Abstract
The protein products of several rad checkpoint genes of Schizosaccharomyces pombe (rad1+, rad3+, rad9+, rad17+, rad26+, and hus1+) play crucial roles in sensing changes in DNA structure, and several function in the maintenance of telomeres. When the mammalian homologue of S. pombe Rad9 was inactivated, increases in chromosome end-to-end associations and frequency of telomere loss were observed. This telomere instability correlated with enhanced S- and G2-phase-specific cell killing, delayed kinetics of γ-H2AX focus appearance and disappearance, and reduced chromosomal repair after ionizing radiation (IR) exposure, suggesting that Rad9 plays a role in cell cycle phase-specific DNA damage repair. Furthermore, mammalian Rad9 interacted with Rad51, and inactivation of mammalian Rad9 also resulted in decreased homologous recombinational (HR) repair, which occurs predominantly in the S and G2 phases of the cell cycle. Together, these findings provide evidence of roles for mammalian Rad9 in telomere stability and HR repair as a mechanism for promoting cell survival after IR exposure.
The DNA damage checkpoint pathways are highly conserved among a wide range of eukaryotes, from Saccharomyces cerevisiae to mammals. Human Rad9 (hRad9) is phosphorylated by ATM (ataxia-telangiectasia [A-T] mutated gene product) in response to DNA damage (7). Together with hRad1 and hHus1, hRad9 forms a nuclear complex that resembles PCNA and is believed to sense DNA damage (60, 65, 66). However, the cellular colocalization of hRad9 with the phosphorylated form of histone H2A variant X (γ-H2AX) after DNA damage is independent of ATM function (19), raising the possibility that hRad9 may also have a direct DNA damage-sensing function. In addition, Yin and coworkers (71) have reported that hRad9, much like p53, can transactivate p21. Loss of DNA damage response elements can result in cancer and other genetic diseases (22, 38). In fact, aberrant expression of Mrad9 or hRad9 has been linked to morphological transformation and cancer (8, 30, 59). Inactivation of Mrad9 in mouse embryonic stem (ES) cells results in an increased frequency of spontaneous chromosomal aberrations (23), and similar findings were observed in human cells with reduced levels of hRad9 (12). However, it is not known exactly how mammalian Rad9 influences genomic instability and whether the mechanism is related to defective DNA double-strand break (DSB) repair.
Interestingly, strains of Schizosaccharomyces pombe mutated in either the rad1+, the hus1+, the rad9+, or the rad17+ gene; S. cerevisiae with mutations in related orthologues; and nematodes altered in mrt-2 (encoding a Rad1 homolog) show telomere shortening; however, S. cerevisiae with mutant Mec3 shows telomere elongation, suggesting that these checkpoint proteins are also involved in telomere maintenance (1, 9, 11, 28, 36). Nabetani and coworkers (35) have found that hRad9, along with hHus1 and hRad1, is colocalized specifically at telomeric DNAs and promyelocytic leukemia bodies in ALT (alternative lengthening of telomeres) cells. Whether these observations are functionally significant in relation to telomere instability is unknown.
Genomic stability is maintained by telomeres since these chromosomal terminal structures protect chromosomes from fusion or degradation, as initially reported by Muller (34) and McClintock (32). Shortening or loss of telomeric repeats or altered telomere chromatin structure is correlated with chromosome end-to-end associations that could lead to genomic instability and gene amplification (10, 14, 39). Increased chromosome end-to-end associations, also known as telomeric associations typically seen at metaphase, have been reported in cells derived from tumor tissues, in senescent cells, in the Thiberge-Weissenbach syndrome, in A-T individuals, and following viral infections (45, 56). Chromosome end-to-end associations have been linked to genomic instability and carcinogenicity (10, 15, 38, 39). Both genetic and epigenetic factors can influence telomere stability (26, 55). For example, telomeric end-to-end fusions are enhanced in cells expressing dominant negative alleles of human telomeric protein TRF2 or overexpressing isoforms of human HP1 proteins (55, 64). Although recent data support a role for hRad9 in the cellular response to DNA damage, it is not clear whether this protein functions in telomere maintenance or DNA damage repair following ionizing radiation (IR) exposure. Here we report that inactivation or reduction in levels of mammalian Rad9 enhances telomere instability, increases cell killing by IR specifically in the S and G2 phases of the cell cycle, and reduces DNA DSB repair by homologous recombination (HR).
MATERIALS AND METHODS
Cells and transfection.
Human 293, GM5823+hTERT, MCF-7, and GM847 cells were maintained by previously published procedures (41, 55). Mouse ES cells were cultured by standard methods (23). Full-length hRad9 cDNA and DNA encoding the hRad9 C-terminal fragment (269 to 391 amino acids [aa]; ΔhRad9) were amplified by reverse transcription-PCR, cloned into pcDNA3.1, and transfected into cells (67). hRad9 small interfering RNA (siRNA) and control Luc siRNA were obtained from Dharmacon Research (Lafayette, CO). The siRNA sequence targeting hRad9 was 5′-AACCCUUGGAGGACGGGCUCU-3′.
Clonogenic assays.
Cells were plated onto 60-mm dishes in 5.0 ml of medium, incubated for 6 h, and subsequently exposed to IR. The number of cells per dish was chosen to ensure that about 50 colonies would survive a particular radiation dose treatment. Cells were exposed to IR in a dose range of 0 to 6 Gy at room temperature. Cells were incubated for 12 or more days and then fixed in methanol-acetic acid (3:1) prior to staining with crystal violet. Only colonies containing >50 cells were counted.
Detection of telomeres and determination of telomere restriction fragment length.
Telomeres in metaphase spreads were detected by fluorescent in situ hybridization (FISH) with a telomere-specific probe (55). To identify association between two human chromosomes, we used centromere and telomere probe combinations. Centromere-specific PNA probes were labeled with fluorescein isothiocyanate (green), and telomere-specific PNA probes were labeled with the fluorochrome Cy3 (red). For terminal restriction fragment (TRF) length analysis, DNA was isolated from exponentially growing cells, the DNA was digested with the restriction enzymes RsaI and HinfI (which do not cut the terminal TTAGGG repeat sequences), and the resulting DNA fragments were separated by agarose gel electrophoresis and then hybridized to a 32P-labeled (TTAGGG)5 probe (13). TRF detection and measurement of TRF lengths were performed as described previously, with ImageQuant version 1.2 (Molecular Dynamics) (13, 33, 54, 55). Nondenaturing in-gel hybridization to determine relative amounts of telomeric single-stranded DNA (G tails) was performed as described previously (33).
Telomerase assay.
Telomerase activity was determined with the telomerase PCR enzyme-linked immunosorbent assay kit (Boehringer Mannheim) as previously described (52). Telomerase activity was determined in triplicate, and negative and positive controls were run with each experiment. As a negative control, an aliquot of each extract was heat inactivated for 10 min at 95°C.
Cell synchronization.
To obtain cell samples enriched at different phases of the cell cycle, exponentially growing populations were fractionated by centrifugal elutriation (44). The cell cycle distribution of the fractionated samples was determined by flow cytometry measurements of DNA content (43, 44), as well as by using a premature chromosome condensation (PCC) protocol (43).
Chromosome studies.
Metaphase chromosomes were prepared by standard procedures (45). Giemsa-stained chromosomes from metaphase spreads were analyzed for chromosome end-to-end associations and aberrations. PCC was used to visualize chromosome damage directly in G1- and G2-phase cell populations. However, after S-phase-specific PCC, chromosomes look pulverized and therefore could not be analyzed for chromosome breaks (43, 44).
Assay for G1, S, and G2 chromosomal aberrations at metaphase and checkpoints.
G1-type chromosomal aberrations were assessed in cells exposed to 3 Gy of IR and incubated for 24 h. Cells were then subcultured, and metaphases were collected (24, 55). S-phase-specific chromosome aberrations were assessed after exponentially growing cells (pulse-labeled with bromodeoxyuridine [BrdU]) were irradiated with 2 Gy of IR. Metaphases were harvested following 4 h of irradiation, and S-phase types of chromosomal aberrations were scored. For G2-specific aberrations, cells were irradiated with 1 Gy and metaphases were collected 45 and 90 min posttreatment (13). Chromosome spreads were prepared after hypotonic treatment of cells, fixed in acetic acid-methanol, and stained with Giemsa (37). The categories of G1-type asymmetrical chromosome aberrations that were scored included dicentrics, centric rings, interstitial deletions/acentric rings, and terminal deletions. S-phase-type chromosome aberrations were assessed by counting both the chromosome and chromatid aberrations per metaphase as previously described (24, 55). G2-type chromosomal aberrations were assessed by counting chromatid breaks and gaps per metaphase as previously described (24, 55). The efficiency of the G1- or S-phase checkpoint was determined as previously described (24). The efficiency of G2 checkpoint control was evaluated by comparing different cell types for mitotic indices and chromatid-type aberrations at metaphase after irradiation (13). Fifty metaphases were scored for each postirradiation time point.
PCC.
The technique of PCC was used to visualize chromosome damage directly in G1- and G2-phase cell populations (43, 44). The procedures employed for mitotic Chinese hamster ovary (CHO) cell accumulation, cell fusion, and chromosome preparations are the same as previously described (43). G1- and G2-phase cell populations were obtained by centrifugal elutriation, placed on ice, irradiated, and fused with mitotic CHO cells to induce PCC. Chromosome damage in G1-phase cells is seen in the G1-phase PCC as an increase in the number of chromosome fragments per cell as previously described (43). For treated cells in the G1 phase, the number of breaks per cell was determined by subtracting the number of chromosome fragments per cell in unirradiated cells from that found in irradiated cells. Chromosome damage in G2-phase cells is seen as an increase in breaks, gaps, and exchanges. For treated cells in the G2 phase, the numbers of chromatid gaps, breaks, and exchanges were determined by subtracting the number of such aberrations per cell in the unirradiated populations from that in irradiated cells.
Antibodies and immunoprecipitation.
Published protocols were used for immunoprecipitation and immunoblotting (21). The following antibodies were used: anti-hRad9 and anti-hRad51from Santa Cruz (Santa Cruz, CA) and ATM, ATM S-1981, Chk2, TRF2, and Chk2 P-T68 provided by K. D. Brown, T. deLange, S. Elledge, M. B. Kastan, and J. Chen.
γ-H2AX immunofluorescence measurements.
