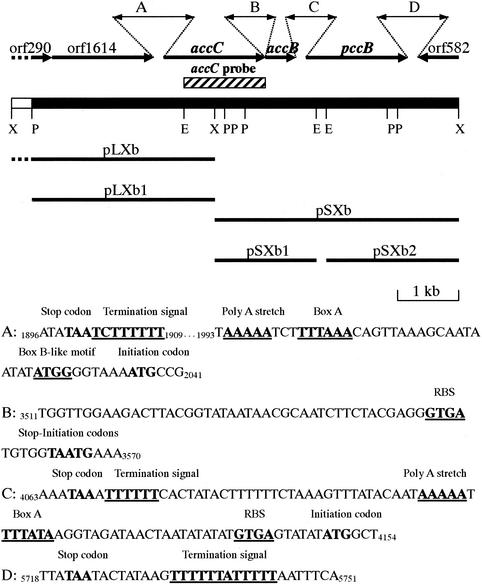
In order to construct a probe for Southern hybridization, degenerated primers were designed based on the N-terminal amino acid sequences of the purified 59- and 20-kDa subunits, which correspond to putative BC and BCCP, respectively. A fragment of approximately 1.6 kb was amplified by PCR when primers of the sense sequence for BC and the antisense sequence for BCCP were used. The size of the PCR fragment was the same as the predicted size of the BC gene, indicating that the BCCP gene follows the BC gene. The PCR fragment was ligated into the pGEM T-Easy vector. Positive clones with the insert were screened by blue-white colony selection and then digested with several restriction enzymes in order to check the restriction map of the partial BC gene. A clone with the suitable insert was sequenced to construct a restriction map. A 1.4-kb EcoRI fragment of the cloned PCR fragment was isolated and labeled with digoxigenin as a hybridization probe for Southern blotting. Specific signals were observed against a 10-kb BamHI fragment, a 2.1-kb EcoRI fragment, a 17-kb SacI fragment, 2.5- and 3.0-kb PstI fragments, and 3.5- and 8.0-kb XbaI fragments. Among these, the two XbaI fragments were selected to be cloned into pUC19 to generate plasmids pLXb (with an 8.0-kb insert) and pSXb (with a 3.5-kb insert) (Fig. 3). Both clones were checked in terms of the insert size and restriction map by single or double digestions with the enzymes mentioned above. The 3.5- and 8.0-kb XbaI fragments were subcloned by digestion with EcoRI and PstI, respectively. Nucleotide sequences of the subcloned fragments were determined from both strands by using synthetic oligonucleotide primers.
FIG. 3.
(Top) Organization of the genes encoding the acyl-CoA carboxylase of A. brierleyi. The accC, accB, and pccB genes encode the BC, BCCP, and CT subunits, respectively. Selected restriction sites are indicated below the filled rectangle (and the open box, representing the partial sequence), as follows: X, XbaI; P, PstI; E, EcoRI. Hatched rectangle, DNA fragment used as a probe for Southern hybridization. Thick bars, cloned and subcloned fragments. (Bottom) Nucleotide sequences of fragments A through D delineated in the diagram above. Stop codons and initiation codons are boldfaced. Termination signals, promoter sequences, and RBSs are underlined.
Acyl-CoA carboxylase genes and sequence analysis.
A 6.4-kb nucleotide sequence from plasmids pLXb and pSXb was determined and analyzed. Four complete open reading frames (ORFs) and two partial ORFs were identified in this region (Fig. 3). A homology search of the databases using the BLAST program showed that the first, partial ORF (orf290; positions 1 to 290) encodes a protein similar to the ATP-binding proteins of ABC transporters. The second ORF (orf1614; positions 283 to 1896) encodes a protein that is 55% identical at the amino acid level with the ABC transporter of S. solfataricus. The third ORF (accC; positions 2036 to 3565) codes for a protein of 509 amino acids. The fourth ORF (accB; positions 3565 to 4068) codes for a protein of 167 amino acids. A homology search of the deduced amino acid sequences of accC and accB revealed that they encode certain BC and BCCP subunits, respectively. The accC and accB gene products had their highest similarities with BC and BCCP of S. tokodaii, 77 and 64%, respectively. The first nucleotide of the initiation codon of accB overlapped with the third nucleotide of the stop codon of accC. Two ATG triplets were found in the same frame of the fifth ORF (pccB; position 4149 or 4176 to 5723), encoding a putative carboxyltransferase (CT) subunit. A typical ribosome-binding site (RBS) preceded the first ATG triplet, as in the putative CT genes of S. tokodaii and S. solfataricus, but the second ATG triplet was not preceded by an RBS, suggesting that the first ATG should be the initiation codon and that pccB encodes a protein of 524 amino acid residues. A homology search of proteins in the databases revealed that the pccB gene product had its highest similarities to the methylmalonyl-CoA decarboxylase (α) subunit of S. tokodaii, with 79% identity, and the CT of S. metallicus, with 78% identity, indicating that the gene product of pccB is certainly CT. The sixth, partial ORF was found downstream of pccB on the complementary strand (orf582; positions 5830 to 6411). The deduced amino acid sequence of this ORF was similar to that of helicase from S. tokodaii or S. solfataricus.