Cells were grown in chamber slides, fixed in 2% paraformaldehyde for 15 min, washed in phosphate-buffered saline (PBS), permeabilized for 5 min on ice in 0.2% Triton X-100, and blocked in PBS with 1% bovine serum albumin. The procedure used for immunostaining is the same as that described previously (46, 53). Briefly, anti-γ-H2AX antibodies (Upstate Biotechnology) were applied to cells, which were then incubated for 1 h, washed with PBS containing 1% bovine serum albumin, and further incubated with Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (Molecular Probes) for 1 h at room temperature. Cells were rinsed with PBS and mounted in Vectashield mounting medium (Vector Laboratories). For detection of cells in S phase, cells were labeled with BrdU for 30 min. BrdU was detected by a monoclonal anti-BrdU antibody (Amersham Pharmacia Biotech) in concert with Alexa Fluor 594-conjugated goat anti-mouse secondary antibody (Molecular Probes). Foci were counted by microscopic and imaging processes. Fluorescence images were captured by using a Zeiss Axioskop 2 mot epifluorescence microscope equipped with a charge-coupled device camera and ISIS software (Metasystems, Altlussheim, Germany). Optical sections through nuclei were captured, and the images were obtained by projection of the individual sections. The results shown are from three independent experiments. Cells with a bubble-like appearance or micronuclei were not considered for γ-H2AX analysis.
In vitro nonhomologous end-joining (NHEJ) assay.
hRad9 was immunodepleted from human 293 cell extracts by anti-hRad9 antibody (54), or cell extracts were made from 293 cells after hRad9 knockdown by siRNA. The end-joining assay was performed as previously described (5, 54).
HR repair measurement.
To determine the chromosomal HR frequency, MCF-7 cells were transfected with 1 to 3 μg of linearized pDR-green fluorescent protein (pDR-GFP) plasmid using Lipofectamine 2000 (Invitrogen Corporation, Carlsbad, CA) (48, 72). Stably transfected cell lines were selected by growth in medium containing 1.0 μg/ml puromycin. Selected pDR-GFP-containing clones were shown to exhibit I-SceI-inducible HR, which produces GFP-positive cells detected by flow cytometry. Fifty-two hours posttransfection with either control siRNA or hRad9 siRNA, the cells were transfected with I-SceI expression vector pCMV3xn-1-SceI or a control cytomegalovirus vector. pGFP, containing the full-length GFP cDNA, was transfected into these cells as a calibration control for GFP-positive cells and to ensure that the transient transfection resulted in equal expression frequencies in the siRNA-treated cells. Seventy-two hours after I-SecI transfection, cells were trypsinized to form a single-cell suspension in Dulbecco modified Eagle medium and subjected to flow cytometric analysis. Two-color (green and propidium red) fluorescence analysis revealed the percentage of green fluorescent cells relative to the total cell number (48, 72). For each analysis, 100,000 cells were processed and each experiment was repeated three times.
RESULTS
hRad9 has been reported to localize on telomeres in cells that maintain telomeres by the telomerase-independent pathway known as ALT (35). To determine whether overexpression of hRad9 or its inactivation influences telomere stability, we overexpressed wild-type hRad9 and a mutant form of hRad9 (hRad9 C-terminal fragment [261 to 391 aa; ΔhRad9]) in cells and monitored telomere fusions at metaphase. We selected three different cell lines to study the effect of ΔhRad9 on telomeres, because in 293 and MCF-7 cells, telomeres are maintained by telomerase, whereas in GM847 cells, telomeres are maintained by the alternative mechanism (ALT). Cells ectopically expressing hRad9 had about threefold more of the encoded protein than did the parental cells (Fig. 1A and data not shown). Expression of ΔhRad9 did not influence levels of endogenous hRad9. Furthermore, the levels of endogenous hRad9 were similar in different phases of the cell cycle and were unaffected by expression of ΔhRad9 (Fig. 1B). 293 cells overexpressing hRad9 exhibited telomere association (TA) frequencies similar to those of control cells, suggesting that an increase in the hRad9 level in these cells has no influence on telomere stability. However, as shown in Table 1, in cells expressing ΔhRad9, there was a significant increase in TA per metaphase (range of 0.65 to 1.1) relative to the frequency observed for the parental cells (range of 0.09 to 0.13). These TAs were random. Mutant ΔhRad9 tagged with HA localized to the nucleus as detected by immunostaining (data not shown) had no influence on endogenous hRad9 levels (Fig. 1A), suggesting that the truncated protein may have a dominant negative effect. Since chromosome end-to-end associations may lead to anaphase bridge formation, the same cells were analyzed for this aberration. For each case, 300 cells at anaphase were examined for bridges. Cells expressing ΔhRad9 displayed a two- to fourfold higher frequency of anaphase bridges in comparison to parental cells (Table 1).
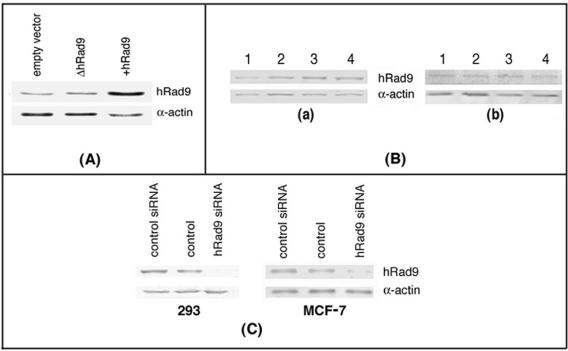
FIG. 1.
Expression of hRad9. (A) hRad9 protein levels were detected by Western analysis with hRad9-specific antibody in human 293 cells. hRad9 levels in cells expressing mutant (Δ) and wild-type hRad9 are shown. Higher levels of the protein were detected in cells engineered to overexpress hRad9, relative to the vector control, but no difference in hRad9 levels was seen in cells ectopically expressing ΔhRad9. (B) hRad9 levels in different phases of the cell cycle as detected in cell extracts of human 293 cells by immunoblotting with anti-hRad9 antibody. Human 293 cells in different phases were enriched by centrifugal elutriation (41). Lane 1 represents an asynchronous population, lane 2 represents a G1-phase-enriched population, lane 3 represents an S-phase-enriched population, and lane 4 represents a G2/M-phase-enriched cell population. (a) hRad9 levels in cells in different phases of the cell cycle. (b) hRad9 levels in cells expressing ΔhRad9 in different phases of the cell cycle. No difference in the levels of hRad9 was seen in different phases of the cell cycle. (C) siRNA-mediated reduction of hRad9 protein in 293 and MCF-7 cells. Cells were transfected with siRNA, and cell lysates were made 48 h posttransfection and examined for hRad9 protein by immunoblotting with anti-hRad9 antibody.
TABLE 1.
Frequencies of chromosomal aberrations in cells with or without functional Rad9a
| Cell type | Chromosome end associations/200 metaphases | Bridges/300 anaphases | Chromosome gaps + breaks/200 metaphases | Chromatid gaps + breaks/200 metaphases |
|---|---|---|---|---|
| 293 | 24 | 16 | 2 | 3 |
| 293 + empty vector | 21 | 13 | 3 | 3 |
| 293 + ΔhRad9 | 78b | 43b | 6 | 17b |
| 293 + hRad9 | 23 | 12 | 3 | 1 |
| 293 (hRad9 siRNA) | 63b | 51b | 5 | 11b |
| 293 (control siRNA) | 19 | 14 | 4 | 2 |
| GM847 | 34 | 19 | 3 | 5 |
| GM847 + empty vector | 31 | 14 | 4 | 2 |
| GM847 + ΔhRad9 | 67b | 39b | 4 | 14b |
| MCF (control siRNA) | 16 | 11 | 2 | 4 |
| MCF (hRad9 siRNA) | 43b | 27b | 6 | 7 |
| GM5823 (A-T) | 68b | 56b | 18b | 12b |
| Mrad9+/+ | 17 | 12 | 4 | 3 |
| Mrad9−/− | 55b | 34b | 6 | 18b |
Chromosome end-to-end associations (telomeric associations), chromosome breaks and gaps, and chromatid breaks and gaps were analyzed at metaphase and bridges at anaphase in cells with or without functional Rad9.
Aberrations in cells expressing ΔhRad9 or having reduced expression of wild-type Rad9 are significantly different from controls (293 or GM847 cells) as assessed by chi-square analysis (P < 0.01). Chromosome aberrations in Mrad9−/− cells are significantly higher compared to those observed in Mrad9+/+ cells.
The presence of ΔhRad9 may produce nonspecific effects that could complicate the interpretation of experimental results. Therefore, we used RNA interference-mediated hRad9 knockdown to confirm independently the dominant negative data. Cells transfected with hRad9-specific siRNA had an 80% reduction in hRad9 levels 48 h after transfection (Fig. 1C). When examined for telomere end associations, these cells gave results similar to those obtained for cells with ΔhRad9 (Table 1).
The chromosome end-to-end association data described above suggest that, due to loss of telomere function, chromosome end associations are formed and that these associations are not resolved in cells devoid of normal levels of Rad9. To further examine how expression of ΔhRad9 or a reduction in hRad9 levels is linked to telomere dysfunction, we determined mean TRF sizes. Southern blotting indicated no significant difference in TRF or G-overhang sizes when comparing DNA derived from cells with or without expression of ΔhRad9 or reduced levels of hRad9 (Fig. 2A). However, this analysis yields an appraisal only of the entire population of TRFs generated and does not monitor the ends of individual chromosomes. We therefore performed FISH for telomeric repeats on metaphase spreads by using a telomere-specific Cy3-labeled (CCCTAA)3 peptide nucleic acid probe. We applied centromere-specific Cy5-labeled FISH to detect human centromeres, which was used to monitor the hybridization for each chromosome and to visualize whether TA has occurred between two chromosomes. Fifty metaphase chromosome spreads (about 2,300 to 3,100 chromosomes) from each cell type were included and analyzed. A significantly higher proportion of chromatid ends in cells with aberrant functional hRad9 levels (about 10% of the telomeres per metaphase) lack telomere-specific fluorescent signals compared to the control cells, in which only <1% of the chromatids do not show telomere signals per metaphase (Fig. 2B). The loss of telomere signals could be a result of breaks adjacent to telomeres or shortened telomere tracts. These observations indicate that some of the chromosome end-to-end fusions observed in Rad9-inactivated cells correlated with loss of telomeric repeats. However, telomere signals were seen in more than 52% of the fusion sites, indicating that a total loss of telomeres is not required for telomere fusions in such cell systems.
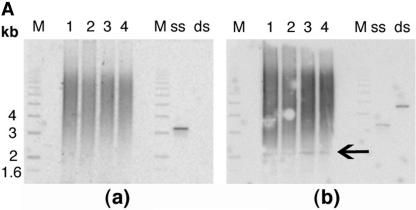
FIG. 2.
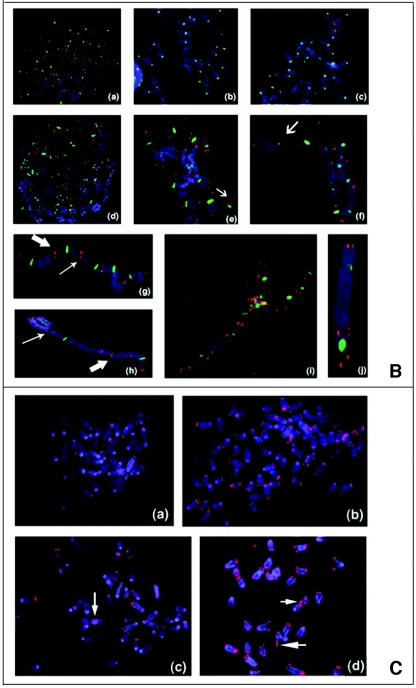
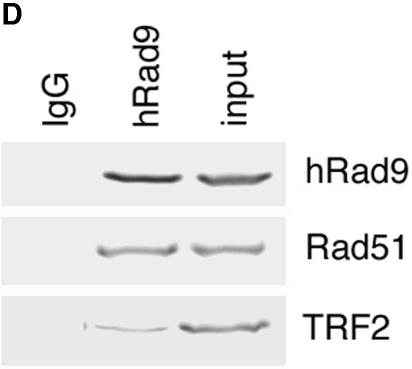
Telomere analysis by single-strand extensions (G tails) and telomere FISH. (A) G-tail sizes in cells with or without hRad9 inactivation. Detection of sizes of TRFs on DNA derived from 293 cells with or without hRad9 knockdown. In part a, nondenaturing in-gel hybridizations to genomic DNA digested with restriction enzymes HinfI and RsaI and with a telomeric repeat probe of the C-rich strand are shown. This method allows visualizing G-strand overhangs on telomeres. Signals were quantified by PhosphorImager analysis and corrected for DNA loading by using the rehybridized gel shown in part b. Lane M, molecular mass standards; lane 1, DNA from 293 cells with empty vector; lane 2, DNA from 293 cells expressing ΔhRad9; lane 3, DNA from 293 cells with hRad9 siRNA; lane 4, DNA from 293 cells with control siRNA; lane ss, denatured plasmid single-stranded DNA containing telomeric repeats (positive control); lane ds, double-stranded plasmid DNA used as a negative control (detected only once the DNA is denatured, as shown in part b). Part a shows the same gel as in part b after denaturing of the DNA in the gel and rehybridization with the same probe. The arrow in part b indicates an internal restriction fragment carrying telomeric repeats that was used to correct for DNA loading. (B) Telomere FISH analysis showing human metaphase chromosomal spreads. Telomere signals are red, and centromere signals are green. Part a, metaphase of 293 cells with empty vector; part b, metaphase of 293 cells with control siRNA; part c, metaphase section of MCF-7 cells with control siRNA; part d, metaphase of 293 cells with ΔhRad9; part e, metaphase section of 293 cells with hRad9 siRNA; part f, metaphase section of MCF-7 cells with hRad9 siRNA; part g, metaphase section of 293 cells with mutant TRF2ΔBΔM showing a chain of chromosomes resulting from single and both TAs; part h, TA with the interstitial region of a chromatin seen in 293 cells expressing mutant TRF2ΔBΔM; part i, metaphase of 293 cells with expression of mutant TRF2ΔBΔM and knockdown of hRad9 (note that almost all chromosomes undergo telomere fusions); part j, representative dicentric chromosome with fused telomeres seen in 293 cells expressing mutant TRF2 and inactivated hRad9. Note chromosome end associations, loss of telomeric signals, and gaps, as indicated by arrows. (C) Mouse metaphase chromosomes. Part a, representative metaphase of Mrad9+/+ cells; part b, Mrad9−/− metaphase showing polyploidy; part c, representative chromosome end association without loss of telomere signal at the association site seen in Mrad9−/− cells (Robertsonian fusion); part d, telomere fusion and loss of telomere signals as indicated by arrows in Mrad9−/− cells. Mrad9−/− ES cells have chromosome end associations, as well as loss of telomeric signals. (D) hRad9 interacts with TRF2 and Rad51. Immunoprecipitation of endogenous hRad9, TRF2, and Rad51 in 293 cells. Cell extracts were immunoprecipitated with anti-hRad9 antibody, followed by immunoblotting with antibodies to TRF2, Rad51, and hRad9. IgG, immunoglobulin G.
To provide more evidence for the influence of mammalian Rad9 on telomere stability, we evaluated mouse ES cells in which both alleles of Mrad9 were inactivated (23). About 4% of Mrad9 knockout cells showed chromosomal aneuploidy, as well as polyploidy (Fig. 2C). Consistent with the influence of hRad9 on telomere stability in human cells, we also found that Mrad9 knockout mouse ES cells had a significantly higher frequency of telomere end associations, as well as loss of telomere signals (Fig. 2C and Table 1). These cells frequently showed Robertsonian fusions, supporting the argument that loss of telomeres results in fusions that lead to metacentric chromosomes (Fig. 2C). Telomere signals were seen in more than 20% of the fusion sites, indicating that total loss of telomeres is not required for telomere fusions (Fig. 2C).
hRad9 interacts with TRF2 and influences telomere stability.
Telomere end association has been correlated with the inactivation of TRF2 (64). Therefore, we tested whether hRad9 interacts with TRF2. Analysis of anti-hRad9 immunoprecipitates with anti-TRF2 antibody demonstrated interaction between the two (Fig. 2D). When cells were exposed to IR, the interaction of hRad9 with TRF2 was unaffected (data not shown). These observations suggest that interactions of hRad9 with TRF2 occur prior to induction of DNA damage by the application of exogenous agents. Since expression of mutant TRF2ΔBΔM is known to induce telomere fusions (60), we further tested whether knockdown of hRad9 in cells expressing mutant TRF2ΔBΔM will affect telomere function. Cells expressing mutant TRF2ΔBΔM had more than 60% of their telomeres dysfunctional. However, cells expressing mutant TRF2ΔBΔM and reduced levels of hRad9 had levels of dysfunctional telomeres similar to those seen in cells expressing only mutant TRF2ΔBΔM (Fig. 2B). We failed to observe any additional telomere dysfunction measured by chromosome end fusions in cells expressing the mutant TRF2ΔBΔM and knocked-down hRad9 levels. These results suggest that hRad9 cannot rescue TRF2-mediated telomere stability.
We next determined whether telomere dysfunction in Rad9-deficient cells was due to abnormal telomerase activity. This activity was compared in vitro in extracts of cells with or without reduced levels of hRad9, and no significant difference was observed (data not shown). Mrad9+/+ and Mrad9−/− cell extracts also had similar levels of telomerase activity (data not shown), suggesting that mammalian Rad9 does not influence telomerase activity, as determined by an in vitro assay.
Recent studies have revealed that Mrad9 knockout ES cells, relative to Mrad9+/+ cells, have a higher frequency of spontaneous chromosomal aberrations (23), which correlates with their telomere instability phenotype. Thus, we determined whether the frequency of spontaneous chromosome and chromatid aberrations was increased in human cells with inactivated hRad9. Again, cells expressing ΔhRad9, as well as those with reduced levels of hRad9 due to the presence of siRNA, displayed a higher frequency of chromatid, as well as chromosome-type, aberrations compared to parental cells (Table 1). No significant increase in these frequencies was observed in cells overexpressing hRad9 (Table 1). Thus, mammalian cells with reduced levels of Rad9 or expression of ΔhRad9 or inactivated Rad9 expressed had frequent loss of telomere signals, enhanced telomere fusions, and higher frequencies of chromatid breaks and gaps, but not of chromosomal breaks and gaps, compared to control cells. Based on these observations of telomere dysfunction and chromosome aberrations in cells with inactivated mammalian Rad9, such genomic instability might be due to defective DNA repair or damage response checkpoints.
hRad9 knockdown or expression of mutant hRad9 enhances S- and G2-phase-specific IR-induced cell killing.
Rad9 forms a complex with Hus1 and Rad1 (hRad9-hHus1-hRad1) called the 9-1-1 complex (73). This complex is loaded onto DNA in response to many different types of genotoxic stress, including IR, UV light, replication inhibitors, and alkylation, suggesting that Rad9 has a role in cellular responses activated by multiple types of DNA damage (73). Although it is known that Mrad9 inactivation results in enhanced cell killing after IR exposure (23), it was not known whether Rad9 inactivation affects IR sensitivity in a specific cell cycle phase.
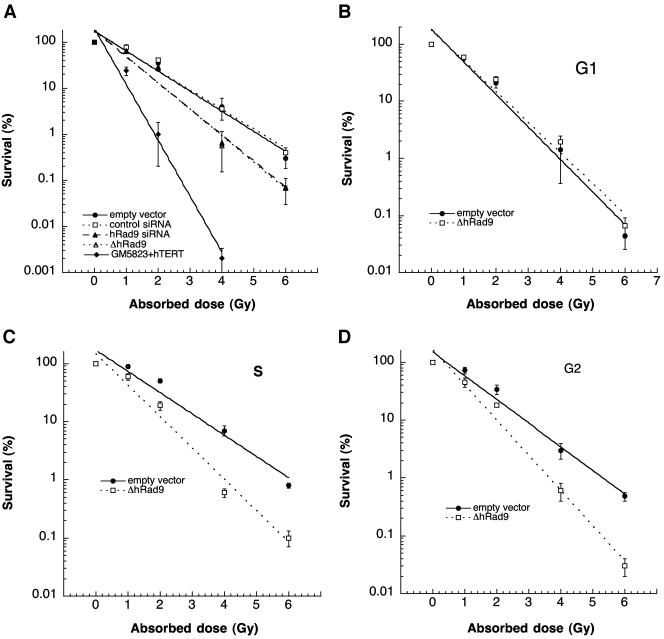
Telomere instability in mammals can correlate with organismal hypersensitivity to IR (18). Since inactivation of mammalian Rad9 influences telomere stability, we determined whether overexpression or inactivation of wild-type hRad9 influences cell survival after exposure to IR at a specific cell cycle phase. Cell survival after IR exposure was determined by using a colony formation assay described previously (13). Cells overexpressing hRad9 did not exhibit any altered sensitivity to IR exposure in comparison to the parental cells; however, cells with hRad9 levels reduced by siRNA, as well as cells expressing ΔhRad9, demonstrated enhanced cell killing after IR exposure (Fig. 3A). To define the precise phase of enhanced cell death after IR exposure, we enriched cells in different phases of the cell cycle by centrifugal elutriation. The G1-phase-enriched cell population had about 91% such cells, the S-phase-enriched population contained greater than 81% cells in S phase, and the G2/M-phase-enriched population contained 72 to 76% cells in that phase (data not shown). The survival characteristics of the cells varied through the cell cycle. Expression of ΔhRad9 did not alter the IR sensitivity of cells in G1 (Fig. 3B). However, S- and G2-phase cells with inactivated hRad9 showed enhanced IR-induced cell killing relative to control cells (Fig. 3C and D). These studies indicate that hRad9 has a specific role in S- and G2-phase cell survival after IR exposure. We further established that enhanced cell killing in S- and G2-phase cells was not due to the change in hRad9 levels since the abundance of this protein was similar in control radioresistant hRad9+ cells in different phases of the cell cycle (Fig. 1B).