The archaeal genes are frequently organized in transcriptional units (47). We therefore searched for the promoter sequences, RBSs, and transcription termination signals of the A. brierleyi acyl-CoA carboxylase genes by comparison with the known sequences of Archaea. The putative strong promoters TTTAAA and TTTATA, which have been identified as the Box A motif, were found approximately 30 nucleotides upstream of the accC and pccB genes, respectively (Fig. 3). The poly(A) sequences that determine the polarity of transcription (5, 65) were also found within 6 nucleotides upstream of the Box A motif (Fig. 3). Another promoter region, which is known as the Box B motif (ATGC [65] or TGC [70]), was not found within the acyl-CoA carboxylase genes, but a Box B-like sequence (ATGG) was observed upstream of the accC gene (Fig. 3). GTGA is the typical RBS or Shine-Dalgarno sequence, which is frequently utilized in Archaea (11, 12). The accB and pccB genes of A. brierleyi were also preceded by this motif with a normal distance as found in Bacteria (Fig. 3). The possible transcription termination signal in Archaea (47, 70), which is a polythymidine stretch, was located immediately downstream of the accB and pccB genes (Fig. 3). The intergenic region between accB and pccB spans 80 nucleotides. This region was found to be AT rich (39.25% A, 41.12% T) when it was compared to the accC (36.93% A, 26.60% T), accB (42.26% A, 24.60% T), and pccB (35.11% A, 28.89% T) coding regions. The result indicates that accC and accB were cotranscribed as an operon, whereas pccB was transcribed as a monocistronic mRNA.
Deduced properties of BC.
The accC gene product (BC) was shown to be composed of 509 amino acids with a calculated molecular mass of 57.1 kDa. The molecular mass matches the SDS-PAGE result (59 kDa). The N-terminal amino acid residues of the 59-kDa subunit of the purified enzyme completely matched the deduced amino acid sequence of accC. Multiple sequence alignment of the BC proteins among three domains of organisms (9 eukaryotes, 14 bacteria, and 7 archaea [see Materials and Methods]) revealed that archaeal amino acid sequences are closer to those from Bacteria than to those from Eucarya. The putative BC of A. brierleyi had 42 to 48% and 28 to 47% sequence identities to the BCs of Bacteria (except in the case of M. tuberculosis) and Eucarya, respectively. Several motifs of conserved amino acid residues in A. brierleyi were similar to known sequences of the other organisms. A10NRGEIA16 is obviously conserved near the N termini of most bacteria and archaea but is not conserved in eukaryotes. The glycine-rich region, which had been proposed to be the ATP-binding site in other biotin enzymes (2, 6, 14, 22, 25, 26, 28, 32, 53, 59, 61, 73), was also found in A. brierleyi as a GGGGVG sequence. Most acetyl-CoA carboxylases from Eucarya, Bacteria, and Archaea share the common sequence GGGGXG, where X is an amino acid that varies depending on the organism. The GGGG(K/R)G sequence can usually be found in Eucarya, whereas GGGG(R/I/K/M)G is present in Bacteria. In Archaea, we obtained the sequence GG(G/A)G(I/V/M/R/T)G, which is more diverse than that for Eucarya or Bacteria. The adjacent amino acid following this sequence is usually a hydrophobic residue (40). However, it was discovered later that the glycine-rich sequence of BC or related enzymes does not participate in the binding of ATP but may play an important role as a lid for the active site of the enzymes (23). Recently, the four active-site residues of BC, K116, K159, H209, and E276, which are highly conserved in the BCs, were shown to be involved in the binding of ATP (60). We also found the residues K116, K160, H210, and E277 in the sequence of A. brierleyi BC.
Deduced properties of BCCP.
The accB gene product (BCCP) was determined to be the biotinylated subunit of A. brierleyi acyl-CoA carboxylase, which is composed of 167 amino acids with a calculated molecular mass of 18.6 kDa. The molecular mass was in good accordance with the results of SDS-PAGE (20 kDa). The N-terminal amino acid sequence indicated that the ATG triplet (positions 3565 to 3567) is the initiation codon of BCCP rather than the TTG triplet (positions 3514 to 3516) or the GTG triplet (positions 3559 to 3561). Multiple sequence alignment of various BCCPs from A. brierleyi, S. metallicus, S. tokodaii, S. solfataricus, Eucarya, and many bacteria (as in the comparison of BCs) showed strong conservation of the amino acid sequence around the biotin-binding site and flanking residues. EAMK35M (position relative to the C terminus) is the typical sequence for conserved amino acid residues around the biotin-binding site in various biotin-containing proteins. The lysine residue (boldfaced) is the biotinylation site (56), which is found 35 amino acid residues away from the C terminus in most biotin enzymes. In the case of A. brierleyi, we found EAMK35S, where the second methionine is replaced by serine. The conserved glycine and proline residues flanking the biotin-binding site (56) were located at positions (counted from lysine as the zero position) P−29, G−16, G−10, and G+11 in the BCCP subunit of A. brierleyi as well as in most of the other organisms. Proline could be found at least once in the range of positions −25 to −36 (53, 59). An Ala-Pro-rich sequence that acts as a mobile spacer to allow the biotinylated portion to move between active sites of the acetyl-CoA carboxylase subunit was not found within the A. brierleyi BCCP.