FIG. 3.
Comparison of gamma ray sensitivity. Dose-response curves are shown for 293 cells with or without hRad9 knockdown after IR exposure. The A-T cell line GM5823+hTERT was used as a control to indicate radiosensitivity. (A) Survival curves for exponentially growing cells. Note that cells with hRad9 siRNA or expression of ΔhRad9 have lower survival after IR treatment. (B to D) Survival through the cell cycle of cells with or without ΔhRad9. Dose-response survival curves for cells in the G1 phase (B), the S phase (C), and the G2 phase (D) are shown. The cells with ΔhRad9 expression have statistically significantly (P < 0.01 as determined by the chi-square test) enhanced cell killing compared to control cells in the S and G2 phases. The values shown are means from three experiments.
Mammalian Rad9 influences the appearance and disappearance of IR-induced γ-H2AX foci.
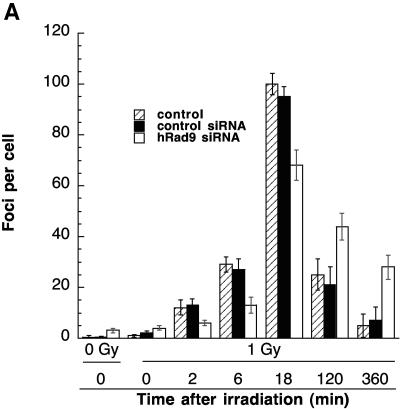
H2AX is associated with nuclear foci containing factors that are essential for DNA repair, replication, and cell cycle regulation (17, 49, 50). H2AX phosphorylation is critical for protecting the genome from spontaneous DSBs, as well as those induced by IR or V(D)J recombination (3, 4). H2AX phosphorylation coincides with the sites of DSBs, irrespective of their origin (6, 29, 47, 51, 69). Since inactivation of Rad9 influences cell survival after IR exposure and hRad9 colocalizes with γ-H2AX at DNA DSBs (19), we examined whether a reduction in hRad9 abundance or activity can influence the appearance of IR-induced γ-H2AX foci. Human 293 cells with reduced hRad9 levels had about three γ-H2AX foci per cell 2 min after irradiation with 1 Gy. Control cells with normal levels of hRad9 had about seven foci per cell under the same conditions (Fig. 4A). Similar results were observed in Mrad9−/− ES cells, where the appearance of γ-H2AX foci was delayed compared to that in control Mrad9+/+ ES cells (data not shown).
FIG. 4.
Appearance of γ-H2AX in cells with or without hRad9. Cells were irradiated and collected at different times postexposure. (A) Human 293 cells were irradiated, and γ-H2AX foci were observed at various times postirradiation. For each time point, 100 cells were analyzed. Each experiment was repeated three times. The mean number of foci is plotted against time. Cells with reduced levels of hRad9 caused by hRad9 siRNA showed fewer γ-H2Ax foci until 5 min postirradiation, and the foci persisted for a longer time. (B) Mouse cells with or without Mrad9 were grown on coverslips, labeled with BrdU for 30 min, irradiated with 1 Gy of gamma rays, and processed for detection of foci as previously described (24, 46). BrdU labeling and γ-H2AX were detected by anti-BrdU and anti-phospho-histone-H2AX antibodies, respectively (Upstate).
To assess the disappearance of γ-H2AX foci, we analyzed human cells 120 and 360 min postirradiation. Furthermore, we determined whether differences in the appearance of γ-H2AX foci after IR exposure are due to cell cycle distribution. To address this question, we labeled the cells for 30 min with BrdU, followed by irradiation, and then examined them for γ-H2AX foci at different times after treatment. Specifically, S-phase Mrad9−/− cells, as well as human cells with inactivated hRad9, have a lower number of γ-H2AX foci compared to parental control cells (Fig. 4B and data not shown). In general, the number of γ-H2AX foci in cells with inactivated mammalian Rad9 was significantly lower until 5 min postirradiation. However, the γ-H2AX focus number was higher at 120 min postirradiation, suggesting that this protein plays a role in DNA damage repair (Fig. 4A).
IR exposure induces higher frequencies of S- and G2-phase-specific chromosomal aberrations in cells with reduced levels of hRad9.
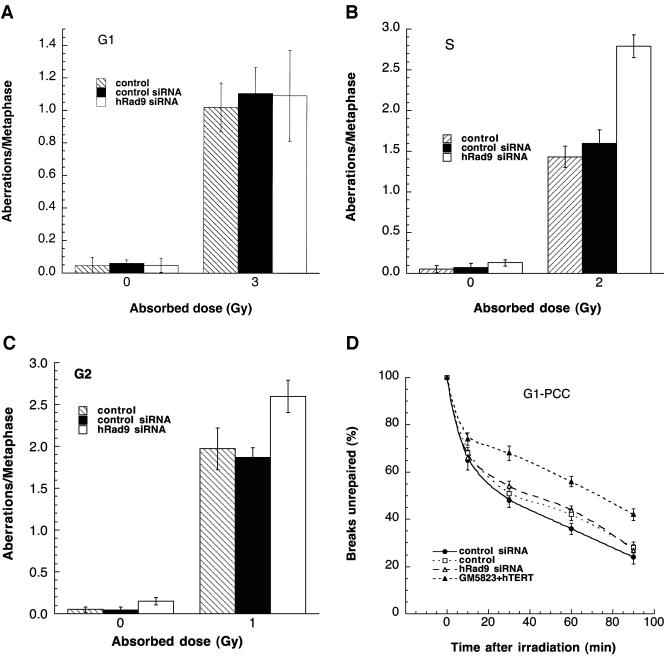
Telomere instability and IR sensitivity are linked to defective DNA repair (27, 38, 54, 55, 70). To determine whether the S- and G2-phase-specific increased cell killing by IR in cells with reduced levels of hRad9 correlated with DNA repair, we compared cell cycle stage-specific IR-induced chromosomal aberrations in cells with or without reduced levels of this protein. We first set out to determine frequencies of chromosomal aberrations induced by IR in G1-, S-, and G2-phase cells. Cell cycle phase-specific chromosome aberrations were ascertained based on the frequency of chromosomal and chromatid-type aberrations observed at metaphase. G1-specific aberrations detected at metaphase are mostly of the chromosomal type and include a high frequency of dicentrics (55). S-phase-type aberrations detected at metaphase are chromosomal, as well as those involving chromatids. G2-type aberrations detected at metaphase are predominantly the chromatid type with the least number of dicentrics. To determine G1-type chromosome damage, cells were treated with 3 Gy and replated 24 h after irradiation, and aberrations were scored at metaphase as previously described (24, 55). No difference in residual IR-induced G1 chromosomal aberrations was seen in metaphase cells with or without reduced levels of hRad9 (Fig. 5A). Furthermore, cells overexpressing hRad9 did not show any difference in G1-phase-type chromosome aberrations relative to parental cells after IR exposure (data not shown). To determine whether defective repair can be documented in cells overexpressing hRad9 or with ΔhRad9 in phases of the cell cycle other than G1, we evaluated S-phase-specific chromosome aberrations. We first determined the time needed for S-phase cells to reach metaphase after IR treatment. Exponentially growing 293 cells were labeled with BrdU for 30 min as previously described (55) and then irradiated with 2 Gy. Anti-BrdU immunostaining was performed to determine when metaphase chromosomes contain BrdU. In these experiments, BrdU-labeled metaphases appeared approximately 3 h postirradiation (data not shown). Thus, cells producing hRad9 protein were treated with 2 Gy of IR and metaphases were collected 3 to 5 h postirradiation. Cells overexpressing hRad9 and collected 3 h postirradiation displayed frequencies of metaphases with chromatid and chromosomal aberrations similar to those of parental cells (data not shown). In contrast, cells with reduced levels of hRad9 had significantly higher frequencies of chromosome and chromatid aberrations after IR exposure compared to parental cells (Fig. 5B). Furthermore, when cells were treated with 1 Gy of gamma rays, hRad9 overexpression had no effect on G2-phase-specific chromosome repair (data not shown), while hRad9 inactivation increased G2-phase-specific chromosome aberrations (Fig. 5C). These observations suggest a role for Rad9 in repairing chromosome damage in the S, as well as in the G2, phase of the cell cycle.
FIG. 5.
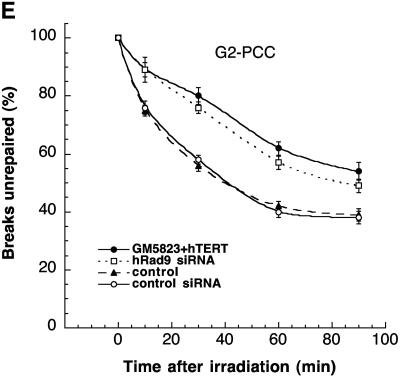
Analysis of chromosome damage and repair in 293 cells with or without wild-type levels of hRad9. (A to C) Chromosomal aberrations analyzed at metaphase. (A) Cells in plateau phase were irradiated with 3 Gy, incubated for 24 h postirradiation, and then subcultured, and metaphases were collected. G1-type aberrations were examined at metaphase. All categories of asymmetric chromosome aberrations were scored: dicentrics, centric rings, interstitial deletions/acentric rings, and terminal deletions. The frequency of chromosomal aberrations was identical for cells with or without wild-type levels of hRad9, indicating normal repair of G1-phase-specific chromosome damage. (B) Cells in exponential phase were irradiated with 2 Gy. Metaphases were harvested at 4 h after irradiation, and S-phase types of chromosomal aberrations were scored. Cells with hRad9 knockdown showed significant differences in chromosomal aberration frequencies compared to control cells (P < 0.05, Student t test). (C) Cells in exponential phase were irradiated with 1 Gy. Metaphases were harvested following irradiation, and G2-type chromosomal aberrations were monitored. Cells with hRad9 knockdown showed significantly higher (P < 0.05, Student t test) frequencies of chromosomal aberrations compared to control cells. (D and E) Chromosome break repair in cells in G1- and G2-phase cells. (D) Chromosome break repair in G1-phase cells. Cells were irradiated with 3 Gy of IR, and chromosome damage was analyzed by a PCC technique. Sixty PCCs were scored for each time point, and the data represent means and standard deviations of two sets of experiments. The mean number of chromosome breaks per cell immediately after irradiation was about 12, and this number was reduced after incubation at 37°C. Cells with or without hRad9 knockdown show no difference in the kinetics or residual levels of breaks. (E) Chromatid gap and break repair in G2-phase cells. Cells were irradiated with 3 Gy of IR, and chromatid gaps and breaks were analyzed by the PCC technique. The mean number of chromosome breaks per cell immediately after irradiation was about 10, and this number was reduced after incubation at 37°C. Forty five PCCs were scored for each time point, and the data are the mean and standard deviation of two sets of triplicate sample experiments. Note that the rate of chromosome repair is slow in cells with Rad9 siRNA, and such cells have higher frequencies of residual chromosome damage. The difference in residual damage in cells with or without wild-type levels of hRad9 is statistically significant, as demonstrated by the Student t test (P < 0.05).
Reduced levels of hRad9 influence the kinetics of G2-phase-specific but not G1-phase-specific chromosome damage repair.
The underlying reasons for the increased aberrations at mitosis observed in cells with inactivated Rad9 could be the inability to modulate cell cycle progression following IR exposure, increased initial levels of genetic damage, and/or decreased efficiency of DNA damage repair. To address whether IR-induced increased initial levels of genetic damage and/or decreased efficiency of repair is responsible, we used PCC to visualize directly chromosome events in interphase cells following irradiation (40). G1 chromosomes are rodlike and unreplicated, whereas G2 chromosomes are replicated and have two chromatids (43). Prematurely condensed chromosomes from S-phase cells appear fragmented (pulverized) even without irradiation; therefore, S-phase cells cannot be compared. G1- and G2-phase cells, with or without hRad9, were enriched by centrifugal elutriation, gamma irradiated, and then allowed to repair the resulting DNA damage for increasing intervals of time prior to the induction of PCC. Subsequently, residual levels of chromosome damage were assessed directly. Cells with or without reduced levels of hRad9 were irradiated with 3 Gy of gamma rays on ice and then incubated at 37°C for different periods of time prior to fusion with mitotic cells to visualize chromosomes. Chromosome fragments were counted at different time points, and the mean additional number of fragments seen immediately after irradiation was about 12 higher than the control. No difference in initial chromosome damage (12 breaks per cell) or the kinetics of chromosome repair was observed in G1-phase cells with or without normal Rad9 function (Fig. 5D), consistent with results observed at metaphase (Fig. 5A). Interestingly, G2-phase cells with Rad9 inactivated had initial chromosome damage (10 breaks) similar to that of control cells; however, hRad9-deficient cells had a higher frequency of unrepaired chromosome breaks and gaps compared to parental cells (Fig. 5E). Such results were consistent with the observations made for cells at metaphase (Fig. 5C) and thus support the argument that hRad9 influences the repair of G2-phase-specific chromosome damage.
Observable chromosome aberrations in cells with reduced levels of hRad9 do not correlate with a deficiency of S or G2 checkpoints.
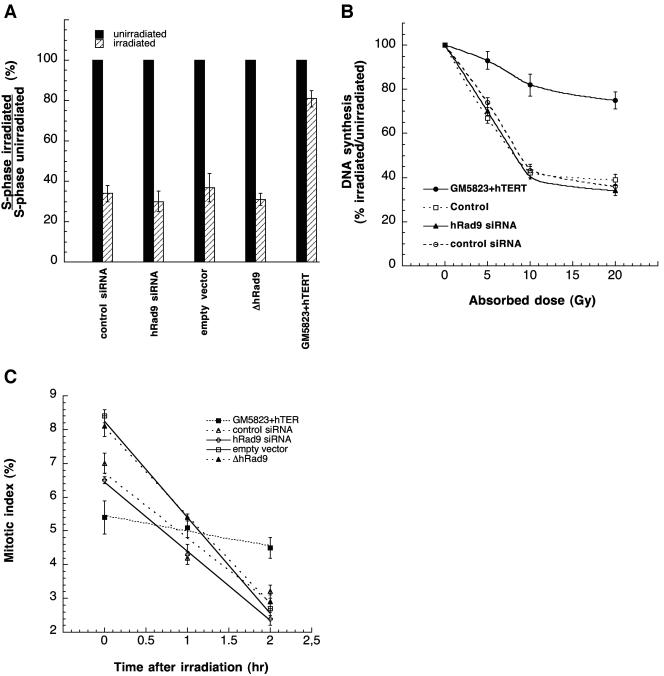
To determine whether the increase in S-phase-specific IR-induced chromosomal damage is due to defective cell cycle checkpoints, we first determined whether mammalian Rad9 inactivation results in loss of the G1 checkpoint. We found that 293 cells with reduced levels of hRad9 caused by siRNA or ΔhRad9 expression showed a significant increase in G1-phase cells with a decrease in S-phase entry after IR exposure, similar to control cells that showed a significant decrease in S-phase entry, indicating G1 arrest (Fig. 6A and data not shown). Similar results were found for Mrad9−/− and Mard9+/+ cells. These results suggest that Rad9 has no (detectable) influence on the G1 checkpoint in these cell systems.
FIG. 6.
Cell cycle checkpoint analysis. (A) Human 293 cells were exposed to 8 Gy of IR. Twelve hours following irradiation, cells were pulse-labeled with BrdU for 30 min. The proportion of S-phase cells was determined. The percentage of S-phase cells in the irradiated culture relative to those in the unirradiated control is shown. The results represent the mean of three experiments. For each independent experiment, 200 cells were examined. Black bars represent unirradiated control cells that enter into S phase, as they are BrdU positive. Dashed bars represent cells that enter into S phase postirradiation, and their percentage is determined by dividing the number of S-phase irradiated cells by the number of S-phase unirradiated cells. (B) S-phase checkpoint was determined as radioresistant DNA synthesis after exposure to IR. 293 cells were irradiated at the doses indicated. The rate of DNA synthesis was determined 1 h postirradiation by pulse-labeling with [3H]thymidine for 30 min. The value of the unirradiated control was set to 100% for each cell type. The mean and standard deviation of triplicate experimental points are shown. (C) Comparison of the mitotic index among cells after treatment with 2 Gy of IR. Populations were examined for the frequency of mitotic cells at different times postirradiation. The mean represents the value from three independent experiments. For each experiment, 200 metaphases were scored. Note that the mitotic index decreased in cells whether or not hRad9 levels were wild type.
Exposure to IR causes a transient inhibition of DNA replication. Cells deficient in ATM function exhibit radioresistant DNA synthesis, an indicator of loss of the S-phase checkpoint (38). Cells expressing ΔhRad9 or cells with siRNA-reduced Rad9 levels, as well as Mrad9−/− cells, did show inhibition of DNA synthesis after IR exposure which was similar to that of the control cells (Fig. 6B and data not shown). To determine whether mammalian Rad9 influences the G2 checkpoint, the efficiency of G2 delay was evaluated by measuring the proportion of cells in mitosis (mitotic index) after irradiation. Cells expressing ΔRad9 or having reduced levels of hRad9 showed no significant difference in the mitotic index after irradiation with 2 Gy of IR compared to the control, suggesting that inactivation of mammalian Rad9 does not lead to a defective G2 checkpoint (Fig. 6C). These results suggest that Rad9 has a critical role in DNA damage repair rather than in IR-induced cell cycle checkpoint control. Such results are consistent with previous studies of Mrad9−/− mouse ES cells, which demonstrate that there is no major problem with IR-induced G2 checkpoint control initiation, only a defect in the prolonged maintenance of the checkpoint, consistent with a defect in DNA repair (23). Thus, the enhanced chromosomal damage seen in mammalian cells with inactivated Rad9 cannot be attributed to defective cell cycle checkpoints, again suggesting that altered DNA repair is the more likely explanation.
IR-induced phosphorylation of ATM, as well as Chk2, is not influenced by knockdown of hRad9.
In mammalian cells, in response to DNA damage, ATM gets autophosphorylated and phosphorylates various proteins that control G1-, S-, and G2-phase cell cycle arrest (16, 38, 57). We examined whether inactivation of hRad9 influences IR-induced ATM phosphorylation at Ser-1981 in 293 cells. ATM phosphorylation by IR was found in cells with or without knockdown of hRad9 (Fig. 7). We also examined whether inactivation of hRad9 influences ATM-mediated, IR-induced phosphorylation of Chk2 in 293 cells. ATM phosphorylates Chk2 on Thr-68 in response to DNA damage (31). Cells with or without reduced levels of hRad9 showed no difference in the IR-induced phosphorylation of Chk2 at Thr-68 (Fig. 7). Since hRad9 function does not influence IR-induced ATM or ATM-mediated Chk2 phosphorylation, such results argue further against a role for hRad9 in cell cycle checkpoint control but thus support a role in DNA damage repair.
FIG. 7.
hRad9 is not required for IR-induced phosphorylation of ATM and Chk2. (A) 293 cells with or without hRad9 knockdown were irradiated with 2 Gy of IR. Cell extracts were immunoblotted with anti-ATM antibodies and anti-p-Ser-1981-ATM antibody. (B) 293 cells with or without hRad9 knockdown were irradiated with 5 Gy of IR. Cell extracts were immunoblotted with anti-Chk2 antibody and anti-p-T68-Chk2 antibodies. No difference in cells with or without hRad9 was observed for IR-induced phosphorylation of ATM or Chk2.
There are two major ways to maintain the ends of chromosomes in yeast and mammalian nuclei: mechanisms related to telomerase and recombination. Without telomerase or recombination, cells enter a state of growth arrest. Since mammalian Rad9 inactivation did not influence telomerase activity, we tested whether hRad9 was involved in NHEJ or HR.
hRad9 has no impact on NHEJ.