Deduced properties of CT.
The pccB gene product (CT) of A. brierleyi was composed of 524 amino acids with a calculated molecular mass of 57.3 kDa. This result is in accordance with the SDS-PAGE result (62 kDa), suggesting that the 62-kDa subunit is the pccB gene product. The alignment of amino acid sequences around the substrate-binding sites of acetyl-CoA carboxylases and propionyl-CoA carboxylases from a diatom (C. cryptica), plant (alfalfa), yeast (S. cerevisiae), bacteria (E. coli, M. tuberculosis, B. subtilis, C. glutamicum, and M. xanthus), and archaea (A. brierleyi, S. solfataricus, and S. metallicus) showed that the archaeal sequences were more similar to the bacterial than to the eukaryotic sequences. The putative CT of A. brierleyi had 19 to 26% sequence identity to the corresponding regions in the diatom, alfalfa, and yeast enzymes, whereas it had 17 to 29% and 20 to 29% identities with the alpha and beta chains of CTs in E. coli, M. tuberculosis, and B. subtilis, respectively. However, A. brierleyi CT was similar to the CTs of C. glutamicum and M. xanthus, with 48 to 53% identity. Multiple sequence alignment of amino acid residues around the acyl-CoA binding site in eukaryotes, bacteria, and archaea (as mentioned above) indicated that the acyl-CoA binding site (2, 33, 53, 59) in the CT subunit of A. brierleyi was Q315NIVVGFARVAGNVIGIVA333 (amino acid positions relative to the N terminus; conserved amino acids are boldfaced), in which three glycine residues (G320, G326, and G330) and one arginine (R323) were highly conserved. This result corresponds to the report that arginine residues could be present in the vicinity of the acyl-CoA binding site of the elongase from etiolated leek seedlings (57). The carboxybiotin binding site of A. brierleyi was found to be G92RTVFAFSQDFTELGGTLGETHANKIGKVYELALKVGAPVIGINDSGGARI142. We found three conserved glycine residues (G92, G107, and G139) in this motif also.
DISCUSSION
It is known that the biotin-containing proteins are rare in organisms; however, acetyl-CoA carboxylase, a member of the biotin enzyme family, is always present in all kinds of organisms, including archaea. Although E. coli (13) and A. brierleyi each have a single biotinylated protein within the cells, the acyl-CoA carboxylase of A. brierleyi has both acetyl-CoA and propionyl-CoA carboxylase activities, in contrast to the case of E. coli, in which only acetyl-CoA carboxylase activity was detected. The presence of a single biotinylated protein in a cell extract (Fig. 1A) of A. brierleyi strongly suggests the bifunctional characteristic of the purified enzyme in the modified 3-hydroxypropionate cycle.
The archeal acyl-CoA carboxylase of A. brierleyi contains three different subunits for the functions of BC, BCCP, and CT. The subunit structure is distinct from those of the other known biotin enzymes. In Eucarya all three functional units are fused into a single peptide, but in Bacteria they can be separated into four subunits (BC, BCCP, CTα, and CTβ). On the other hand, two nonidentical subunits (BC-BCCP and CT) were found in all propionyl-CoA carboxylases from both Bacteria and Eucarya. Moreover, the kinetic parameters of the purified enzyme indicated that acyl-CoA carboxylase had the specific activities of acetyl-CoA and propionyl-CoA carboxylases equally, and the Km for acetyl-CoA was only 1.6 times higher than the Km for propionyl-CoA. Both propionyl-CoA carboxylase from Nocardia mediterranei (15) and acyl-CoA carboxylase from M. tuberculosis, M. bovis (46), and Propionibacterium shermanii (62) also had acetyl-CoA and propionyl-CoA carboxylase activities. Therefore, our results would suggest that the position of the A. brierleyi acyl-CoA carboxylase should be between those of propionyl-CoA carboxylase and the bacterial type of acetyl-CoA carboxylase and also that the enzyme should be named as an acyl-CoA carboxylase.
Due to the fact that A. brierleyi is a thermoacidophilic archaeon that grows autotrophically at 70°C and pH 2.0, the acyl-CoA carboxylase from this archaeon is a thermophilic enzyme and has maximum enzymatic activity at a slightly acidic pH, in contrast to the other acetyl-CoA carboxylases. Most acetyl-CoA carboxylases from mesophilic bacteria have their pH optima at a slightly alkaline pH (46, 52, 62, 69) and are sensitive to temperatures in the range from 35 to 45°C. The specific requirement of a divalent cation for enzymatic activities is similar to those reported for several organisms such as the diatom (52), maize leaf (44), and P. shermanii (62), which require Mg2+ or Mn2+. Moreover, we found that Co2+ can be used as a divalent cation in the carboxylation reaction, as is seen in P. shermanii (62). It is interesting that the maximum enzymatic activity of acetyl-CoA carboxylase was obtained when the concentration of Mg2+ was higher than that of ATP. These phenomena are in accordance with the notion of Nikolau and Hawke (44) that the MgATP complex is the substrate of acetyl-CoA carboxylase, while free Mg2+ acts as an activator, but free ATP is an inhibitor, of the enzyme. However, the activity of A. brierleyi acetyl-CoA carboxylase was inhibited by Mg2+ at concentrations higher than 5 mM. This result might be due to the mechanism of enzyme depolymerization by an excess of Mg2+ (4). Acetyl-CoA carboxylases from many sources require K+ for enzyme activation (44, 46, 58) or enzyme stability (52). In contrast to the findings of those reports, we found that the acetyl-CoA carboxylase activity of A. brierleyi was inhibited by NaCl or KCl, as observed in the acetyl-CoA carboxylase from the rainbow trout liver (36). Rainwater and Kolattukudy (46) reported that K+ stimulates the activities of acetyl-CoA and propionyl-CoA carboxylases, whereas the chloride ion inhibits propionyl-CoA carboxylase. It might be possible that A. brierleyi acyl-CoA carboxylase was also inhibited by the chloride ion, since NaCl and KCl gave similar results with respect to inhibition of acetyl-CoA carboxylase. Most acetyl-CoA carboxylases from animals such as the rainbow trout (36) and rat (67) were activated by citrate to form a polymeric state of the enzyme (4). However, we found that citrate does not activate the acyl-CoA carboxylase of A. brierleyi as it does the nematode (38), yeast (39), and N. mediterranei (15) enzymes. In contrast, A. brierleyi acyl-CoA carboxylase was inhibited by citrate even at concentrations lower than 5.0 mM. The inhibition of acyl-CoA carboxylase by citrate was probably due to formation of a complex with Mg2+ (44, 67).
The ATP concentration in an assay mixture was assumed to be a major factor in controlling the ratio of the enzymatic activity of acetyl-CoA carboxylase to that of propionyl-CoA carboxylase when the substrate concentration was fixed (data not shown). In the purification step of the enzyme (Table 1), the specific activity of acetyl-CoA carboxylase was slightly higher than that of propionyl-CoA carboxylase because the concentration of ATP used was 2.0 mM. After optimization of the assay mixture, ATP was used at 1.0 mM, which gave higher activity for propionyl-CoA carboxylase than for acetyl-CoA carboxylase. Interestingly, A. brierleyi acyl-CoA carboxylase had a Km for propionyl-CoA that was only 1.6 times lower than its Km for acetyl-CoA. This result differs from those for other organisms, in which the Km for propionyl-CoA was 3- to 14-fold lower than the Km for acetyl-CoA (15, 17, 27, 39, 46). These findings imply the real in vivo bifunctional characteristics of acetyl-CoA carboxylase and propionyl-CoA carboxylase in the modified 3-hydroxypropionate cycle.
The inhibition of acetyl-CoA and propionyl-CoA carboxylase activities of A. brierleyi by malonyl-CoA, methylmalonyl-CoA, succinyl-CoA, and CoASH is similar to the reported findings for the acetyl-CoA carboxylase which was isolated from a rat (42). However, the propionyl-CoA carboxylase of M. xanthus was able to carboxylate succinyl-CoA (27), and CoASH was reported to be an activator of acetyl-CoA carboxylase from spinach. Interestingly, CoASH had negative and positive effects on the activities of acetyl-CoA carboxylases although the enzymes were isolated from the same source (42, 72) but were different in the preparations. Palmitoyl-CoA, which is known to be a specific inhibitor of most acetyl-CoA carboxylases (42, 52, 67, 72), did not inhibit the activity of acetyl-CoA carboxylase or propionyl-CoA carboxylase but instead had a slightly stimulative effect on both activities. Yeh et al. (72) reported that palmitoyl-CoA inhibits CoA binding to the carboxylase at the CoA-binding site. In A. brierleyi, acyl-CoA carboxylase does not function as a key enzyme in fatty acid biosynthesis; therefore, it is reasonable that acyl-CoA derivatives such as palmitoyl-CoA do not affect the enzymatic activity. Although the binding motifs for palmitoyl-CoA in acetyl-CoA carboxylase have not been clarified yet, we presume that such binding motifs are missing in the amino acid sequences in A. brierleyi.
We have cloned and sequenced the genes encoding acyl-CoA carboxylase from a genomic library of A. brierleyi. The BC, BCCP, and CT subunits are encoded by the accC, accB, and pccB genes, respectively. These genes are clustered on the same strand. Although the organization of acetyl-CoA carboxylase genes varies in bacteria (6, 25, 26, 32-34), the accC and accB genes are usually adjacent to each other, except for Anabaena sp., where the genes are not linked and are separated by at least several kilobases (14). In E. coli, P. aeruginosa, and B. subtilis, the distances between these two genes are 10, 17, and 15 nucleotides, respectively, whereas the genes overlap by 1 nucleotide in A. brierleyi and S. metallicus (10) and by 7 nucleotides in S. tokodaii. Transcriptional analysis experiments revealed that the accC and accB genes in E. coli (32), P. aeruginosa (6), and B. subtilis (34) form a two-cistronic operon and are cotranscribed. We presume that accC and accB are cotranscribed but pccB is not in A. brierleyi, since a transcription termination signal was found immediately downstream of the accB gene. The intergenic region between the accB and pccB genes of A. brierleyi is AT rich, as observed for the E. coli homologue, which exhibits the characteristics of bent structure and slow migration on PAGE gels (43).