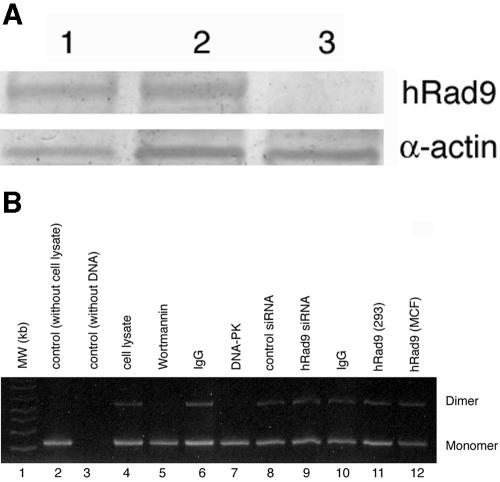
The aforementioned results established a role for hRad9 in S- and G2-phase-specific DNA damage repair. It has been suggested that the cell cycle could influence DSB repair since NHEJ acts predominantly in G1 and HR acts in S and G2 (61). To determine whether the hRad9 protein is directly involved in DNA end joining, we used a well-characterized in vitro assay that involves the rejoining of DNA ends of a plasmid in 293 cell extracts as previously described (5, 54). Plasmid pUC19 was digested with SalI, and DNA end joining was performed as previously described (5, 54). hRad9 in 293 cell extracts was depleted by immunoprecipitation with hRad9 antibody. Complete immunodepletion of hRad9 was confirmed by Western analysis (Fig. 8A). hRad9-immunodepleted extracts or extracts from Rad9 siRNA-treated cells were used to assay DNA end-joining activity. No difference in this activity was found in extracts with or without hRad9 (Fig. 8B). These results suggest that hRad9 does not have a role in the DNA end-joining process. However, these observations did not rule out a role for hRad9 in chromosomal recombination.
FIG. 8.
hRad9 is not essential for NHEJ. Depletion of hRad9 was achieved by two different approaches: immunodepletion and use of hRad9 siRNA. (A) Immunodepletion of hRad9 from whole 293-cell extracts as determined by immunoblotting. The extracts were precipitated with protein A-Sepharose beads only (lane 1), with preimmune serum (lane 2), and with hRad9-depleted whole-cell extract (depleted with hRad9 antibody) (lane 3). (B) Ethidium bromide-stained agarose gels show the results of end-joining assays. Plasmid puc19 DNA was linearized by digestion with SalI. The positions of monomer and dimer plasmids are indicated to the right of the gel. Lane 1, molecular weight (MW) markers; lane 2, only plasmid DNA; lane 3, only cell lysate; lane 4, cell lysate and DNA; lane 5, cell lysate with wortmannin and DNA; lane 6, immunoglobulin G (IgG) depleted in cell lysate and then added DNA; lane 7, DNA protein kinase (DNA-PK) depleted in cell lysate and then added DNA; lane 8, cell lysates of cells with control siRNA and plasmid DNA; lane 9, cell lysate from cells with hRad9 siRNA and plasmid DNA; lane 10, immunoglobulin G (IgG) depleted in cell lysate and DNA; lane 11, hRad9 depleted in 293 cell lysates and DNA; lane 12, hRad9 depleted in MCF-7 cells and DNA.
hRad9 interacts with hRad51 and influences HR repair.
The defect in chromosome repair found in hRad9 knockdown cells in S and G2 suggested a role for hRad9 in HR since this process predominates in cells in these phases of the cell cycle, when sister chromatids are available as a template for repair. It is known that the hRad9-hHus1-hRad1 complex is loaded onto damaged DNA in an hRad17-dependent manner (73) and hRad9 colocalizes with γ-H2AX at those sites (19). hRad9 also colocalizes with telomeric DNAs and promyelocytic leukemia bodies in ALT cells (35). ALT cells have abundant telomeric circles, suggesting that frequent t-loop HR events could promote rolling-circle replication of telomeres in the absence of telomerase. Wang and coworkers (68) demonstrated that t-loop deletion by HR influences mammalian telomeres. A key feature of HR is DNA strand invasion, which is catalyzed by the Rad51 protein coating single-stranded DNA; however, the exact roles of the many accessory proteins that facilitate this process are not known. Rad51D is involved in the maintenance of telomeres. Rad51 can form distinct nuclear foci in response to IR exposure (63). These foci can be visualized by microscopy and are thought to be sites where repair reactions take place (62). Since hRad9 influences telomere stability, as well as DNA damage repair, we determined whether hRad9 interacts with Rad51.
Analysis of anti-hRad9 immunoprecipitates with anti-hRad51 antibody demonstrated interaction between the two (Fig. 2D). When cells were exposed to IR, the association of hRad9 with hRad51 was unaffected (data not shown). These observations suggest that interaction of hRad9 with hRad51, as well as with TRF2, occurs prior to induction of DNA damage by the application of exogenous agents.
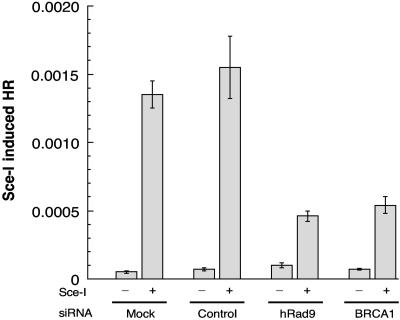
The interaction of hRad9 with hRad51 suggests that hRad9 may have a role in HR. Therefore, we measured HR by examining the frequency of reconstitution of a GFP reporter gene within a chromosomally integrated plasmid substrate in cells with or without reduced levels of hRad9. Following I-SceI transfection, cells containing the pDR-GFP substrate demonstrated a >80-fold increase in the number of GFP-positive members compared with cells containing the control vector, indicating that almost all of the GFP-positive cells resulted from DSB-induced recombination (Fig. 9). pDR-GFP cells treated with hRad9 siRNA had a three- to fourfold reduction in the number of GFP positives in comparison to control transfected cells (t test, P < 0.001), providing strong evidence that hRad9 affects the HR pathway for DNA DSB repair.
FIG. 9.
hRad9 knockdown influences HR. Impaired I-SceI-induced HR in hRad9-deficient MCF-7 cells was found. HR was measured by dual-color flow cytometric detection of GFP-positive cells. HR frequencies are shown with (+) or without (−) I-SceI induction for untreated cells, for cells treated with control siRNA, and for cells treated with hRad9-specific siRNA. hRad9-deficient and BRCA1-deficient cells show similar reductions in HR. The knockdown of BRCA1 by BRCA1 siRNA in MCF-7 cells has been described previously (72). The results presented are the mean and standard error from four independent experiments.
DISCUSSION
The function of mammalian Rad9, a gene involved in sensing DNA damage, was examined in the context of chromosome stability and DNA repair in mammalian cells. Human cells in which hRad9 was inactivated by expression of ΔhRad9, or levels of the protein were reduced by siRNA, or mouse cells with Mrad9 deleted (23) were used. A cDNA fragment encoding aa 269 to 391 of hRad9 was expressed in different human cells, which were subsequently examined for various end points. We found that the C-terminal fragment of hRad9 (aa 269 to 391) demonstrated dominant negative activity for almost all of the assays employed. Thus, expression of this fragment might block the interaction of wild-type hRad9 with other proteins and thus has a dominant negative function. This function of the hRad9 fragment is consistent with the previously described observation of Wang and coworkers (67), who reported that the C-terminal region of hRad9 interacts with other proteins. We found that, indeed, the frequencies of chromosomal abnormalities, such as chromosome end-to-end associations and chromosomal breaks, were higher in cells with inactivated mammalian Rad9. These chromosomal aberrations appear to involve dysfunctional telomeres, as a significant number of cells showed TAs. Thus, the results suggest that abrogating the function of mammalian Rad9, which is implicated in mediating DNA damage-sensing checkpoints, increases the frequency of unrepaired chromosomal aberrations that can lead to an increase in the number of telomere fusions. This mechanism might contribute to the inviability of mouse embryonic fibroblasts when Mrad9 is inactivated (23). The frequencies of spontaneous, as well as IR-induced, chromosomal aberrations were dramatically higher in cells with inactivated Rad9 relative to those in controls, suggesting that loss of telomere function occurred by breakage near telomeres, rather than by a telomerase-based mechanism. This is supported by our finding that knockdown of hRad9 does not influence telomerase activity (data not shown). hRad9 forms a heterotrimer with Hus1 and Rad1, which is loaded onto DNA by Rad17-mediated mechanisms, and these proteins are evolutionarily conserved. Interestingly, studies with S. pombe, S. cerevisiae, and Caenorhabditis elegans orthologues also indicate roles for these proteins in telomere metabolism (1, 11, 25, 28, 36).
It is important to note that in cells with both alleles of Rad9 inactivated, frequent chromosome breakage with complete loss of telomeric repeats in some chromosomes, as well as chromosome end fusions, occurred under normal growth conditions in the absence of exogenous genotoxic challenges (Fig. 2 and Table 1). The fact that the G1 checkpoint was operative in cells with reduced levels of Rad9 is supported by the observations that irradiation induced comparable frequencies of cells in G1 and S phase compared to those cells with normal levels of hRad9 (Fig. 6A). Furthermore, cells with reduced levels of Rad9 did not show any defect in the S-phase-specific cell cycle checkpoint, as such cells did not differ from control cells for radioresistant DNA synthesis (Fig. 6B). The mitotic index after irradiation was identical in cells with or without functional Rad9, indicating that mammalian Rad9 inactivation does not influence the G2 checkpoint in the cell lines studied (Fig. 6C). hRad9 knockdown had no influence on IR-induced ATM Ser-1981 phosphorylation or Chk2 Thre-68 phosphorylation, supporting the argument that mammalian Rad9 has a minimum role in cell cycle checkpoint control postirradiation. In contrast, there was a clear and significant difference in the chromosome aberration frequency if log-phase cells were irradiated, and mitoses shortly thereafter were monitored; at 4 h postirradiation, there was a 1.8-fold difference in the frequencies of S-phase aberrations/metaphase between cells with and without Rad9 fully functional (Fig. 5B). Furthermore, when G2-induced aberrations were monitored after irradiation, cells with reduced levels of Rad9 showed higher frequencies of aberrations in the same time span compared to controls, indicating very inefficient repair (Fig. 5C). Utilizing the PCC technique, we found that there was no difference in the kinetics of repair or the residual number of chromosome breaks in G1-phase cells with or without reduced levels of hRad9 (Fig. 5D); however, cells deficient in Rad9 had a higher frequency of chromosome gaps and breaks in the G2 phase (Fig. 5E). Consistent with these data, survival studies demonstrated that cells in the S and G2 phases were sensitive to irradiation if Rad9 was inactivated (Fig. 3C and D). These data, taken together, reinforce the idea that in cells with hRad9 knockdown, chromosomal damage that might occur during the S or G2 phase could be due to defective chromosome repair (Fig. 5B, C, and E). In cells lacking Rad9 protein, cell cycle arrest after DNA damage is operative and the cells do not enter mitosis exceptionally early. Our study reveals that as a consequence of this inability to appropriately repair, such cells enter mitosis with a higher frequency of chromosomal aberrations.
hRad9 is rapidly retained at sites of damaged DNA and colocalizes with the phosphorylated form of H2AX (γ-H2AX) (19). In addition, we found quantitative differences in the kinetics of appearance and disappearance of γ-H2AX foci and kinetics of repair of G2 chromosomes (Fig. 4 and 5E), dependent upon whether cells do or do not have functional mammalian Rad9. These results provide strong evidence that mammalian Rad9 plays a critical role in repair of DNA damage.