Although some properties of A. brierleyi acyl-CoA carboxylase were shown to be different from those of the other acetyl-CoA carboxylases, the enzymes share common features at the molecular level, as shown by the conservation of amino acid sequences. Since the enzyme is the biotin- and ATP-dependent carboxylase, which requires a divalent cation (Mg2+), bicarbonate, and acetyl-CoA (or propionyl-CoA) for the reaction, the conserved motifs of these substrates were found within the amino acid sequence. The ATP-binding motif and the biotin carboxylation site were proposed to be located on the BC subunit; however, the exact position of the biotin carboxylation site is still unclear (31).
The biotin-binding site on the BCCP subunit of A. brierleyi acyl-CoA carboxylase is strongly conserved, as is found in the other biotin enzymes such as acetyl-CoA carboxylase (6, 32, 45, 50, 53), propionyl-CoA carboxylase (8, 30), transcarboxylase (56), 3-methylcrotonoyl-CoA carboxylase (61), and methylmalonyl-CoA decarboxylase (7, 19). The amino acid residues flanking the biotin-binding site are also conserved, and it has been proposed that this region might be involved in recognition of the specific lysine for the biotinylation reaction by the biotin ligase (56). In contrast to the biotinylated subunit of acetyl-CoA carboxylases from E. coli (32), P. aeruginosa (6), and B. subtilis (34), including the methylmalonyl-CoA decarboxylase of Veillonella parvula (19) and Propionigenium modestum (7), the A. brierleyi BCCP does not contain an Ala-Pro-rich sequence. The presence of this sequence in the BCCP subunit results in an unusual behavior of the protein, slower migration on SDS-PAGE gels (32). However, the molecular mass of BCCP of A. brierleyi by SDS-PAGE (20 kDa) corresponds to the molecular mass of the deduced amino acid sequence (18.6 kDa), which reinforces the above discussion.
The carboxybiotin binding motif and the acyl-CoA binding motif are located on the CT subunit of A. brierleyi, unlike the CT subunit in most bacteria, which is usually composed of two nonidentical chains and has its substrate-binding sites located in different chains (25, 33). Alignment of amino acid sequences around the acyl-CoA binding site in Eucarya, Bacteria, and Archaea indicated that the carboxybiotin binding site occupies a larger number of amino acid residues than the acyl-CoA binding site. It seems likely that the substrate-binding sites of the CT subunit are less conserved than those of the BC and BCCP subunits. It is possible that there are variations of the substrate-binding pocket in the CT subunits that are specific for their acyl-CoA substrates (16), since the E. coli acetyl-CoA carboxylase recognized only acetyl-CoA as the substrate, but A. brierleyi acyl-CoA carboxylase catalyzed carboxylation of both acetyl-CoA and propionyl-CoA. Therefore, the bifunctional characteristic of acyl-CoA carboxylase from A. brierleyi may be reflected by the difference in the conserved amino acid residues in the CT subunit.
Although the three-dimensional structures of BC (68) and BCCP of E. coli (3, 49, 71) have been elucidated, the crystal structure of the CT or holoenzyme of acyl-CoA carboxylase (which has propionyl-CoA carboxylase activity) has not yet been reported. We are interested in analyzing the crystal structure of the bifunctional A. brierleyi acyl-CoA carboxylase in further studies.
Acknowledgments
We thank Miho Aoshima for advice and helpful suggestions. This work was supported in part by a grant-in-aid for science research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Alber, B. E., and G. Fuchs. 2002. Propionyl-coenzyme A synthase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J. Biol. Chem. 277:12137-12143. [DOI] [PubMed] [Google Scholar]
- 2.Al-Feel, W., S. S. Chirala, and S. J. Wakil. 1992. Cloning of the yeast FAS3 gene and primary structure of yeast acetyl-CoA carboxylase. Proc. Natl. Acad. Sci. USA 89:4534-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Athappilly, F. K., and W. A. Hendrickson. 1995. Structure of the biotinyl domain of acetyl-coenzyme A carboxylase determined by MAD phasing. Structure 3:1407-1419. [DOI] [PubMed] [Google Scholar]
- 4.Beaty, N. B., and M. D. Lane. 1983. The polymerization of acetyl-CoA carboxylase. J. Biol. Chem. 258:13051-13055. [PubMed] [Google Scholar]
- 5.Bell, S. D., and S. P. Jackson. 1998. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol. 6:222-228. [DOI] [PubMed] [Google Scholar]
- 6.Best, E. A., and V. C. Knauf. 1993. Organization and nucleotide sequences of the genes encoding the biotin carboxyl carrier protein and biotin carboxylase protein of Pseudomonas aeruginosa acetyl coenzyme A carboxylase. J. Bacteriol. 175:6881-6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bott, M., K. Pfister, P. Burda, O. Kalbermatter, G. Woehlke, and P. Dimroth. 1997. Methylmalonyl-CoA decarboxylase from Propionigenium modestum—cloning and sequencing of the structural genes and purification of the enzyme complex. Eur. J. Biochem. 250:590-599. [DOI] [PubMed] [Google Scholar]
- 8.Browner, M. F., F. Taroni, E. Sztul, and L. E. Rosenberg. 1989. Sequence analysis, biogenesis, and mitochondrial import of the α-subunit of rat liver propionyl-CoA carboxylase. J. Biol. Chem. 264:12680-12685. [PubMed] [Google Scholar]
- 9.Burns, B. P., S. L. Hazell, and G. L. Mendz. 1995. Acetyl-CoA carboxylase activity in Helicobacter pylori and the requirement of increased CO2 for growth. Microbiology 141:3113-3118. [DOI] [PubMed] [Google Scholar]
- 10.Burton, N. P., T. D. Williams, and P. R. Norris. 1999. Carboxylase genes of Sulfolobus metallicus. Arch. Microbiol. 172:349-353. [DOI] [PubMed] [Google Scholar]
- 11.Condò, I., A. Ciammaruconi, D. Benelli, D. Ruggero, and P. Londei. 1999. cis-acting signals controlling translational initiation in the thermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 34:377-384. [DOI] [PubMed] [Google Scholar]
- 12.Dennis, P. P. 1997. Ancient ciphers: translation in Archaea. Cell 89:1007-1010. [DOI] [PubMed] [Google Scholar]
- 13.Fall, R. R., A. W. Alberts, and P. R. Vagelos. 1975. Analysis of bacterial biotin proteins. Biochim. Biophys. Acta 379:496-503. [DOI] [PubMed] [Google Scholar]
- 14.Gornicki, P., L. A. Scappino, and R. Haselkorn. 1993. Genes for two subunits of acetyl coenzyme A carboxylase of Anabaena sp. strain PCC 7120: biotin carboxylase and biotin carboxyl carrier protein. J. Bacteriol. 175:5268-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gygax, D., R. Stalder, and J. Nüesch. 1984. Propionyl-CoA carboxylase in Nocardia mediterranei. FEMS Microbiol. Lett. 23:211-216. [Google Scholar]
- 16.Hector, M. L., and R. R. Fall. 1976. Multiple acyl-coenzyme A carboxylases in Pseudomonas citronellolis. Biochemistry 15:3465-3472. [DOI] [PubMed] [Google Scholar]
- 17.Henrikson, K. P., and S. H. G. Allen. 1979. Purification and subunit structure of propionyl coenzyme A carboxylase of Mycobacterium smegmatis. J. Biol. Chem. 254:5888-5891. [PubMed] [Google Scholar]
- 18.Huber, H., and K. O. Stetter. 2001. Order III Sulfolobales, p. 198-210. In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y.
- 19.Huder, J. B., and P. Dimroth. 1993. Sequence of the sodium ion pump methylmalonyl-CoA decarboxylase from Veillonella parvula. J. Biol. Chem. 268:24564-24571. [PubMed] [Google Scholar]
- 20.Hügler, M., C. Menendez, H. Schägger, and G. Fuchs. 2002. Malonyl-coenzyme A reductase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J. Bacteriol. 184:2404-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishii, M., T. Miyake, T. Satoh, H. Sugiyama, Y. Oshima, T. Kodama, and Y. Igarashi. 1997. Autotrophic carbon dioxide fixation in Acidianus brierleyi. Arch. Microbiol. 166:368-371. [DOI] [PubMed] [Google Scholar]
- 22.Jitrapakdee, S., G. W. Booker, A. I. Cassady, and J. C. Wallace. 1996. Cloning, sequencing and expression of rat liver pyruvate carboxylase. Biochem. J. 316:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazuta, Y., E. Tokunaga, E. Aramaki, and H. Kondo. 1998. Identification of lysine-238 of Escherichia coli biotin carboxylase as an ATP-binding residue. FEBS Lett. 427:377-380. [DOI] [PubMed] [Google Scholar]
- 24.Kelly, D. P., L. A. Chambers, and P. A. Trudinger. 1969. Cyanolysis and spectrophotometric estimation of trithionate in mixture with thiosulfate and tetrathionate. Anal. Chem. 41:898-901. [Google Scholar]
- 25.Kiatpapan, P., H. Kobayashi, M. Sakaguchi, H. Ono, M. Yamashita, Y. Kaneko, and Y. Murooka. 2001. Molecular characterization of Lactobacillus plantarum genes for β-ketoacyl-acyl carrier protein synthase III (fabH) and acetyl coenzyme A carboxylase (accBCDA), which are essential for fatty acid biosynthesis. Appl. Environ. Microbiol. 67:426-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura, Y., R. Miyake, Y. Tokumasu, and M. Sato. 2000. Molecular cloning and characterization of two genes for the biotin carboxylase and carboxyltransferase subunits of acetyl coenzyme A carboxylase in Myxococcus xanthus. J. Bacteriol. 182:5462-5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura, Y., T. Kojyo, I. Kimura, and M. Sato. 1998. Propionyl-CoA carboxylase of Myxococcus xanthus: catalytic properties and function in developing cells. Arch. Microbiol. 170:179-184. [DOI] [PubMed] [Google Scholar]