The association of hRad9 with TRF2 provides further evidence that hRad9 may have a role in maintaining the stability of telomeres. Although knockdown of hRad9 has a minimal role in TRF2 association with telomeres (data not shown) and inactivation of hRad9 and TRF2 did not rescue a telomere instability phenotype (Fig. 2), it is possible that TRF2 might be mediating DNA damage signaling through Rad9. This notion is further supported by the data indicating that inactivation of hRad9 influences HR (Fig. 9) but not NHEJ (Fig. 8).
Our results demonstrate that hRad9 can influence telomere stability and HR. However, it is not clear exactly how the protein functions in these processes. It is possible that hRad9 modulates chromatin structure, an early essential step for HR. This is supported by our demonstration that hRad9 inactivation delays the appearance of IR-induced γ-H2AX foci. Although the data support a role for Rad9 in chromatin remodeling, especially at telomeres, it is not clear whether the protein can influence chromatin structure beyond this limited region. Such a general chromatin alteration should influence repair in all phases of the cell cycle, as has been observed in ATM null cells (42). However, our results do not support this activity for hRad9 since inactivation of the protein resulted in an enhanced defect in chromosome repair confined only to the S and G2 phases of the cell cycle. Alternatively, Rad9 may interact with and influence the activity of proteins that have a role in HR repair, which occurs predominantly in S- and G2-phase cells. This is supported by our demonstration of a physical interaction between hRad9 and hRad51. In fact, Shinohara and coworkers (58) demonstrated that S. cerevisiae orthologues of some of the hRad9 checkpoint pathway-related proteins interact with Rad51 for repair of DSBs in meiosis via recombination. Furthermore, Grushcow and coworkers (20), as well as Aylon and Kupiec (2), showed that the S. cerevisiae equivalents of hRad9, hRad1, hHus1, and hRad17 pathway members are involved in recombination partner choice. In addition, we observed the delayed appearance of γ-H2AX foci in S-phase cells after IR exposure of Mrad9−/− relative to Mrad9+/+ cells. Moreover, the high frequency of chromatid gaps and breaks in hRad9-inactivated cells supports a role for the protein in HR repair. Inactivation of mammalian Rad9 could thus contribute, at least in part, to the radiosensitivity, genomic instability, and loss of telomeres observed by mediating the abrogation of HR repair.
Acknowledgments
This investigation was supported by NIH grants (NS34746, CA10445), the Department of Defense, the A-T Children's Society, and funds from Radiation Oncology, Washington University School of Medicine, to T.K.P.; NIH grants CA89816 and GM52493 to H.B.L.; and a Canadian Cancer Society grant (013235) to R.J.W.
We thank W. Wright for critical comments on and suggestions for the manuscript. We thank K. D. Brown, M. B. Kastan, S. J. Elledge, T. deLange, and J. Chen for providing several reagents that were used in this study.
REFERENCES
- 1.Ahmed, S., and J. Hodgkin. 2000. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403:159-164. [DOI] [PubMed] [Google Scholar]
- 2.Aylon, Y., and M. Kupiec. 2005. Cell cycle-dependent regulation of double-strand break repair: a role for the CDK. Cell Cycle 4:259-261. [PubMed] [Google Scholar]
- 3.Bassing, C. H., K. F. Chua, J. Sekiguchi, H. Suh, S. R. Whitlow, J. C. Fleming, B. C. Monroe, D. N. Ciccone, C. Yan, K. Vlasakova, D. M. Livingston, D. O. Ferguson, R. Scully, and F. W. Alt. 2002. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl. Acad. Sci. USA 99:8173-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassing, C. H., W. Swat, and F. W. Alt. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell 109(Suppl.):S45-S55. [DOI] [PubMed] [Google Scholar]
- 5.Baumann, P., and S. C. West. 1998. DNA end-joining catalyzed by human cell-free extracts. Proc. Natl. Acad. Sci. USA 95:14066-14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, H. T., A. Bhandoola, M. J. Difilippantonio, J. Zhu, M. J. Brown, X. Tai, E. P. Rogakou, T. M. Brotz, W. M. Bonner, T. Ried, and A. Nussenzweig. 2000. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science 290:1962-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, M. J., Y. T. Lin, H. B. Lieberman, G. Chen, and E. Y. Lee. 2001. ATM-dependent phosphorylation of human Rad9 is required for ionizing radiation-induced checkpoint activation. J. Biol. Chem. 276:16580-16586. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, C. K., L. W. Chow, W. T. Loo, T. K. Chan, and V. Chan. 2005. The cell cycle checkpoint gene Rad9 is a novel oncogene activated by 11q13 amplification and DNA methylation in breast cancer. Cancer Res. 65:8646-8654. [DOI] [PubMed] [Google Scholar]
- 9.Corda, Y., V. Schramke, M. P. Longhese, T. Smokvina, V. Paciotti, V. Brevet, E. Gilson, and V. Geli. 1999. Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat. Genet. 21:204-208. [DOI] [PubMed] [Google Scholar]
- 10.Counter, C. M., A. A. Avilion, C. E. LeFeuvre, N. G. Stewart, C. W. Greider, C. B. Harley, and S. Bacchetti. 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11:1921-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahlen, M., T. Olsson, G. Kanter-Smoler, A. Ramne, and P. Sunnerhagen. 1998. Regulation of telomere length by checkpoint genes in Schizosaccharomyces pombe. Mol. Biol. Cell 9:611-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang, T., S. Bao, and X. F. Wang. 2005. Human Rad9 is required for the activation of S-phase checkpoint and the maintenance of chromosomal stability. Genes Cells 10:287-295. [DOI] [PubMed] [Google Scholar]
- 13.Dhar, S., J. A. Squire, M. P. Hande, R. J. Wellinger, and T. K. Pandita. 2000. Inactivation of 14-3-3sigma influences telomere behavior and ionizing radiation-induced chromosomal instability. Mol. Cell. Biol. 20:7764-7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ducray, C., J. P. Pommier, L. Martins, F. D. Boussin, and L. Sabatier. 1999. Telomere dynamics, end-to-end fusions and telomerase activation during the human fibroblast immortalization process. Oncogene 18:4211-4223. [DOI] [PubMed] [Google Scholar]
- 15.Dynek, J. N., and S. Smith. 2004. Resolution of sister telomere association is required for progression through mitosis. Science 304:97-100. [DOI] [PubMed] [Google Scholar]
- 16.Elledge, S. J. 1996. Cell cycle checkpoints: preventing an identity crisis. Science 274:1664-1672. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Capetillo, O., A. Celeste, and A. Nussenzweig. 2003. Focusing on foci: H2AX and the recruitment of DNA-damage response factors. Cell Cycle 2:426-427. [PubMed] [Google Scholar]
- 18.Goytisolo, F. A., E. Samper, J. Martin-Caballero, P. Finnon, E. Herrera, J. M. Flores, S. D. Bouffler, and M. A. Blasco. 2000. Short telomeres result in organismal hypersensitivity to ionizing radiation in mammals. J. Exp. Med. 192:1625-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greer, D. A., B. D. Besley, K. B. Kennedy, and S. Davey. 2003. hRad9 rapidly binds DNA containing double-strand breaks and is required for damage-dependent topoisomerase II beta binding protein 1 focus formation. Cancer Res. 63:4829-4835. [PubMed] [Google Scholar]
- 20.Grushcow, J. M., T. M. Holzen, K. J. Park, T. Weinert, M. Lichten, and D. K. Bishop. 1999. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics 153:607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta, A., G. G. Sharma, C. S. Young, M. Agarwal, E. R. Smith, T. T. Paull, J. C. Lucchesi, K. K. Khanna, T. Ludwig, and T. K. Pandita. 2005. Involvement of human MOF in ATM function. Mol. Cell. Biol. 5:5292-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartwell, L. H., and M. B. Kastan. 1994. Cell cycle control and cancer. Science 266:1821-1828. [DOI] [PubMed] [Google Scholar]
- 23.Hopkins, K. M., W. Auerbach, X. Y. Wang, M. P. Hande, H. Hang, D. J. Wolgemuth, A. L. Joyner, and H. B. Lieberman. 2004. Deletion of mouse Rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol. Cell. Biol. 4:7235-7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunt, C. R., D. J. Dix, G. G. Sharma, R. K. Pandita, A. Gupta, M. Funk, and T. K. Pandita. 2004. Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol. Cell. Biol. 4:899-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia, X., T. Weinert, and D. Lydall. 2004. Mec1 and Rad53 inhibit formation of single-stranded DNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics 166:753-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lippman, Z., A. V. Gendrel, M. Black, M. W. Vaughn, N. Dedhia, W. R. McCombie, K. Lavine, V. Mittal, B. May, K. D. Kasschau, J. C. Carrington, R. W. Doerge, V. Colot, and R. Martienssen. 2004. Role of transposable elements in heterochromatin and epigenetic control. Nature 430:471-476. [DOI] [PubMed] [Google Scholar]
- 27.Lombard, D. B., K. F. Chua, R. Mostoslavsky, S. Franco, M. Gostissa, and F. W. Alt. 2005. DNA repair, genome stability, and aging. Cell 120:497-512. [DOI] [PubMed] [Google Scholar]
- 28.Longhese, M. P., V. Paciotti, H. Neecke, and G. Lucchini. 2000. Checkpoint proteins influence telomeric silencing and length maintenance in budding yeast. Genetics 155:1577-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahadevaiah, S. K., J. M. Turner, F. Baudat, E. P. Rogakou, P. de Boer, J. Blanco-Rodriguez, M. Jasin, S. Keeney, W. M. Bonner, and P. S. Burgoyne. 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27:271-276. [DOI] [PubMed] [Google Scholar]