- 28.Kondo, H., K. Shiratsuchi, T. Yoshimoto, T. Masuda, A. Kitazono, D. Tsuru, M. Anai, M. Sekiguchi, and T. Tanabe. 1991. Acetyl-CoA carboxylase from Escherichia coli: gene organization and nucleotide sequence of the biotin carboxylase subunit. Proc. Natl. Acad. Sci. USA 88:9730-9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Lamhonwah, A.-M., F. Guan, and R. A. Gravel. 1987. Sequence homology around the biotin-binding site of human propionyl-CoA carboxylase and pyruvate carboxylase. Arch. Biochem. Biophys. 254:631-636. [DOI] [PubMed] [Google Scholar]
- 31.Levert, K. L., R. B. Lloyd, and G. L. Waldrop. 2000. Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin? Biochemistry 39:4122-4128. [DOI] [PubMed] [Google Scholar]
- 32.Li, S.-J., and J. E. Cronan, Jr. 1992. The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J. Biol. Chem. 267:855-863. [PubMed] [Google Scholar]
- 33.Li, S.-J., and J. E. Cronan, Jr. 1992. The genes encoding the two carboxyltransferase subunits of Escherichia coli acetyl-CoA carboxylase. J. Biol. Chem. 267:16841-16847. [PubMed] [Google Scholar]
- 34.Marini, P., S.-J. Li, D. Gardiol, J. E. Cronan, Jr., and D. de Mendoza. 1995. The genes encoding the biotin carboxyl carrier protein and biotin carboxylase subunits of Bacillus subtilis acetyl coenzyme A carboxylase, the first enzyme of fatty acid synthesis. J. Bacteriol. 177:7003-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 36.McKim, J. M., Jr., H. W. Schaup, K. Marien, and D. P. Selivonchick. 1989. Isolation and identification of acetyl-CoA carboxylase from rainbow trout (Salmo gairdneri) liver. Lipids 24:187-192. [DOI] [PubMed] [Google Scholar]
- 37.Menendez, C., Z. Bauer, H. Huber, N. Gad'On, K.-O. Stetter, and G. Fuchs. 1999. Presence of acetyl coenzyme A (CoA) carboxylase and propionyl-CoA carboxylase in autotrophic Crenarchaeota and indication for operation of a 3-hydroxypropionate cycle in autotrophic carbon fixation. J. Bacteriol. 181:1088-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer, H., B. Nevaldine, and F. Meyer. 1978. Acyl-coenzyme A carboxylase of the free-living nematode Turbatrix aceti. Biochemistry 17:1822-1827. [DOI] [PubMed] [Google Scholar]
- 39.Mishina, M., T. Kamiryo, A. Tanaka, S. Fukui, and S. Numa. 1976. Acetyl-coenzyme-A carboxylase of Candida lipolytica. Eur. J. Biochem. 71:295-300. [DOI] [PubMed] [Google Scholar]
- 40.Möller, W., and R. Amons. 1985. Phosphate-binding sequences in nucleotide-binding proteins. FEBS Lett. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 41.Moss, J., and M. D. Lane. 1971. The biotin-dependent enzymes. Adv. Enzymol. Relat. Areas Mol. Biol. 35:321-442. [DOI] [PubMed] [Google Scholar]
- 42.Moule, S. K., N. J. Edgell, A. C. Borthwick, and R. M. Denton. 1992. Coenzyme A is a potent inhibitor of acetyl-CoA carboxylase from rat epididymal fat-pads. Biochem. J. 283:35-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muramatsu, S., and T. Mizuno. 1989. Nucleotide sequence of the fabE gene and flanking regions containing a bent DNA sequence of Escherichia coli. Nucleic Acids Res. 17:3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikolau, B. J., and J. C. Hawke. 1984. Purification and characterization of maize leaf acetyl-coenzyme A carboxylase. Arch. Biochem. Biophys. 228:86-96. [DOI] [PubMed] [Google Scholar]
- 45.Norman, E., K. A. L. de Smet, N. G. Stoker, C. Ratledge, P. R. Wheeler, and J. W. Dale. 1994. Lipid synthesis in mycobacteria: characterization of the biotin carboxyl carrier protein genes from Mycobacterium leprae and M. tuberculosis. J. Bacteriol. 176:2525-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rainwater, D. L., and P. E. Kolattukudy. 1982. Isolation and characterization of acyl coenzyme A carboxylases from Mycobacterium tuberculosis and Mycobacterium bovis, which produce multiple methyl-branched mycocerosic acids. J. Bacteriol. 151:905-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reiter, W.-D., P. Palm, and W. Zillig. 1988. Transcription termination in the archaebacterium Sulfolobus: signal structures and linkage to transcription initiation. Nucleic Acids Res. 16:2445-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reverdatto, S., V. Beilinson, and N. C. Nielsen. 1999. A multisubunit acetyl coenzyme A carboxylase from soybean. Plant Physiol. 119:961-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts, E. L., N. Shu, M. J. Howard, R. W. Broadhurst, A. Chapman-Smith, J. C. Wallace, T. Morris, J. E. Cronan, Jr., and R. N. Perham. 1999. Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy. Biochemistry 38:5045-5053. [DOI] [PubMed] [Google Scholar]
- 50.Rodríguez, E., and H. Gramajo. 1999. Genetic and biochemical characterization of the α and β components of a propionyl-CoA carboxylase complex of Streptomyces coelicolor A3(2). Microbiology 145:3109-3119. [DOI] [PubMed] [Google Scholar]
- 51.Rodríguez, E., C. Banchio, L. Diacovich, M. J. Bibb, and H. Gramajo. 2001. Role of an essential acyl coenzyme A carboxylase in the primary and secondary metabolism of Streptomyces coelicolor A3(2). Appl. Environ. Microbiol. 67:4166-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roessler, P. G. 1990. Purification and characterization of acetyl-CoA carboxylase from the diatom Cyclotella cryptica. Plant Physiol. 92:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roessler, P. G., and J. B. Ohlrogge. 1993. Cloning and characterization of the gene that encodes acetyl-coenzyme A carboxylase in the alga Cyclotella cryptica. J. Biol. Chem. 268:19254-19259. [PubMed] [Google Scholar]
- 54.Sakoda, M., and K. Hiromi. 1976. Determination of the best-fit values of kinetic parameters of the Michaelis-Menten equation by the method of least squares with the Taylor expansion. J. Biochem. 80:547-555. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 56.Samols, D., C. G. Thornton, V. L. Murtif, G. K. Kumar, F. Carl Haase, and H. G. Wood. 1988. Evolutionary conservation among biotin enzymes. J. Biol. Chem. 263:6461-6464. [PubMed] [Google Scholar]
- 57.Santarelli, X., S. Chevalier, C. Cassagne, and R. Lessire. 1998. Arginyl residues are involved in acyl-CoA binding to the elongase from etiolated leek seedlings. Biochim. Biophys. Acta 1391:357-366. [DOI] [PubMed] [Google Scholar]
- 58.Schulz, T. K. F., M. V. Duin, and D. I. Zandee. 1983. Propionyl-CoA carboxylase from the sea mussel Mytilus edulis L.: some properties and its role in the anaerobic energy metabolism. Mol. Physiol. 4:215-230. [Google Scholar]
- 59.Shorrosh, B. S., R. A. Dixon, and J. B. Ohlrogge. 1994. Molecular cloning, characterization, and elicitation of acetyl-CoA carboxylase from alfalfa. Proc. Natl. Acad. Sci. USA 91:4323-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sloane, V., C. Z. Blanchard, F. Guillot, and G. L. Waldrop. 2001. Site-directed mutagenesis of ATP binding residues of biotin carboxylase. J. Biol. Chem. 276:24991-24996. [DOI] [PubMed] [Google Scholar]
- 61.Song, J., E. S. Wurtele, and B. J. Nikolau. 1994. Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of 3-methylcrotonoyl-CoA carboxylase: identification of the biotin carboxylase and biotin-carrier domains. Proc. Natl. Acad. Sci. USA 91:5779-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stirling, L. A., P. M. Ahmad, and F. Ahmad. 1981. Acyl coenzyme A carboxylase of Propionibacterium shermanii: detection and properties. J. Bacteriol. 148:933-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strauss, G., and G. Fuchs. 1993. Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3-hydroxypropionate cycle. Eur. J. Biochem. 215:633-643. [DOI] [PubMed] [Google Scholar]
- 64.Tanabe, T., K. Wada, T. Okazaki, and S. Numa. 1975. Acetyl-coenzyme-A carboxylase from rat liver. Eur. J. Biochem. 57:15-24. [DOI] [PubMed] [Google Scholar]
- 65.Thomm, M., and G. Wich. 1988. An archaebacterial promoter element for stable RNA genes with homology to the TATA box of higher eukaryotes. Nucleic Acids Res. 16:151-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Toh, H., H. Kondo, and T. Tanabe. 1993. Molecular evolution of biotin-dependent carboxylases. Eur. J. Biochem. 215:687-696. [DOI] [PubMed] [Google Scholar]
- 67.Trumble, G. E., M. A. Smith, and W. W. Winder. 1995. Purification and characterization of rat skeletal muscle acetyl-CoA carboxylase. Eur. J. Biochem. 231:192-198. [DOI] [PubMed] [Google Scholar]
- 68.Waldrop, G. L., I. Rayment, and H. M. Holden. 1994. Three-dimensional structure of the biotin carboxylase subunit of acetyl-CoA carboxylase. Biochemistry. 33:10249-10256. [DOI] [PubMed] [Google Scholar]
- 69.Watanabe, F., Y. Nakano, and S. Kitaoka. 1988. Subcellular location and some properties of propionyl-coenzyme A carboxylase in Euglena gracilis Z. Comp. Biochem. Physiol. 89B:565-568. [Google Scholar]
- 70.Wich, G., H. Hummel, M. Jarsch, U. Bär, and A. Böck. 1986. Transcription signals for stable RNA genes in Methanococcus. Nucleic Acids Res. 14:2459-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yao, X., D. Wei, J. C. Soden, M. F. Summers, and D. Beckett. 1997. Structure of the carboxy-terminal fragment of the apo-biotin carboxyl carrier subunit of Escherichia coli acetyl-CoA carboxylase. Biochemistry 36:15089-15100. [DOI] [PubMed] [Google Scholar]
- 72.Yeh, L.-A., C.-S. Song, and K.-H. Kim. 1981. Coenzyme A activation of acetyl-CoA carboxylase. J. Biol. Chem. 256:2289-2296. [PubMed] [Google Scholar]
- 73.Zhang, J., W.-L. Xia, K. Brew, and F. Ahmad. 1993. Adipose pyruvate carboxylase: amino acid sequence and domain structure deduced from cDNA sequencing. Proc. Natl. Acad. Sci. USA 90:1766-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]