- 30.Maniwa, Y., M. Yoshimura, V. P. Bermudez, T. Yuki, K. Okada, N. Kanomata, C. Ohbayashi, Y. Hayashi, J. Hurwitz, and Y. Okita. 2005. Accumulation of hRad9 protein in the nuclei of nonsmall cell lung carcinoma cells. Cancer 103:126-132. [DOI] [PubMed] [Google Scholar]
- 31.Matsuoka, S., G. Rotman, A. Ogawa, Y. Shiloh, K. Tamai, and S. J. Elledge. 2000. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. USA 97:10389-10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClintock, B. 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 26:234-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McElligott, R., and R. J. Wellinger. 1997. The terminal DNA structure of mammalian chromosomes. EMBO J. 16:3705-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller, H. 1938. The remaking of chromosomes. Collecting Net 13:181-195. [Google Scholar]
- 35.Nabetani, A., O. Yokoyama, and F. Ishikawa. 2004. Localization of hRad9, hHus1, hRad1, and hRad17 and caffeine-sensitive DNA replication at the alternative lengthening of telomeres-associated promyelocytic leukemia body. J. Biol. Chem. 279:25849-25857. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura, T. M., B. A. Moser, and P. Russell. 2002. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics 161:1437-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandita, T. K. 1983. Effect of temperature variation on sister chromatid exchange frequency in cultured human lymphocytes. Hum. Genet. 63:189-190. [DOI] [PubMed] [Google Scholar]
- 38.Pandita, T. K. 2003. A multifaceted role for ATM in genome maintenance. Expert Rev. Mol. Med. 2003:1-21. [DOI] [PubMed] [Google Scholar]
- 39.Pandita, T. K. 2001. The role of ATM in telomere structure and function. Radiat. Res. 156:642-647. [DOI] [PubMed] [Google Scholar]
- 40.Pandita, T. K., V. Gregoire, K. Dhingra, and W. N. Hittelman. 1994. Effect of chromosome size on aberration levels caused by gamma radiation as detected by fluorescence in situ hybridization. Cytogenet. Cell Genet. 67:94-101. [DOI] [PubMed] [Google Scholar]
- 41.Pandita, T. K., E. J. Hall, T. K. Hei, M. A. Piatyszek, W. E. Wright, C. Q. Piao, R. K. Pandita, J. C. Willey, C. R. Geard, M. B. Kastan, and J. W. Shay. 1996. Chromosome end-to-end associations and telomerase activity during cancer progression in human cells after treatment with alpha-particles simulating radon progeny. Oncogene 13:1423-1430. [PubMed] [Google Scholar]
- 42.Pandita, T. K., and W. N. Hittelman. 1992. The contribution of DNA and chromosome repair deficiencies to the radiosensitivity of ataxia-telangiectasia. Radiat. Res. 131:214-223. [PubMed] [Google Scholar]
- 43.Pandita, T. K., and W. N. Hittelman. 1992. Initial chromosome damage but not DNA damage is greater in ataxia telangiectasia cells. Radiat. Res. 130:94-103. [PubMed] [Google Scholar]
- 44.Pandita, T. K., H. B. Lieberman, D. S. Lim, S. Dhar, W. Zheng, Y. Taya, and M. B. Kastan. 2000. Ionizing radiation activates the ATM kinase throughout the cell cycle. Oncogene 19:1386-1391. [DOI] [PubMed] [Google Scholar]
- 45.Pandita, T. K., S. Pathak, and C. R. Geard. 1995. Chromosome end associations, telomeres and telomerase activity in ataxia telangiectasia cells. Cytogenet. Cell Genet. 71:86-93. [DOI] [PubMed] [Google Scholar]
- 46.Pandita, T. K., C. H. Westphal, M. Anger, S. G. Sawant, C. R. Geard, R. K. Pandita, and H. Scherthan. 1999. Atm inactivation results in aberrant telomere clustering during meiotic prophase. Mol. Cell. Biol. 9:5096-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petersen, S., R. Casellas, B. Reina-San-Martin, H. T. Chen, M. J. Difilippantonio, P. C. Wilson, L. Hanitsch, A. Celeste, M. Muramatsu, D. R. Pilch, C. Redon, T. Ried, W. M. Bonner, T. Honjo, M. C. Nussenzweig, and A. Nussenzweig. 2001. AID is required to initiate Nbs1/γ-H2AX focus formation and mutations at sites of class switching. Nature 414:660-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierce, A. J., R. D. Johnson, L. H. Thompson, and M. Jasin. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13:2633-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pilch, D. R., O. A. Sedelnikova, C. Redon, A. Celeste, A. Nussenzweig, and W. M. Bonner. 2003. Characteristics of γ-H2AX foci at DNA double-strand breaks sites. Biochem. Cell Biol. 81:123-129. [DOI] [PubMed] [Google Scholar]
- 50.Riballo, E., M. Kuhne, N. Rief, A. Doherty, G. C. Smith, M. J. Recio, C. Reis, K. Dahm, A. Fricke, A. Krempler, A. R. Parker, S. P. Jackson, A. Gennery, P. A. Jeggo, and M. Lobrich. 2004. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to γ-H2AX foci. Mol. Cell 16:715-724. [DOI] [PubMed] [Google Scholar]
- 51.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 52.Sawant, S. G., V. Gregoire, S. Dhar, C. B. Umbricht, S. Cvilic, S. Sukumar, and T. K. Pandita. 1999. Telomerase activity as a measure for monitoring radiocurability of tumor cells. FASEB J. 13:1047-1054. [DOI] [PubMed] [Google Scholar]
- 53.Scherthan, H., M. Jerratsch, S. Dhar, Y. A. Wang, S. P. Goff, and T. K. Pandita. 2000. Meiotic telomere distribution and Sertoli cell nuclear architecture are altered in Atm- and Atm-p53-deficient mice. Mol. Cell. Biol. 0:7773-7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma, G. G., A. Gupta, H. Wang, H. Scherthan, S. Dhar, V. Gandhi, G. Iliakis, J. W. Shay, C. S. Young, and T. K. Pandita. 2003. hTERT associates with human telomeres and enhances genomic stability and DNA repair. Oncogene 22:131-146. [DOI] [PubMed] [Google Scholar]
- 55.Sharma, G. G., K. K. Hwang, R. K. Pandita, A. Gupta, S. Dhar, J. Parenteau, M. Agarwal, H. J. Worman, R. J. Wellinger, and T. K. Pandita. 2003. Human heterochromatin protein 1 isoforms HP1Hsα and HP1Hsβ interfere with hTERT-telomere interactions and correlate with changes in cell growth and response to ionizing radiation. Mol. Cell. Biol. 3:8363-8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shay, J. W., and W. E. Wright. 2005. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis 26:867-874. [DOI] [PubMed] [Google Scholar]
- 57.Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3:155-168. [DOI] [PubMed] [Google Scholar]
- 58.Shinohara, M., K. Sakai, T. Ogawa, and A. Shinohara. 2003. The mitotic DNA damage checkpoint proteins Rad17 and Rad24 are required for repair of double-strand breaks during meiosis in yeast. Genetics 164:855-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smilenov, L. B., H. B. Lieberman, S. A. Mitchell, R. A. Baker, K. M. Hopkins, and E. J. Hall. 2005. Combined haploinsufficiency for ATM and RAD9 as a factor in cell transformation, apoptosis, and DNA lesion repair dynamics. Cancer Res. 65:933-938. [PubMed] [Google Scholar]
- 60.St Onge, R. P., C. M. Udell, R. Casselman, and S. Davey. 1999. The human G2 checkpoint control protein hRAD9 is a nuclear phosphoprotein that forms complexes with hRAD1 and hHUS1. Mol. Biol. Cell 10:1985-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takata, M., M. S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto, H. Utsumi, Y. Yamaguchi-Iwai, A. Shinohara, and S. Takeda. 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17:5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tarsounas, M., A. A. Davies, and S. C. West. 2004. RAD51 localization and activation following DNA damage. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359:87-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tarsounas, M., P. Munoz, A. Claas, P. G. Smiraldo, D. L. Pittman, M. A. Blasco, and S. C. West. 2004. Telomere maintenance requires the RAD51D recombination/repair protein. Cell 117:337-347. [DOI] [PubMed] [Google Scholar]
- 64.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 65.Venclovas, C., and M. P. Thelen. 2000. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 28:2481-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Volkmer, E., and L. M. Karnitz. 1999. Human homologs of Schizosaccharomyces pombe rad1, hus1, and rad9 form a DNA damage-responsive protein complex. J. Biol. Chem. 274:567-570. [DOI] [PubMed] [Google Scholar]
- 67.Wang, L., C. L. Hsu, J. Ni, P. H. Wang, S. Yeh, P. Keng, and C. Chang. 2004. Human checkpoint protein hRad9 functions as a negative coregulator to repress androgen receptor transactivation in prostate cancer cells. Mol. Cell. Biol. 4:2202-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang, R. C., A. Smogorzewska, and T. de Lange. 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119:355-368. [DOI] [PubMed] [Google Scholar]
- 69.Ward, I. M., and J. Chen. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276:47759-47762. [DOI] [PubMed] [Google Scholar]
- 70.Wong, K. K., S. Chang, S. R. Weiler, S. Ganesan, J. Chaudhuri, C. Zhu, S. E. Artandi, K. L. Rudolph, G. J. Gottlieb, L. Chin, F. W. Alt, and R. A. DePinho. 2000. Telomere dysfunction impairs DNA repair and enhances sensitivity to ionizing radiation. Nat. Genet. 26:85-88. [DOI] [PubMed] [Google Scholar]
- 71.Yin, Y., A. Zhu, Y. J. Jin, Y. X. Liu, X. Zhang, K. M. Hopkins, and H. B. Lieberman. 2004. Human RAD9 checkpoint control/proapoptotic protein can activate transcription of p21. Proc. Natl. Acad. Sci. USA 101:8864-8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, J., Z. Ma, A. Treszezamsky, and S. N. Powell. 2005. MDC1 interacts with Rad51 and facilitates homologous recombination. Nat. Struct. Mol. Biol. 12:902-909. [DOI] [PubMed] [Google Scholar]
- 73.Zou, L., D. Cortez, and S. J. Elledge. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16:198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]